SUMMARY
The last few decades have seen the emergence of computational neuroscience as a mature field where researchers are interested in modeling complex and large neuronal systems and require access to high performance computing machines and associated cyber infrastructure to manage computational workflow and data. The neuronal simulation tools, used in this research field, are also implemented for parallel computers and suitable for high performance computing machines. But using these tools on complex high performance computing machines remains a challenge because of issues with acquiring computer time on these machines located at national supercomputer centers, dealing with complex user interface of these machines, dealing with data management and retrieval. The Neuroscience Gateway is being developed to alleviate and/or hide these barriers to entry for computational neuroscientists. It hides or eliminates, from the point of view of the users, all the administrative and technical barriers and makes parallel neuronal simulation tools easily available and accessible on complex high performance computing machines. It handles the running of jobs and data management and retrieval. This paper shares the early experiences in bringing up this gateway and describes the software architecture it is based on, how it is implemented, and how users can use this for computational neuroscience research using high performance computing at the back end. We also look at parallel scaling of some publicly available neuronal models and analyze the recent usage data of the neuroscience gateway.
Keywords: computational neuroscience, science gateway, high performance computing, NEURON
1. INTRODUCTION
Computational neuroscience has seen tremendous growth in recent years. This is evident from the large number of publications, in prestigious neuroscience journals, that are more and more based on modeling. In the last two decades, this has motivated development of simulation tools such as NEURON [1], GENESIS3 [2], MOOSE [3], NEST [4], PyNN [5] and Brian [6], which are mature and written for parallel computers. Complex neuroscience problems, which involve network models, optimization or exploration of high-dimensional parameter spaces, require access to high performance computing (HPC), data storage, and complex workflow. Yet accessing and using HPC resources remain difficult because of the need to write yearly allocation proposals for acquiring computer time on national supercomputer centers, understand complex HPC architecture, learn complex OS/software, install neuronal software optimally, learn policies and batch environments, manage data transfer/retrieval, and understand remote authentication issues. As a solution to this problem, we have developed a community infrastructure layer, that is, a science gateway [7, 8], which provides a set of applications that are made available via a portal and is customized for researchers from a specific domain science. It allows researchers to use national resources and hides the complexities associated with using those resources. Our science gateway is specifically meant for computational neuroscientists, and it abstracts away technical and administrative details associated with using HPC for neuronal simulations. As a result, this allows neuroscience researchers easy access to best neuroscience simulation and analysis packages running on large-scale HPC resources. The neuroscience gateway (NSG) [9] was initially made available to few selected users, in the third quarter of 2012, and these users helped us to test the NSG before it was opened to all researchers. NSG was opened to all users at the end of December 2012. Since then, we continue to add more neural simulation tools. This extended paper is based on the conference paper [10] and describes the NSG software architecture, how it is implemented, the impact it has had until now, and the future plan.
2. MOTIVATION AND BACKGROUND
Most computational neuronal modeling projects start ‘small’, and many stay ‘small’, in the sense of being accommodated by individual desktop systems, but many eventually outstrip the speed and/or storage capabilities of local hardware. This is most often the case with projects that involve complex models (especially large-scale networks) or complex protocols (often involving learning rules), or require high-dimensional optimization or parameter space exploration. Such projects have a tremendous potential to use cyber infrastructure (CI), but only a very few neuroscientists have been able to perform simulations on extreme scale HPC machines [11–13]. There is a broad consensus that the wider computational neuroscience community needs access to complex CI and HPC resources. Those who lack such access are at a significant disadvantage relative to the very few who have it. This disparity is particularly telling in light of the fact that the widely used simulation tools such as NEURON, GENESIS3, MOOSE, NEST, Brian, and PyNN have been implemented on and released for parallel hardware, including HPC machines, for several years now. The typical neuroscientist—even one who is engaged in computational modeling—has limited compute resources, usually only a few desktop machines. The investigator whose project requires HPC machines must write a yearly proposal for allocation of supercomputer time. In the USA, these are peer reviewed for computer time on National Science Foundation (NSF)’s HPC machines, and there is no guarantee that full requested resource will be approved. If the proposal succeeds, the next task is to install simulation software (NEURON, GENESIS 3, MOOSE, NEST, PyNN, Brian) optimally on the supercomputer, then apply the batch software process to configure and run the model on the cluster, and finally deal with output data retrieval and storage issues. These steps involve many vexing details that not only are time consuming and require knowledge of HPC and information technology (IT), but also differ significantly from facility to facility. Investigators who want to use neuronal software on HPC resources directly have to deal with these issues themselves. The entire process constitutes a barrier that impedes effective utilization of HPC resources and also distracts neuroscientists from their primary research goals. We believe that many of these issues are encountered by neuroscientists in other parts of the world such as in Europe and Asia. Our proposed solution enables the entire community of computational neuroscientists to access NSF (and other organization) funded national HPC resources transparently through a common, convenient interface that is already configured for optimal use and operates within a common spatial and semantic framework. The benefit of this infrastructure can be extended to many more end users, allowing investigators to focus on their research and fostering collaboration.
In recent years, user-friendly scientific gateways have been developed successfully for many research fields and have resulted in tremendous adoption of CI and HPC by the broader user community of those fields. A few examples of such gateways in the USA include the nanoHUB gateway [14] for nanotechnology, the CIPRES gateway [15] for phylogenetics research, the GRIDCHEM [16] gateway for computational chemistry, and the ROBETTA [17] gateway for protein structure prediction. In the USA, most of these gateways are funded by the NSF for specific scientific domains and primarily utilize NSF’s Extreme Science and Engineering Discovery Environment (XSEDE) [18] or Open Science Grid (OSG) [19] for accessing HPC resources. Similarly in Europe, PRACE [20] and EGI [21] provide HPC and high throughput computing resources for users. In the USA, in addition to NSF-funded science gateways, there are gateways that are funded by other organizations such as the US Department of Energy (DOE) at various DOE laboratories.
Similarly, many gateways and gateway development frameworks exist in Europe and Asia. Liferay [22], EUMEDGRID [23], Catania Science Gateway Framework [24], INDICATE [25], and Apache Airavata [26] are some other frameworks available to build science gateways and provide provision for customization. EGI database [27] lists many applications, frameworks, and workflows that are available for building science gateways.
Example of neuroscience-related science gateways include the neuGRID [28], the Blue Brain project portal [29], the DECIDE gateway [30], the Charite grid portal [31], and other neuroscience gateways [32–35] dealing with neuroimaging and data analysis. It should be noted that most of these neuroscience-related science gateways deal with brain imaging analysis primarily focusing on MRI data, ultrasound data, image segmentation, statistical analysis of PET images, processing of EEG data, and visualization. These are useful to clinical oriented neuroscientists and medical researchers dealing directly with people with neurodegenerative diseases. Many of these gateways, such as the Charite grid, deal with secure and efficient data management and pseudonymization of data and again are geared towards clinical and medical brain image processing, whereas the NSG is focused on providing neuronal simulation tools and software (e.g., NEURON, GENESIS, Brian, NEST, and PyNN), which are used to simulate neuronal network models, neuronal spikes, and so on. As a result, the NSG is providing different research codes for computational neuroscientists who primarily include non-clinical researchers trying to model neuronal activities that have impact on fundamental understanding of neuronal functions and eventually and indirectly help understanding brain-related diseases.
3. NEUROSCIENCE GATEWAY ARCHITECTURE
The NSG architecture design is based on the Workbench Framework (WF) [36,15], which has been very successful in building the CIPRES phylogenetics gateway as well as other gateways such as the Portal for Petascale Lifescience Applications and Research (PoPLAR) gateway [37] for bioinformatics researchers. The WF is a very mature framework and was implemented at the San Diego Supercomputer Center (SDSC), by SDSC researchers, and programmers. In the succeeding text, we describe the various software architecture components of the WF in Section 3.1, their functional adaptation specifically for the NSG in Section 3.2, the NSG workflow, from user’s point of view, in Section 3.3, and discuss the computer time allocation-related issues in Section 3.4.
3.1. Underlying architecture
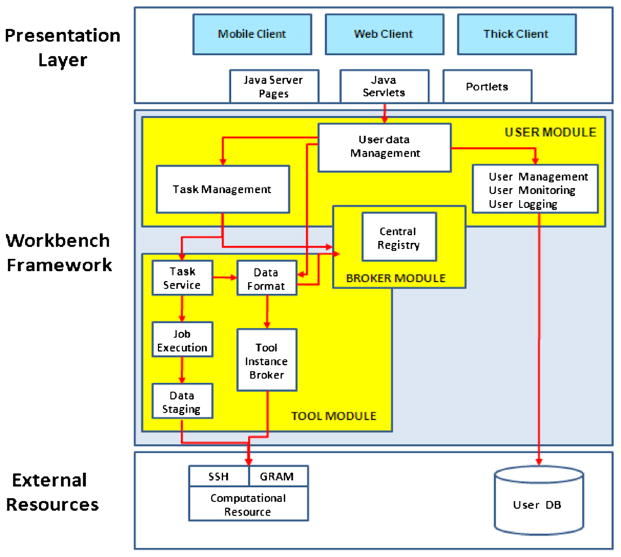
The WF is a software development kit (SDK) designed to generically deploy computational jobs and database searches to a generic set of computational resources and databases, respectively. The WF contains modules to manage submission of jobs to various computational resources and modules to manage queries on data resources. The higher level schematic of the WF architecture, as described in a diagram by the WF project developers [15] is shown in Figure 1. The modules in the WF are as follows.
Figure 1.
Workbench framework (from WF Reference [15]).
3.1.1. Presentation layer
The WF presentation layer accesses SDK capabilities through the J2EE front controller pattern, which involves only two Java classes. As a result, the WF is neutral with respect to interface access. The presentation layer provides lightweight access through a web browser and preserves flexibility for alternative access routes and adopts an architecture on the basis of Linux, Apache Tomcat, MySQL, and Java Struts2. The browser interface is easy to use and supports data and task management in user-created folders. The complexity of gateway layer is hidden from users through this interface that also allows users to create a login-protected personal account. Registered users can store their data and records of their activities for a defined period. Uploaded user data are checked for format compatibility. Users can also manually specify data types and formats.
3.1.2. User module
The user module passes user-initiated queries and tasks from the interface to the executive portions of the infrastructure via data and task management modules. It also stores all user data and task information in a MySQL database. It supports individual user roles, permitting the assignment of individual user accounts, the sharing of data between accounts, and selective access to tools and data sources that may be proprietary. Mapping user information takes places at this layer that helps track the individual usage on computational resources.
3.1.3. Broker module
The broker module provides access to all application-specific information in a central registry. This registry contains information about all data types required as input and output for each application along with the formats accepted by each tool. Concepts and concept relationships are formulated in XML files and read by central registry API implementing classes. Defining tools and data types in a single location allows adding new tools and data types with no impact on the functioning of the application outside the registry.
3.1.4. Tool module
The tool module manages the translation of tasks submitted by users into command lines and submission of the command line strings along with user data for execution by appropriate compute engines. The tool module handles data formatting for jobs and job staging. It also keeps track of which tools can be run on which computational resources and the status of those resources. The design allows great flexibility in determining what assets a science gateway can access for job execution. Computational resources can be added through editing the tool resource configuration file, and the application can send command line scripts and receive output via essentially any well-defined protocol such as the unix command line, web services, SSH, DRMMA, GRAM, and gsissh.
3.1.5. External resources
The generic design of the WF architecture supports access to a wide variety of computational resources and databases, whether local or remote. Access can be accomplished through a combination of individual mechanisms, including SSH, GRAM/Globus, SOAP, and ReST services.
3.2. Neuroscience gateway adaptation
The phylogenetics gateway CIPRES is built on the basis of the WF. CIPRES is one of the most successful gateways and handles large number of users. WF was modified for the CIPRES gateway. From here on, we refer to this modified version of the WF as the CIPRES-workbench framework (CIPRES-WF). To build the NSG, we decided to start from the CIPRES-WF. The adaptation of CIPRES-WF to NSG was carried out with the idea of hiding all the complexities associated with accessing and using an HPC resource such as job submission, input data transfer, choosing of machine specific HPC parameters, and output retrieval and storage
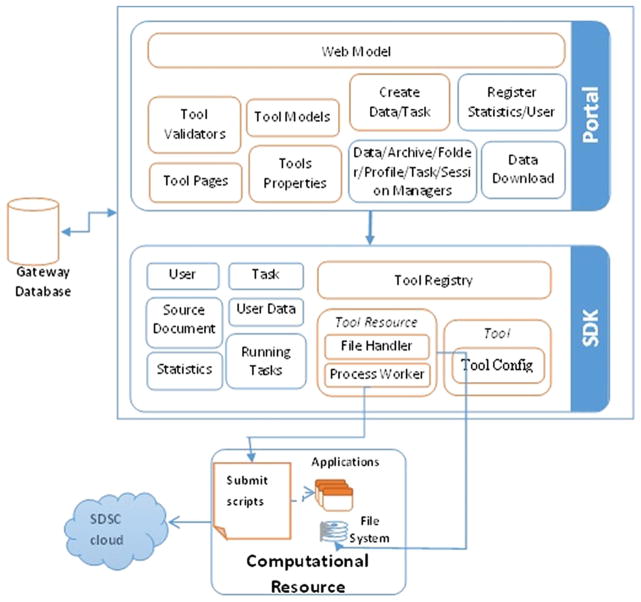
As shown in Figure 1, each module has many components essential to the functioning of a gateway. The adaptation of the CIPRES-WF to the functional NSG is shown in Figure 2. It outlines the NSG architecture highlighting some of the key components essential to the functioning of NSG. Among those components, the ones that required modifications for the NSG are highlighted in purple, whereas the other boxes are left with blue. The implementation includes enhancement and modification to the CIPRES-WF based on the needs of the neuroscience community. The portal and SDK together make up the framework for the NSG. The portal layer in Figure 2 maps to the presentation layer in Figure 1, and the SDK layer in Figure 2 maps to the overall workbench framework in Figure 1. We list in the succeeding text the key adaptations carried out to the existing CIPRES-WF for the NSG.
Figure 2.
Neuroscience gateway architecture.
Addition of uuencode/uudecode functionality to support upload of input files in zip format in the portal. This is performed in the portal layer in the create data function.
Modification of job submission environment to accommodate compilation of the NEURON code in the computational resource. The submit scripts on the computational resource were modified to accommodate this.
Storing of output file per session in SDSC’s Cloud storage on computational resource. This is performed by modifying submit scripts to copy the output results to cloud.
Automatic deletion of unused user files based on time length of inactivity in the portal. This is performed in the portal layer interacting with the gateway database.
Defining computational neuronal tools in the PISE XML format and interfacing with the portal in SDK. This is performed in the SDK layer in the tool module.
Mounting the HPC cluster’s home directory on a newly acquired disk space to eliminate GridFTP errors.
Hardware needed for setting up the NSG utilizes SDSC’s reference VM server, MySQL, Cloud storage, and web services. Some of the core functional implementation changes are discussed in the succeeding text.
3.2.1. Access
A web browser serves as the entry point to the NSG portal. The web browser offers a simple interface, which allows users to upload input file or neuronal models, specify neuronal code specific input files, and specify the job submission parameters such as number of cores or nodes, expected wall clock time for job completion. Users are able to monitor the status of submitted jobs and extract output files from the user-friendly portal. The access interface is a part of the portal layer as shown in Figure 2.
Although the community gateway account is used for job submission, individual user accounts are necessary to keep track of usage and access. Some of the neuronal simulation tools, such as NEURON, require that users be able to use the higher order calculator (HOC) [38] programming language as a scripting language for neuronal models and compile a HOC code via NSG. This is different from most other science gateways where users run precompiled codes only. Because of the possibility of malicious or incorrect use or handling of HOC codes, which poses a security concern, direct user registration on the NSG is not allowed. Users are required to fill out a form with identity information, which allows NSG administrators to validate the user manually (‘vetting’) prior to creating their account. Once registered, NSG can track each individual user’s access and usage, as well as can enforce NSG-specified usage policies. The account information and usage statistics are stored in the NSG’s MySQL database at SDSC.
3.2.2. Installation of computational neuroscience tools
Currently NEURON 7.2, NEURON 7.3, PGENESIS 2.3, NEST, Brian, and PyNN have been installed on SDSC’s Trestles HPC machine and are being installed on Texas Advanced Computing Center’s (TACC) [39] Lonestar HPC machine. These codes are available through the NSG for the neuroscience community. Additional neuronal tools are installed on the basis of users’ needs. In the SDK, the tools are defined in the PISEXML format. The tools and the resource are then added in the SDK in the tool registry module.
3.2.3. User-input file and job submission
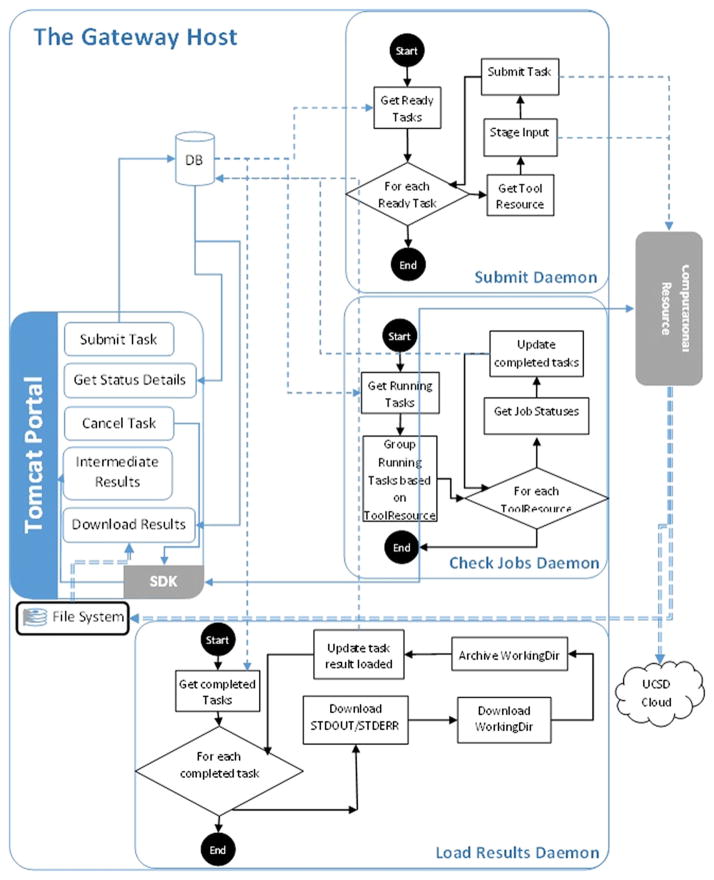
Most neuroscience computational models usually have more than one input file from sources such as ModelDB [40]. To accommodate this requirement, we have added the capability for NSG users to upload input file in a zip format. The CIPRES-WF did not have the capability of uploading binary files, and it also uses precompiled executable to run their job. For the NSG, we had to add and implement the functionality to uuencode the uploaded zip file and to uudecode the zip file on the computational resource during the staging of the input. In Figure 2, the portal layer handles this functionality. The NSG allows compilation and running of user’s code based on the requirement of the neuroscience application (e.g., NEURON, GENESIS 3, MOOSE, NEST, Brian, and PyNN). NEURON allows custom C++ code to be used for new equations within a particular model. To accommodate this, we created a mechanism to collect all such codes and compile them as a part of the job submission process. Job scripts are automatically created and submitted. Once a job completes, the working directory is compressed along with the input files, job submission scripts, and output files, and are transferred to SDSC’s Cloud storage. The compressed file is also made available for immediate download through the NSG portal. File staging is handled via the Java CoG Kit GridFTP API and Java runtime exec of Globus ‘gsissh’ is used to remotely run commands. Figure 3 shows the event-driven interaction flow diagram for the NSG. Figure 3 shows the daemons responsible for submitting a job, monitoring job status, and retrieving the output. The daemons also help keep track of job and usage monitoring by updating the database. The submit daemon is called when a task is created and submitted. While the job is processing on the HPC cluster, an intermediate results view option is available in the portal that gives a snapshot of the working directory that was created in the backend HPC cluster. Advanced users can look at the intermediate results folder to see if their job has started or if any output file has been written. Another notable feature is the ability to clone a job on the portal. Users are able to clone their jobs, and this is helpful when they want to submit a job with the same input file but vary the parameters such as number of cores or the wall clock time. Ability to do parameter sweep studies will be provided in the future. NSG portal occasionally faced issues related to using GridFTP or the NFS server on the remote HPC cluster. During file staging from the portal to the remote cluster or while copying the results back, the task would fail either because there are too many open GridFTP connections on the remote cluster or the NFS home directory on the HPC cluster is slow because of multiple users using it. Addressing this issue requires restarting autofs by system administrator of the HPC cluster. To avoid this, we mounted the HPC cluster’s home directory on a new disk space meant specifically for the NSG.
Figure 3.
Event-driven interaction flow.
3.2.4. Storage and data retrieval
The output data, generated as a result of simulations using the NSG, are saved as a zip file and are made available on the portal. Email notification is sent to the users when a job completes. This is handled by a curl command in the job submission script, which notifies a servlet in the Web application when the job finishes. In case of curl failure, two daemon processes named ‘checkJobsD’ and ‘loadResultsD’ check to see which jobs have finished and transfer the result to the NSG. The check jobs and load results flowcharts are shown in Figure 3. The NSG also moves the data from the HPC resource’s scratch disk space to SDSC’s Cloud archival storage and employs a storage policy based on data last accessed. User’s data are allowed to be stored in NSG’s disk space as long as the user is using NSG actively. If there is no activity for a year, the user’s data will be deleted after the user is informed of such inactivity.
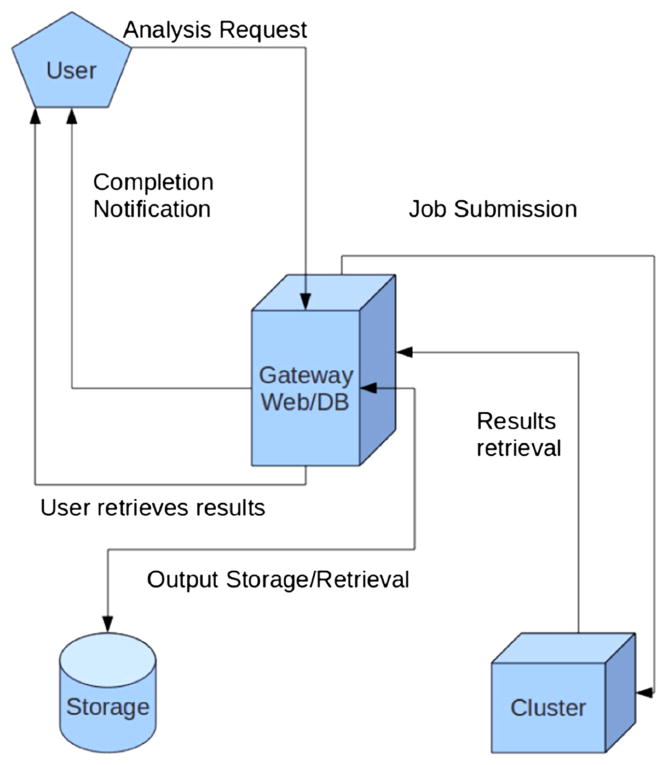
3.3. User workflow
Figure 4 shows at a high level and from a neuroscience user’s point of view how the flow of work will appear as a simple environment. It should be noted that our goal was to provide a simple workflow as far as the users are concerned and hide all the complexities from them. The NSG user workflow consists of the following steps:
Figure 4.
User’s view of the neuroscience gateway flow of work.
User logs into the NSG.
User uploads input data.
User chooses particular neuronal software.
User requests simulation run on an HPC resource.
NSG frontend sends input data and job request to the remote HPC machine.
NSG retrieves output data from the remote machine.
NSG notifies users of job (completion) status.
NSG provides user information about output data location and retrieval process.
3.4. Allocation and policies
Initial allocation, called Startup allocation within the XSEDE allocation process (XRAC) [41], was obtained for 50,000 core hours on SDSC’s Trestles and 50,000 core hours on TACC’s Lonestar machines. Each core hour is called a service unit (SU) as a part of XSEDE terminology. We also obtained community gateway account on both Trestles and Lonestar. The initial allocation of 100,000 core hours was used up by early 2013 and, an additional 200,000 core hours was requested and approved by the XSEDE allocation committee. This allocated time was also completely used up by the NSG users by mid-September 2013. Even when the allocated hours were almost fully spent, users requested to keep the NSG functional, and we were able to use an SDSC guest account that, among other things, allows users to use HPC resources under special circumstances. The NSG PIs and CO-PIs dealt with acquiring computer time and thus shielding the users from the complexities associated with getting allocation. Subsequent to this, an allocation request for 2.3 million core hours was submitted to the XRAC review committee in early October 2013. This estimate was based on usage during the third quarter of 2013 and from direct input from selected users. Although we do not have any usage data of what users did prior to the availability of the NSG, the usage demonstrates that there is certainly a need of such a science gateway for neuronal simulations on HPC resources. This allocation proposal was favorably reviewed, and in December 2014, we were awarded 1.6 million core hours for the 2014 calendar year. We consider this as a good recognition of the NSG as the national scale XRAC review process is rigorous and award of HPC core hours is very competitive and selective. We will continue to submit yearly XRAC allocation requests such that the NSG always has time on HPC resources.
Users of the community gateway account abide by the policies set by the NSG administrators. Currently, we allow 5000 core hours per user per year. On the basis of the total amount of computer time acquired every year for the NSG and the total number of NSG users, we will decide what percentage of the total time can be allocated freely to each user and monitor their usage. Because not all the users consume the same amount of resource, we try to obtain an estimate from the users, and on the basis of their average usage and job size, the amount allocated to them will vary. The NSG also has the capability to allow users to run jobs with their own acquired allocation. For assessment of impact to identifiable members of the community, individual gateway user’s tag will be propagated through the batch system, such that the final job accounting reconciliation process will report quantitative usage by those individual gateway users. All user activities such as job submission statistics and usage are logged in the database. As a part of the user support environment, a ticketing system, monitored by NSG developers, is in place and is used to keep track of user questions and provide immediate assistance.
4. ANALYSIS AND USER ACTIVITIES
In this section, we discuss three topics that are part of most science gateways. Our focus for NSG has been to provide easy access to HPC resources for computational neuroscientists. The parallel performance of neuronal models is important in this regard as we expect our users to run parallel models using HPC resources. The performance of parallel models is also important when we write XRAC allocation proposals justifying the total time we request for each year. Associated with this is the analysis of usage statistics of the allocated time by NSG users. As a part of the user activities, we also discuss some of the educational projects performed by undergraduate and high school students using the NSG.
4.1. Parallel scaling performance
The parallel scaling performance results for few of the publicly available neuronal models on HPC resources are discussed in this section. Parallel scaling is defined as the plot of speedup versus number of cores. Speedup on N cores is defined as the time taken to run on one core divided by the time taken to run the same case on N cores. Parallel scaling will very much depend on the specific parallel neuronal models that will be developed as a part of research by the individual computational neuroscientists and run via the NSG on HPC resources. We decided to run two parallel neuronal models using the NEURON software because most of our users are currently using this software for their research. The idea behind the scaling study is to show that depending on size of the parallel model, the NEURON code will scale to different number of cores [42]. It should be noted that the other neuronal tools, currently provided by the NSG, are also capable of running parallel models.
The neuronal simulation software NEURON, installed on both SDSC’s Trestles and TACC’s Lonestar machine, is a flexible and powerful simulation environment for modeling individual neurons and network of neurons. It provides tools for conveniently building, managing, and using models that are numerically sound and computationally effIcient. It is particularly suitable for problems that involve cells with complex anatomical and biophysical properties. NEURON is a parallel code and can execute parallel simulations using both MPI and OpenMP implementations. Most of the NSG usage so far has been using the parallel NEURON code. Two publically available models from ModelDB [40] were run on SDSC’s Trestles cluster and TACC’s Lonestar cluster to study the parallel scaling effects as example benchmark problems.
Figure 5 describes the scaling of biophysically realistic neural modeling of the MEG mu rhythm [43] system starting from four cores. This model deals with variations in cortical oscillations and has been correlated with attention, working memory, and stimulus detection. Figure 6 describes the scaling of the large-scale model of the olfactory bulb [44] starting from 24 cores. The olfactory bulb case models sparse distributed representation of odors in a large-scale olfactory bulb circuit. Typically, NSG users will run models starting on few cores and scale up, as the memory requirement may not make it feasible to run models on a single core. Following this, we have also showed results, in Figures 5 and 6, starting from few cores. Figure 5 shows that the MEG mu rhythm model scales to about 128 cores, and Figure 6 shows that the olfactory bulb model scales to about 384 cores. These results are expected as the MEG mu rhythm model is a smaller model, that is, having less number of neuronal cells, compared with the olfactory bulb model. It should be noted that the time taken to do these benchmark runs was at most around 20 h and so an insignifIcant part of the total allocation utilized by the users as reported in Section 3.4
Figure 5.
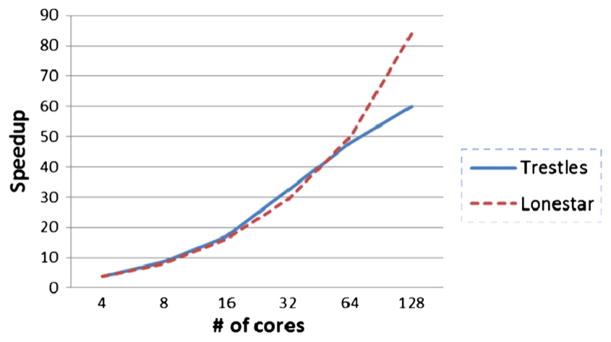
MEG mu rhythm model speedup.
Figure 6.
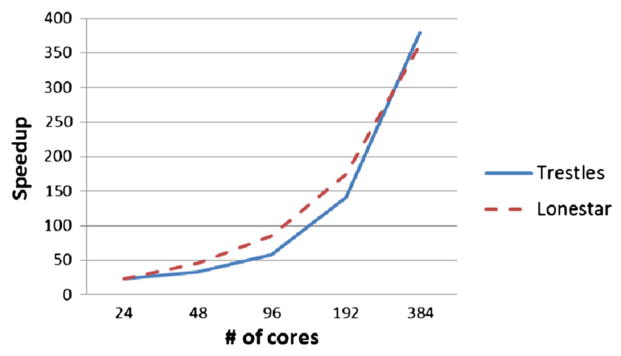
OlfactoryBulb_2013 model speedup.
4.2. User activity—education projects
An example of user activities is work performed by students using NSG. A high school student created a tutorial on multiple sclerosis using the NEURON code. The tutorial is now available on the NSG website [45]. As a part of a separate project, an undergraduate student from the University of California San Diego performed parallel scaling study of various models, available from ModelDB, on HPC resources located at SDSC. In this study, it was shown that consistency of a model was not affected by running on varying number cores [46].
The Neuroscience Information Framework (NIF) [47] is a dynamic inventory of Web-based neuroscience resources such as data, materials, and tools—accessible via any computer connected to the internet. NIF enables discovery and access to public research data and tools worldwide via open source, networked environment. The NSG team and the NIF joined forces, to utilize the computational capabilities provided by the NSG with the semantic knowledge layer provided by the NIF environment. The ultimate goal is to provide an API for NIF and thus enabling NIF users to submit computational jobs directly from the NIF website. To achieve this, the overall integration is to be performed in a series of small steps where each step addresses certain aspect of interaction between the two systems and provides clear measurable results. The first integration step was completed with the help of two high school student interns who utilized a simple strategy to answer the question—which models will run ‘off the shelf’ in a parallel computing environment. NIF provided search and filtering capability on the metadata schema of several model repositories, including ModelDB, to find eligible computational models. The students then downloaded the models from the repository and submitted all the models using the NSG to run on SDSC’s Trestles machine. The result set consisted of a set of attributes including a flag for the ability to run the model, type of error generated, run time, number of nodes used, output files of each model, and other basic metadata such as date. The data were put up on a server, and the metadata that students maintained in a spreadsheet was made available to the NIF crawler and was ingested into the data stores at NIF. A view was built on the basis of this metadata, which links to the raw output of models and can inform NIF users, if the model is expected to run, as is, on the NSG as shown in Figure 7. Future work involves developing standard APIs between the two systems to automate operations.
Figure 7.
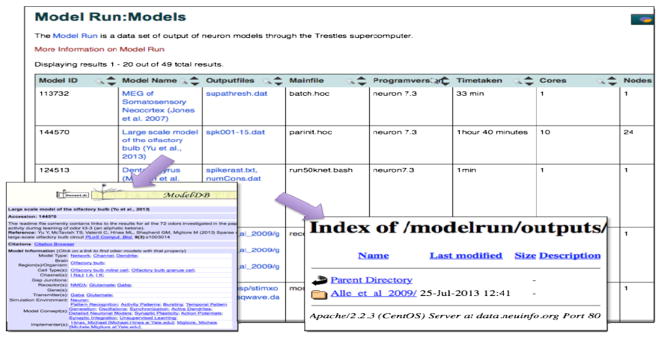
ModelDB runs using neuroscience gateway on the Neuroscience Information Framework website.
4.3. User base and usage
Users were added, following the ‘vetting’ process, to the NSG since the end of December 2012. Within the first 10-month period, we had 110 users, out of which 65% are from USA and the rest are from Europe and Asia, and this distribution is shown in Figure 8. Forty-five users attended the first NSG workshop, held at SDSC in March 2013, and this was simultaneously broadcasted over the Web for remote attendees. Of the 45 attendees, 10 attended the workshop on-site at SDSC and 35 users attended virtually. In addition, we have also presented three webinars in 2013 to explain the functionalities of the NSG.
Figure 8.
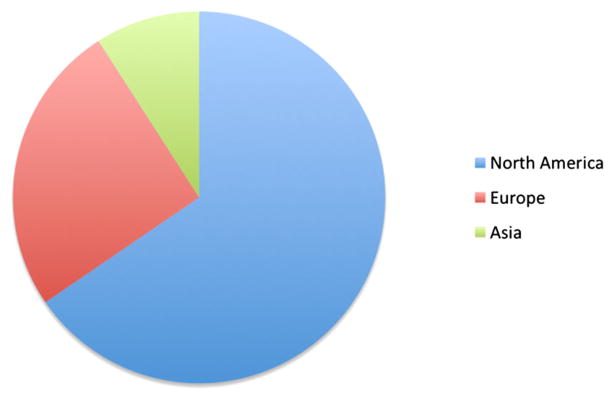
Distribution of location of neuroscience gateway users.
As mentioned in Section 3.4, a total of 300,000 core hours has been consumed by NSG users since the beginning of the friendly user period in December 2012 and till mid-September 2013 as shown in Figure 9. During the first quarter of 2013, usage was low, as is expected of any new science gateways because users were just becoming aware of this new gateway. There was one exception in January 2013 when a single user used 30,000 core hours, and we consider it an exception as the per-user basis usage limit was not properly in place then. During the second quarter of 2013, we observed that more users were starting to use the NSG but still mostly to test and run small jobs to become familiar with the gateway. During the third quarter of 2013, users started to employ the NSG in a more consistent way for actual neuroscience simulations, driven by their research needs. We believe that a realistic steady state usage was reached during the July to mid-September period of 2013. As mentioned in Section 3.4, our initial allocation of 300,000 core hours was used up by mid-September 2013, and we resorted to SDSC guest allocation to keep NSG functional until new XRAC allocation was received for the 2014 calendar year.
Figure 9.
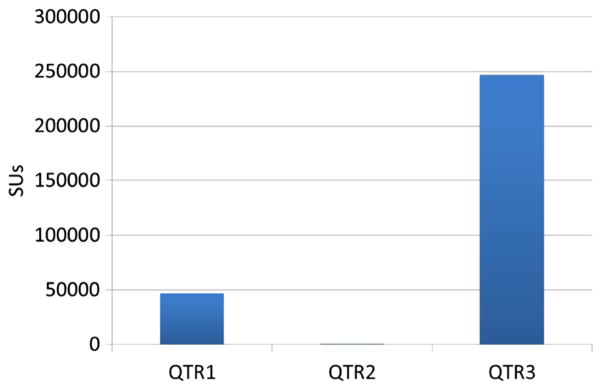
Service units used for quarters 1–3, 2013.
Usage during the period from July to September 15, 2013, that is, over two and half months, was 246,353 core hours as can be seen from Figures 9 and 10. Because we ran out of all allocated core hours by September 15, 2013, it is the cutoff date chosen in Figure 10. In Figure 11, we show the total number of jobs during each month of the third quarter of 2013 and average job size.
Figure 10.
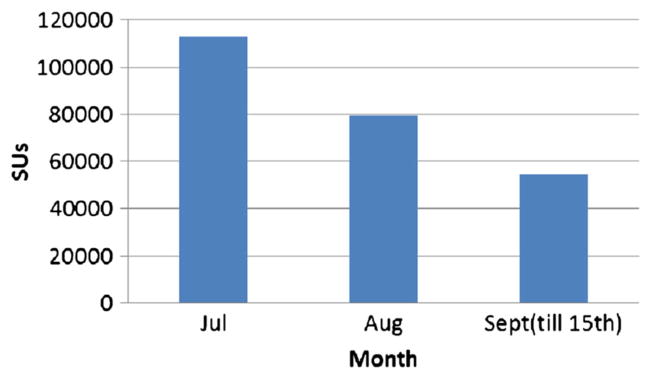
The 2013 third quarter service unit consumption.
Figure 11.
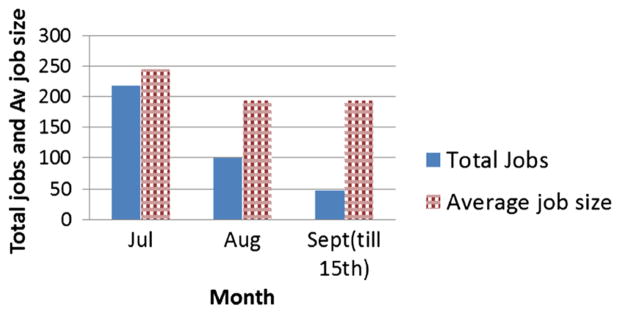
The 2013 third quarter jobs and job size.
As stated earlier, because of the inherent way neuronal models are created by computational neuroscience users, scaling of models is very much dependent on the particular parallel model being run by the users and will vary from user to user.
As shown in scaling plots in Figures 5 and 6 for two benchmark models, the results using NEURON on Trestles and Lonestar are scaling well to the number of cores of average job sizes as shown in Figure 11. It should be noted from Figure 11 that the total number of jobs submitted in August and 15 days of September 2013 are consistent and show about 100 jobs a month. In July, the job count is higher from that by about 100 jobs as two high school students, participating in July as summer student interns, submitted these additional ~100 jobs as a part of their summer internship work to test various publicly available neuronal models. This usage of the initial 300,000 core hours demonstrates the interest and initial success of the project. Now that we have received 1.6 million core hours, for the 2014 calendar year, we are providing this computer time to NSG users. This alleviates the need for the NSG users to deal with the allocation process directly and is one of the benefits of a science gateway.
An important metrics of the NSG is publications being performed by neuroscience researchers and for which the NSG was used and acknowledged. As the NSG is being used in a consistent way for research primarily from July 2013, we believe that publications are expected to come out by the third quarter of 2014, that is, about a year after the researchers have started to use the NSG, and this is the feedback we have received from the users. So although at this point (early 2014), we cannot list publications, where the research was performed using NSG, we expect to list that in the later part of 2014 from the NSG website.
5. SUMMARY AND FUTURE WORK
The main thrust of this paper was to share the early experiences associated with bringing up a domain science-specific (in this case, computational neuroscience) science gateway. We have discussed our experience in adapting the CIPRES-WF for the NSG and how utilizing a proven and existing software base was beneficial in bringing up the NSG in a short period.
As mentioned earlier, from the very outset of developing the NSG, we decided to use the CIPRES-WF’s code base as our starting point. The key reasons for choosing CIPRES-WF and adapting it to create the NSG are as follows:
The WF is a well-established framework for science gateways and which has been developed over the past 10 years by experienced software developers and researchers.
The CIPRES-WF has been successfully used for building gateways for phylogenetics community and other domain sciences such as bioinformatics.
CIPRES-WF developers are researchers at SDSC, and as a result, we were able to obtain expert help when needed.
Reuse of existing NSF-funded software was considered a good practice.
Any additions or modifications performed as a part of the NSG implementation will be contributed back to the WF and can be adopted by future gateway developers.
Bringing up the NSG to early production took about 2 months of time using approximately 75% of equivalent staff effort. In reality, three staff members shared the work load and spent approximately 40%, 20%, and 15% of effort, respectively, over the 2-month period. Although there was overlapping work by all of the three staff members, one of the staff members spent time primarily in NSG software implementation, the second staff member spent time primarily in installing, testing, and benchmarking the neuronal software on HPC resources, and the third staff member dealt with writing allocation proposal to acquire time on HPC resources and other coordination efforts. We also received guidance and advice from our other collaborators at UCSD and Yale University.
All of the IT resources, such as VM servers and SDSC Cloud storage, which are used for the NSG project, are located at SDSC, and this was beneficial to the project. Reusability of software played a key role and helped us to save lot of time and effort. As a result, we were able to make the portal available to computational neuroscientists within a relatively short period since the initiation of the project. We believe that this discussion will give other potential gateway developers some general idea about the effort and time needed to bring up a science gateway that has similar requirements and general characteristics as the NSG and specially if they are able and interested to use the WF for their domain specific science gateway.
As a part of the future work, we plan to integrate a programmatic interface module, which will provide an interface layer for external neuroscience frameworks such as ModelDB, NIF, and NeuroConstruct [48], and will allow mapping of user requests directly from these external frameworks to the NSG. The module will translate user requirements and authentication to the NSG interface. The external framework would format its request in XML for job processing, status query, and output data retrieval. REST API will also be incorporated to provide programmatic access to NSG. An interface for model sharing with data provenance will be provided. Users who are willing to share their models or output will be able to do so in a collaborative environment. Validated data models will be provided for educational purposes.
Acknowledgments
This work was supported in part by the National Science Foundation awards DBI (Division of Biological Infrastructure) 1146949 and DBI 1146830, and by NIH awards NS011613 and DC09977. The work was also supported by computer allocation time provided by the NSF funded XSEDE organization on SDSC’s Trestles and TACC’s Lonestar HPC resources and by XSEDE Extended Collaborative Support Services (ECSS) program that allowed Terri Schwartz (SDSC) to collaborate with the NSG team. The authors would like to thank Mark Miller (SDSC), PI of the CIPRES-WF software, for providing advice and guidance as the NSG was being implemented. The authors would also like to thank Nancy Wilkins-Diehr (SDSC) and Suresh Marru (Indiana University) for helpful discussions, and UCSD undergraduate student Prithvi Sundar, West View High School, San Diego students Debleena Sengupta and Daniel Guan, and Canyon Crest Academy High School student Evan Sheng for their summer internship project work. The authors would like to thank Saminda Wijeratne (Indiana University) for his help with some of the figures.
References
- 1.Hines ML, Carnevale NT. Translating network models to parallel hardware in NEURON. Journal of Neuroscience Methods. 2008;169:425–455. doi: 10.1016/j.jneumeth.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.http://genesis-sim.org/
- 3.http://moose.sourceforge.net/
- 4.http://www.nest-initiative.uni-freiburg.de/index.php/Software:About_NEST
- 5.http://neuralensemble.org/trac/PyNN/
- 6.http://brainsimulator.org
- 7.Wilkins-Diehr N, Gannon D, Klimeck G, Oster S, Pamidighantam S. TeraGrid science gateways and their impact on science. IEEE Computer. 2008;41(11):32–41. [Google Scholar]
- 8.https://www.xsede.org/gateways-overview
- 9.http://www.nsgportal.org
- 10.Sivagnanam S, Majumdar A, Yoshimoto K, Astakhov V, Bandrowski A, Martone ME, Carnevale NT. IWSG, volume 993 of CEUR Workshop Proceedings. 2013 CEUR-WS.org.
- 11.Ananthanarayanan R, Esser S, Simon HD, Modha DS. SC09 Proceedings; Nov 14–20, 2009; Portland, Oregon, UCA. ACM; 2009. 978-1-60558-744-8/09/112009. [Google Scholar]
- 12.Kumar S, Heidelberger P, Chen D, Hines M. Optimization of applications with non-blocking neighborhood collectives via multi-sends on the blue gene/P supercomputer. 24th IEEE International Parallel and Distributed Processing Symposium; 2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markram H. The blue brain project. Nature Reviews Neuroscience. 2006;7:153–160. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- 14.http://www.nanohub.org
- 15.Miller M, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. Gateways Computing Environments Workshop (GC); 2010; New Orleans, LA. Nov 14, 2010. pp. 1–8. [Google Scholar]
- 16.https://www.gridchem.org
- 17.http://robetta.bakerlab.org/
- 18.http://www.xsede.org
- 19.http://www.opensciencegrid.org
- 20.http://www.prace-ri.eu/
- 21.https://www.egi.eu/
- 22.http://www.liferay.com/
- 23.http://www.eumedgrid.eu/
- 24.http://www.catania-science-gateways.it/en
- 25.http://indicate-gw.consorzio-cometa.it/
- 26.http://airavata.apache.org/
- 27.https://appdb.egi.eu/
- 28.https://neugrid4you.eu/
- 29.http://bluebrain.epfi.ch/
- 30.Ardizzone V, Barbera R, Calanducci A, Fargetta M, Ingra E, Porro I, La Rocca G, Monforte S, Ricceri R, Rotondo R, Scardaci D, Schenone A. The DECIDE science gateway. Journal of Grid Computing. 2012;10 (4):689–707. [Google Scholar]
- 31.Wu J, Siewert R, Hoheisel A, Falkner J, Strauß O, Berberovic D, Krefting D. The Charité Grid Portal: user-friendly and secure access to grid-based resources and services. Journal of Grid Computing. 2012;10(4):709–724. [Google Scholar]
- 32.Glatard T, Lartizien C, Gibaud B, Ferreira da Silva R, Forestier G, Cervenansky F, Alessandrini M, Benoit-Cattin H, Bernard O, Camarasu-Pop S, Cerezo N, Clarysse P, Gaignard A, Hugonnard P, Liebgott H, Marache S, Marion A, Montagnat J, Tabary J, Friboulet D. A virtual imaging platform for multi-modality medical image simulation. Medical Imaging, IEEE Transactions on Medical Imaging. 2013;32(1):110–118. doi: 10.1109/TMI.2012.2220154. [DOI] [PubMed] [Google Scholar]
- 33.Dinov I, Lozev K, Petrosyan P, Liu Z, Pierce J, Zamanyan A, Chakrapani S, Van Horn J, Stott Parker D, Magsipoc R, Leung K, Gutman B, Woods R, Toga A. Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PLoS ONE. 2010;5(9):e13070. doi: 10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frisoni GB, Redolfi A, Manset D, Rousseau ME, Toga A, Evans AC. Virtual imaging laboratories for marker discovery in neurodegenerative diseases. Nature Reviews Neurology. 2011;7(8):429–38. doi: 10.1038/nrneurol.2011.99. [DOI] [PubMed] [Google Scholar]
- 35.Shahand S, Benabdelkader A, Huguet J, Jaghouri M, Santcroos M, al Mourabit M, Groot PFC, Caan MWA, van Kampen AHC, Olabarriaga SD. A data-centric science gateway for computational neuroscience. In proceedings of Interenational Workshop on Science Gateways; Zurich. June 3–5, 2013. [Google Scholar]
- 36.Miller M, Pfeiffer W, Schwartz T. CIPRES science gateway: a community resource for phylogenetic analyses. TeraGrid’; 11, July 18–21; Salt Lake City. 2011. [Google Scholar]
- 37.poplar.nics.tennessee.edu/locing!input.action
- 38.Kernighan BW, Pike R. The Unix Programming Environment. Prentice Hall; Englewood Cliffs, N.J: 1984. [Google Scholar]
- 39.www.tacc.utexas.edu
- 40.http://senselab.med.yale.edu/modeldb
- 41.https://www.xsede.org/allocations
- 42.Migliore M, Cannia C, Lytton WW, Markram H, Hines ML. Parallel network simulations with NEURON. Journal of Computational Neuroscience. 2006;21:110–119. doi: 10.1007/s10827-006-7949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.http://senselab.med.yale.edu/modeldb/ShowModel.asp?model=136803
- 44.http://senselab.med.yale.edu/modeldb/showmodel.asp?model=144570
- 45.http://www.nsgportal.org/ed-index.html
- 46.Bandrowski AE, Sivagnanam S, Yoshimoto K, Astakhov V, Majumdar A. Performance of parallel neuronal models on the Triton cluster. Society for Neuroscience Annual Meeting; Washington D.C. Nov 12–16, 2011. [Google Scholar]
- 47.www.neuinfo.org
- 48.http://www.neuroconstruct.org/