Abstract
The phosphatidylinositol 3 kinase (PI3K)/AKT pathway is genetically targeted in more pathway components and in more tumor types than any other growth factor signaling pathway, and thus is frequently activated as a cancer driver. More importantly, the PI3K/AKT pathway is composed of multiple bifurcating and converging kinase cascades, providing many potential targets for cancer therapy. Renal cell carcinoma (RCC) is a high-risk and high-mortality cancer that is notoriously resistant to traditional chemotherapies or radiotherapies. The PI3K/AKT pathway is modestly mutated but highly activated in RCC, representing a promising drug target. Indeed, PI3K pathway inhibitors of the rapalog family are approved for use in RCC. Recent large-scale integrated analyses of a large number of patients have provided a molecular basis for RCC, reiterating the critical role of the PI3K/AKT pathway in this cancer. In this review, we summarizes the genetic alterations of the PI3K/AKT pathway in RCC as indicated in the latest large-scale genome sequencing data, as well as treatments for RCC that target the aberrant activated PI3K/AKT pathway.
Keywords: PI3K, AKT, mTOR, Renal cell carcinoma, Targeted therapy
Introduction
The phosphatidylinositol 3 kinase (PI3K)/AKT pathway is activated by gene mutations and copy number alterations (CNAs) in more pathway components and in more tumor types than any other signaling pathways involved in cancer (Brugge et al., 2007). A catalytic subunit of PI3K, p110α, is the most commonly targeted oncogene in cancer. More importantly, genetic mutations result in activation of the PI3K/AKT pathway, which is composed of multiple bifurcating and converging kinase cascades, representing a “target-rich” environment for cancer therapy. Not surprisingly, inhibitors of PI3K isoforms, AKT, mammalian target of rapamycin (mTOR), and other components in the pathway are being actively pursued for targeted cancer therapy.
Kidney cancer, which is increasing in incidence, is the ninth most common cancer worldwide; about 337,860 new cases were diagnosed in 2012 (Jonasch et al., 2014). In the United States, an estimated 14,000 of the 64,000 new cases of kidney cancer diagnosed in 2014 resulted in patient death, making kidney cancer a high-risk and high-mortality cancer type. Most kidney cancers are renal cell carcinoma (RCC), including the major subtype clear cell RCC (ccRCC), which accounts for most kidney cancer-related deaths. Treatment for ccRCC in the United States is currently leading a revolution from traditional chemotherapy and radiotherapy to molecularly targeted therapy. Seven targeted drugs have been approved by the US Food and Drug Administration since 2005, some of which directly or indirectly target the PI3K/AKT pathway. In this review, we summarize the alterations of the PI3K/AKT pathway that occur in RCC, with an emphasis on its primary components, PI3K and AKT, and we discuss the development of drugs targeting aberrant activation of the PI3K/AKT pathway in RCC.
The PI3K/AKT pathway
PI3K and upstream regulators
PI3K is a family of lipid kinases that phosphorylate the 3-hydroxyl group of the inositol ring of phosphatidylinositol (PtdIns) lipids in the plasma membrane (Fruman et al., 1998). PI3K is divided into three classes (class I, II, and III) according to their different structures and lipid substrate preferences (Vanhaesebroeck et al., 2010). Class I PI3K is the most studied and best understood group because it plays an important role in cancer. Class I PI3Ks are heterodimers of a 110-kDa catalytic subunit (p110) and a regulatory subunit (i.e., p85). There are four p110 isoforms (p110α, p110β, p110γ, and p110δ) produced from different genes and seven regulatory subunits (p85α, p85β, p55α, p55γ, p50α, p101, and p87) produced by a combination of alternative start codons and different gene in mammals (Vanhaesebroeck et al., 2010). The regulatory subunits bind to the p110 catalytic subunits stabilizing the PI3K protein heterodimers, inhibiting the kinase activity under basal conditions and directing PI3K to upstream regulators for activation.
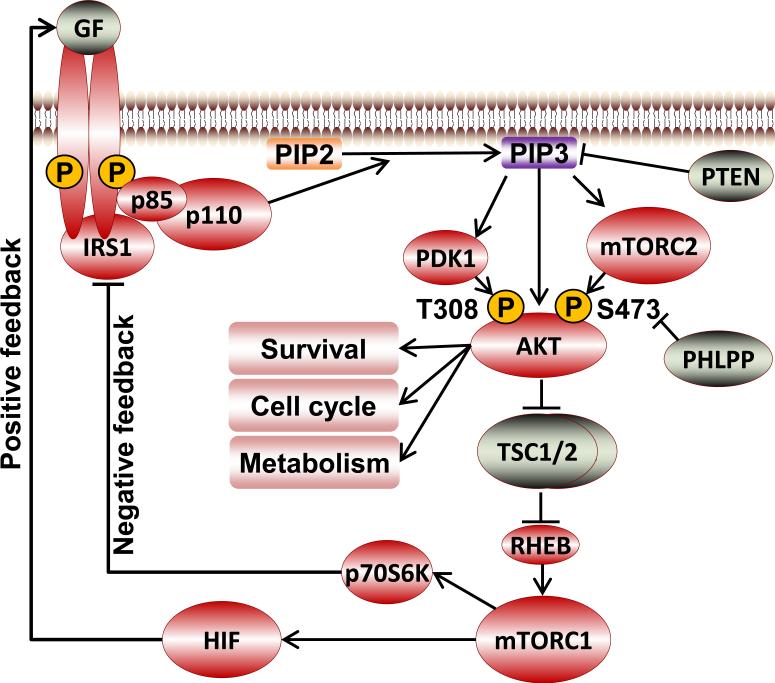
PI3K is normally activated by extracellular signals in physiologic conditions (Fig. 1). A variety of stimuli, including growth factors, cytokines, and hormones, may activate PI3K. Growth factors, such as epidermal growth factor (EGF), platelet-derived growth factor (PDGF), or insulin-like growth factor (Auger et al., 1989; Ruderman et al., 1990), bind to the N-terminal extracellular region of their corresponding transmembrane receptor tyrosine kinases (RTKs), resulting in autophosphorylation of tyrosine residues on the cytoplasmic regions of the RTKs and in linker molecules. PI3K is then recruited to the RTKs through interactions between the p85 SH2 domains and phospho-Tyr residues on members of the RTK complex, which leads to allosteric activation of PI3K. In addition to RTKs, G protein coupled receptors are another large group of classic upstream regulators that activate PI3K, most commonly p110β (Fruman and Rommel, 2014). In addition, small GTPases such as Ras and RAB5 can activate PI3K (Fruman and Rommel, 2014) directly and indirectly.
Fig. 1.
Schematic diagram of the PI3K/AKT pathway.
Activated PI3K phosphorylates phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] to form phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3] on the inner cell membrane, which propagates activation signals to downstream molecules. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) serves as a lipid phosphatase to convert PtdIns(3,4,5)P3 back to PtdIns(4,5)P2, turning off the pathway (Hennessy et al., 2005; Gewinner et al., 2009). Inositol polyphosphate-phosphatase type II (INPP4B) converts PtdIns(3,4)P2 to PtdIns(3)P1, attenuating PI3K signaling as well as redirecting signaling to different pleckstrin homology (PH) domain containing proteins (Gewinner et al., 2009). A key mediator responding to the PtdIns(3,4,5)P3 signal generated by PI3K is the serine/threonine kinase AKT. However, it is important to note that there are multiple PI3K downstream targets in addition to AKT that can initiate particular signaling cascades independent of AKT and also converge on the same downstream cascades.
AKT and downstream effectors
The serine/threonine kinase AKT (also known as protein kinase B), comprising a group of three isoforms (AKT1, AKT2, and AKT3) in mammals, is a key propagator of PI3K signaling. In the canonical PI3K/AKT activation model, AKT and its upstream kinase, 3-phosphoinositide-dependent protein kinase-1 (PDK1), are recruited to the inner cell membrane via interactions of their PH domains with PtdIns(3,4,5)P3 generated by PI3K, which initiates AKT phosphorylation at Thr308 (based on AKT1 amino acid sequence unless designated otherwise) in the activation T-loop by PDK1 (Alessi et al., 1997). mTOR complex 2 (mTORC2) and other potential kinases phosphorylate AKT at Ser473 in the regulatory hydrophobic domain, resulting in optimal activation (Sarbassov et al., 2005; Bozulic et al., 2008;). The phosphorylated active AKT then translocates from the cell membrane to other cell compartments to phosphorylate multiple downstream substrates to fulfill AKT functions (Andjelkovic et al., 1997) (Fig. 1).
AKT activation and stability are elaborately regulated by multiple layers of phosphorylation. In addition to Thr308 and Ser473, phosphorylation of which is required for optimal activation, 31 other residues of AKT1 have been experimentally identified using mass spectrometry or site-specific approaches as potential sites for phosphorylation, including 11 serine residues, 14 threonine residues, and 6 tyrosine residues (http://www.phosphosite.org) (Hornbeck et al., 2012). AKT2 has 22 identified phosphorylation sites, and AKT3 has 18. The number of potential phosphorylation sites of AKT is expected to grow further, considering the total number of serine, threonine, and tyrosine residues in AKT (e.g., 71 in AKT1).
The regulation, stoichiometry, and functions of these phosphorylation sites are only beginning to be elucidated. For example, co-translational phosphorylation at Thr450 is required for proper folding and stability of AKT (Ikenoue et al., 2008; Oh et al., 2010). Phosphorylation at Thr305, Thr312, and Tyr474 has been shown to contribute to optimal AKT activation. Thr72 and Ser246 have been proposed to be trans-autophosphorylated, whereas Thr34, Thr450, and Tyr176 phosphorylation is likely mediated by upstream kinases, including atypical protein kinase C, c-Jun N-terminal kinases, and Ack1 (Mao et al., 2000; Powell et al., 2003; Mahajan et al., 2010). Furthermore, phosphorylation of AKT is isoform-specific. For example, AKT1 Ser129, but not the equivalent AKT2 Ser131, is phosphorylated by the casein kinase 2, contributing to AKT1-specific substrate recognition (Girardi et al., 2014) and potentially to differential functions of AKT1 and AKT2. We have also shown that the pattern of phosphorylation events is markedly different between AKT1 and AKT2 under basal and ligand-induced conditions in multiple cell types (Guo et al., 2013). Six detectable formats of AKT1 with different pI values, but only three detectable formats of AKT2, are present at basal conditions, representing complex combinations of phosphorylation of different sites on individual AKT molecules (Guo et al., 2013). Following insulin stimulation, a large percentage of AKT1 is phosphorylated at Thr308 and Ser473. In contrast, only very little AKT2 is phosphorylated at the equivalent sites (Guo et al., 2013).
Activated AKT phosphorylates a large number of substrates controlling almost every aspect of physiologic and pathologic cellular functions, including cell survival, growth, metabolism, tumorigenesis, and metastasis (Brazil et al., 2004; Manning and Cantley, 2007) (Fig. 1). A critical downstream signaling branch is AKT-mediated activation of mTOR complex 1 (mTORC1), which leads to protein translation and lipid or nucleotide synthesis. AKT phosphorylates and inhibits tuberous sclerosis (TSC) complex 1/2 (Cai et al., 2006), a GTPase-activating protein for the Ras-related small G protein RHEB; therefore, AKT phosphorylation activates RHEB, which in turn activates mTORC1 (Fig. 1) (Manning and Cantley, 2003). AKT also promotes mTORC1 activation by phosphorylating and inhibiting an mTORC1 component, 40KD proline-rich AKT1 substrate 1 (Haar et al., 2007). mTORC1 phosphorylates a variety of substrates, including p70 ribosomal S6 kinase (p70S6K) and eIF4E-binding proteins, which promotes anabolic syntheses (Manning and Cantley, 2003; Fruman and Rommel, 2014;). The mTORC1 signaling branch is also dominantly and negatively regulated by energy-sensing AMP-activated protein kinase, which phosphorylates and activates TSC1/2, therefore inhibiting mTORC1 (Hay and Sonenberg, 2004; Sengupta et al., 2010). In addition, activation of mTOR is dependent on the presence of adequate nutrients and in particular amino acids. Furthermore, MAP kinases and p90RSK phosphorylate a number of the same targets in the mTOR pathway as do AKT isoforms. Thus, activation of the mTOR cascade represents the integration of signaling from multiple upstream cascades. Activation of mTORC1 can inhibit the PI3K/AKT pathway through a negative feedback loop mediated by p70S6K phosphorylation of insulin receptor substrate 1 (O'Reilly et al., 2006; Tremblay et al., 2007).
Genetic alterations of the PI3K/AKT pathway in cancer
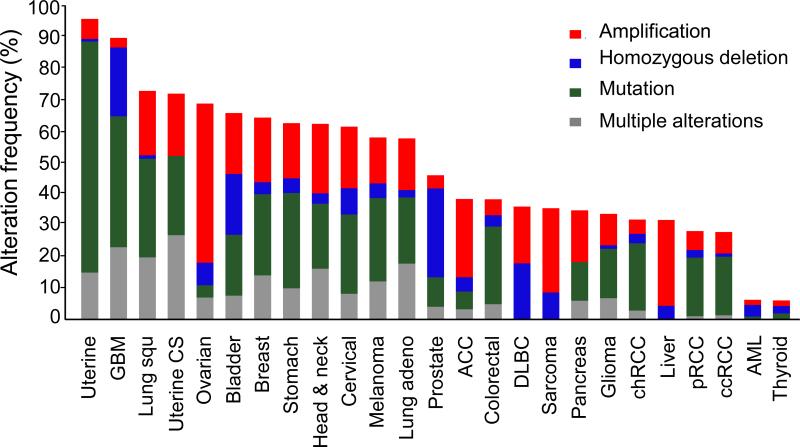
The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) has characterized more than 11,000 cancer patient samples using large-scale genome sequencing coupled with epigenomic, transcriptomic, and proteomic analyses; these data have greatly advanced our understanding of the molecular basis of cancer. Data are available from TCGA or other websites providing analysis tools for large datasets, such as cBioPortal (www.cbioportal.org) (Cerami et al., 2012; Gao et al., 2013) and The Cancer Proteome Atlas (www.TCPAportal.org) (Li et al., 2013). We compiled a panel of 20 representative components of the PI3K/AKT pathway (GNB2L1, EGFR, PIK3CA, PIK3R1, PIK3R2, PTEN, PDPK1, AKT1, AKT2, AKT3, FOXO1, FOXO3, MTOR, RICTOR, TSC1, TSC2, RHEB, AKT1S1, RPTOR, and MLST8), modified from the PI3K pathway genes summarized in the TCGA ccRCC study (The Cancer Genome Atlas Research, 2013), and surveyed the genetic alterations of the panel components in the latest provisional TCGA datasets of all 25 cancer types currently available in cBioPortal. Genetic alterations of the panel components were identified in every cancer lineage. The mutation and CNA rates ranged from 6% in thyroid carcinoma to a striking 95% in endometrioid carcinoma (Fig. 2). The average genetic alteration rate was about 50% across all cancer types.
Fig. 2.
Genetic alteration frequency of 20 representative components of the PI3K/AKT pathway across 25 cancer types in The Cancer Genome Atlas (TCGA) database at cBioPortal.
We surveyed the mutation and copy number alteration rates of the panel components in the latest provisional TCGA datasets at cBioPortal, including uterine corpus endometrial carcinoma (Uterine), glioblastoma multiforme (GBM), lung squamous cell carcinoma (Lung squ), uterine carcinosarcoma (Uterine CS), ovarian serous cystadenocarcinoma (Ovarian), bladder urothelial carcinoma (Bladder), breast invasive carcinoma (Breast), stomach adenocarcinoma (Stomach), head and neck squamous cell carcinoma (Head & neck), cervical squamous cell carcinoma and endocervical adenocarcinoma (Cervical), skin cutaneous melanoma (Melanoma), lung adenocarcinoma (Lung adeno), prostate adenocarcinoma (Prostate), adrenocortical carcinoma (ACC), colorectal adenocarcinoma (Colorectal), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), sarcoma (Sarcoma), pancreatic adenocarcinoma (Pancreas), brain lower grade glioma (Glioma), kidney chromophobe (chRCC), liver hepatocellular carcinoma (Liver), kidney renal papillary cell carcinoma (pRCC), kidney renal clear cell carcinoma (ccRCC), acute myeloid leukemia (AML), and thyroid carcinoma (Thyroid).
Genetic alterations were identified in every layer of the PI3K/AKT signaling cascade. More importantly, many of these genetic alterations have been independently demonstrated by experimental manipulation to be oncogenic. For example, at the layer of PI3K, PIK3CA gene coding for the catalytic subunit p110α has been found to be frequently mutated in cancer, underscoring the importance of this isoform (Samuels et al., 2004; Engelman, 2009; Fruman and Rommel, 2014). PIK3CA was first found to be highly amplified in ovarian cancer, implicating its role as an oncogene (Shayesteh et al., 1999). High-frequency somatic mutations of PIK3CA were then identified in colorectal cancer, glioblastoma, gastric cancer, breast cancer, and lung cancer (Samuels et al., 2004). Strikingly, most of the mutations were observed in two small, highly conserved clusters in the helical and kinase domains of p110α (Samuels et al., 2004), displaying a pattern of activation mutations. Many additional studies revealed PIK3CA mutations in almost all cancer types, including ovarian, head and neck, cervical, endometrial, and kidney cancers (Campbell et al., 2004; Murph et al., 2008). The recurrent mutations of PIK3CA were demonstrated to be oncogenic (Samuels et al., 2004; Kang et al., 2005; Samuels et al., 2005). Intriguingly, PIK3CA mutations are generally mutually exclusive with mutations in other members of the pathway (Stemke-Hale et al., 2008) with the exception of endometrial cancer and a subset of bowel cancers where mutations in multiple pathway members are common (Liang et al., 2012).
Traditionally, PIK3CA was regarded as the second most mutated oncogene in cancer after KRAS gene. However, the latest TCGA data indicate that PIK3CA is actually the most altered oncogene targeted by mutations and amplifications across all cancers (Lawrence et al., 2014), and PIK3CA mutation is particularly prevalent in endometrial cancer (57%), lung squamous cell carcinoma (48%), cervical cancer (42%), invasive breast cancer (39%), head and neck cancer (35%), colon cancer (30%), and ovarian cancer (30%; www.cbioportal.org). Likewise, frequent mutations of PIK3R1 gene coding for the p85 regulatory subunit, have also been identified in TCGA, particularly in endometrial cancer (34%), glioblastoma (11%), uterine carcinosarcoma (11%), and bladder urothelial carcinoma (7.8%; www.cbioportal.org) (Cheung et al., 2011). Oncogenic mutations of p85α may target PI3K/AKT pathway activation by activating p110 (Jaiswal et al., 2009) or inhibiting PTEN (Cheung et al., 2011). Recently, neomorphic truncation mutations of p85α have been discovered to promote the mitogen-activated protein kinase pathway in cancer, revealing a new category of oncogenic functions for p85α mutations (Cheung et al., 2014).
PTEN, the lipid phosphatase that counteracts the PI3K pathway, is the second most mutated or deleted tumor suppressor after p53 across all cancers. Frequent mutations and deletions of PTEN have been found in various cancers, including endometrial cancer (67%), glioblastoma (42%), and prostate cancer (22%). In addition to these genetic alterations, PTEN is also targeted by promoter methylation, transcriptional inhibition, microRNA, and posttranslational modifications, resulting in downregulation of PTEN functions and subsequently upregulation of PI3K/AKT pathway signaling in cancer (Song et al., 2012). INPP4B is another emerging lipid phosphatase tumor suppressor frequently mutated or deleted in multiple cancer types (Hennessy et al., 2005; Gewinner et al., 2009; Agoulnik et al., 2011).
At the layer of AKT, the core component of the PI3K/AKT pathway, amplification and mutations of AKT isoforms have been identified in multiple cancer types, although not at high levels. Amplification of AKT2 was first identified in ovarian cancer (Cheng et al., 1992) and then in multiple other cancers, including pancreatic, gastric, liver, and lung cancers (Cheng et al., 1996; Hers et al., 2011). The latest TCGA data show that AKT amplifications are prevalent in ovarian (21%), uterine (20%), invasive breast (18%), liver (15%), and bladder (12%) cancers. Furthermore, amplification is more common in AKT2 or AKT3 than in AKT1 (Cohen, 2013) (www.cbioportal.org), suggesting that AKT contributions to cancer are isoform-specific.
Although AKT is a central member of the highly mutated PI3K/AKT pathway, somatic AKT mutations occur at relatively low frequencies and were discovered only recently. E17K mutation was first identified in AKT1 in breast, colorectal, and ovarian cancers as an oncogenic mutation (Carpten et al., 2007) (Stemke-Hale et al., 2008), and was then identified in more cancer types and in the AKT3 isoform (Davies et al., 2008). This mutation enhances the association of AKT with the cell membrane, promoting AKT activation (Carpten et al., 2007).
At the layer downstream of AKT, mutations in TSC1/TSC2 have been identified but are relatively rare (Lawrence et al., 2014). RHEB, an activator of mTORC1, is overexpressed in liver, lung, bladder, stomach, colorectal, and breast cancers (Lu et al., 2010). Elevated RHEB was sufficient to induce constitutive mTORC1 pathway activation and skin tumors in mouse models, indicating that RHEB is a bona fide oncogene (Lu et al., 2010). More recently, a recurring activation mutation (Y35N) was identified in endometrial cancer and ccRCC (Lawrence et al., 2014), validating the oncogenic role of RHEB in the PI3K/AKT pathway.
In summary, most components of the PI3K/AKT pathway harbor genetic alterations that activate the pathway across cancer lineages. Next, we summarize the genetic alterations of the PI3K/AKT pathway specifically in the three major types of RCC.
Genetic alterations of the PI3K/AKT pathway in RCC
Kidney cancer, which is increasing in incidence worldwide, falls into the class of cancers with a high risk of death; an estimated 14,000 of the 64,000 newly diagnosed patients in the United States in 2014 died of the disease. RCC accounts for about 90% of cases of kidney cancer. Other minor types of kidney cancer include renal pelvis carcinoma and Wilms tumor (Bhatt and Finelli, 2014), which are not discussed in this review. There are three major histologic subtypes of RCC, including ccRCC (accounting for 75%–80% of cases of RCC), papillary RCC (pRCC, 10%– 15%), and chromophobe RCC (chRCC, 4%–5%), as well as many other minor RCC subtypes (Jonasch et al., 2014). We focus on the three major RCC subtypes in this review.
The PI3K/AKT pathway in ccRCC
ccRCC is the most common type of RCC, accounting for the vast majority of kidney cancer deaths (Jonasch et al., 2014). ccRCC is notoriously resistant to traditional chemotherapies and radiotherapies. Fortunately, recent rapid advances in genomic and proteomic studies have revealed much of the molecular basis of ccRCC and, therefore, provided new therapeutic opportunities for this challenging disease. The von Hippel Lindau (VHL) gene is mutated in 80%–90% of cases of ccRCC (Nickerson et al., 2008), indicating that loss of VHL function is a key underlying driver (Jonasch et al., 2012). VHL serves as the substrate recognizing subunit for a ubiquitin E3 ligase complex that mediates ubiquitination and subsequent degradation of hypoxia-inducible factors (HIFs) (Maxwell et al., 1999). HIF upregulation due to VHL loss plays a critical role in ccRCC tumorigenesis. However, loss of VHL function alone is not sufficient for ccRCC initiation (Frew and Moch, 2015). Other genetic or epigenetic events are required for ccRCC to initiate or progress.
Another group of frequently altered genes in ccRCC is the components of the PI3K/AKT pathway. In the TCGA ccRCC dataset, the overall genetic alteration rate of the 20 representative PI3K/AKT pathway panel components in our analysis was 27.7% (Fig. 3). Most of the components were identified with genetic alterations, including GNB2L1 amplification (6%), PIK3CA amplifications or mutations (5%), PTEN deletions or mutations (5%), or MTOR mutations (6%), in a largely mutually exclusive manner (Fig. 3) (The Cancer Genome Atlas Research, 2013). MTOR mutations are highly clustered in small regions in ccRCC, conferring mTOR hyperactivation (Grabiner et al., 2014). Seven AKT mutations (~2%) were identified, including a known E40K activation mutation in AKT1 (Sun et al., 2001) and an E17K activation mutation in AKT3. Three recurrent Y35N mutations in RHEB were identified, consistent with activation of the PI3K/AKT pathway (Lawrence et al., 2014).
Fig. 3.
Genetic alterations of 20 representative components of the PI3K/AKT pathway in clear cell renal cell carcinoma.
We surveyed the mutations and copy number alterations of the PI3K/AKT panel components in the latest provisional TCGA dataset of ccRCC at cBioPortal. Small grey rectangles represent patients. Genes and alteration rates are indicated to the left.
Similar genetic alteration rates of the PI3K pathway in ccRCC were identified in other large-scale integrated analyses (Sato et al., 2013). Note that we surveyed only the genetic alterations of 20 representative PI3K/AKT pathway components in ccRCC. The true genetic alteration rate of the pathway including all components is likely higher. Furthermore, considering the dysregulation of the PI3K/AKT pathway at epigenetic, posttranscriptional, and posttranslational levels, the aberrant pathway activation is likely more prevalent in ccRCC. Integrated genomic and proteomic studies have shown a convergence upon PI3K/AKT/mTOR activation through a variety of mechanisms in ccRCC (Akbani et al., 2014) (Fisher et al., 2014). Furthermore, high intratumor genomic heterogeneity has been detected in ccRCC (Gerlinger et al., 2012; Gerlinger et al., 2014); therefore, current mutation and CNA data represent only a small fraction of the full spectrum of genetic alterations in a given tumor. The frequency of the PI3K/AKT pathway activation in ccRCC could be substantially higher than that indicated by currently available data.
The VHL/HIF and PI3K/AKT pathways cross talk extensively in a large signaling network, contributing to ccRCC. First, HIF upregulation due to VHL loss promotes expression of a variety of growth factors, including EGF, PDGF, and vascular endothelial growth factor (VEGF) (Jonasch et al., 2012), which in turn may activate the PI3K/AKT pathway through RTKs. The subsequent activation of mTORC1 and mTORC2 then promotes HIF expression (Bernardi et al., 2006; Toschi et al., 2008), therefore forming a positive feedback loop resulting in constitutive activation of the signaling network (Fig. 1). This is consistent with the observation that PI3K/AKT signaling is usually activated in ccRCC (Li et al., 2013; Akbani et al., 2014). Although the overall mutation rate of the PI3K/AKT pathway in ccRCC is relatively low compared with many other cancer types (Fig. 2), the overall activation of AKT in ccRCC is high among all cancer types, as indicated by high phosphorylation levels of AKT and AKT substrates (Li et al., 2013; Akbani et al., 2014). Therefore, the VHL/HIF and PI3K/AKT pathways are closely connected, forming a large signaling network contributing to ccRCC.
In addition to alterations in the VHL/HIF and PI3K/AKT pathways, high frequencies of gene mutations or deletions of PBRM1 (36%), SETD2 (15%), BAP1 (13%), and KDM5C (7%) have also been identified in the ccRCC TCGA dataset and other studies (Varela et al., 2011; The Cancer Genome Atlas Research, 2013). Notably, all of these genes play important roles in chromatin remodeling, indicating that dysregulation of chromatin remodeling is a generalizable event in ccRCC. The potential role of the VHL/HIF and PI3K/AKT pathways in the regulation of chromatin remodeling in ccRCC remains largely unknown, and would be an interesting topic to explore.
The PI3K/AKT pathway in pRCC
pRCC is the second most common subtype of RCC (Twardowski et al., 2014). In the TCGA pRCC dataset, the overall genetic alteration rate of the PI3K/AKT pathway panel components was 28% (Fig. 2), including amplifications of GNB2L1, PDK1, and RPTOR and mutations of PTEN and the PI3K subunits, many of which are known to activate the signaling pathway.
pRCC is further divided into two types, type 1 and type 2, on the basis of different cell morphologies (Jonasch et al., 2014). Studies of hereditary pRCC revealed that type 1 pRCC is associated with mutations of the mesenchymal epithelial transition (MET) gene. MET is a transmembrane receptor for hepatocyte growth factor, which is upstream of the PI3K/AKT pathway. Activation mutations of MET found in type 1 pRCC lead to increased proliferation, invasion, and metastases, owing at least in part to activation of the PI3K/AKT pathway (Twardowski et al., 2014). Kinases activated by gene fusions are found in most cancer types, including pRCC (Stransky et al., 2014). MET fusions to other genes, leading to MET activation, were identified in multiple pRCC samples, demonstrating an alternative mechanism of activation in addition to gene amplifications and mutations (Stransky et al., 2014).
Type 2 pRCC was found to be associated with mutations in fumarate hydratase (FH) (Tomlinson, 2002). FH is a tricarboxylic acid cycle enzyme that catalyzes the conversion from fumarate to malate. Loss of FH function owing to mutations results in accumulation of fumarate, leading to upregulation of HIF1α (Isaacs, 2005), which mimics the loss of VHL in ccRCC.
The PI3K/AKT pathway in chRCC
Compared with ccRCC, chRCC is a rare subtype of RCC; only 3,000 new cases are diagnosed in the United States annually (Davis et al., 2014). However, rare cancer can offer a unique opportunity to identify cancer drivers that are not found in common cancers. Although only 66 patient samples are currently available in the chRCC TCGA dataset, the PI3K/AKT pathway has been shown to be significantly targeted genetically; the 20 representative panel components are altered in 32% of patients (21 out of 66; Fig. 1). PTEN is the most mutated or deleted component, occurring in 11% of patients. Mutations and CNAs have also been identified in multiple other components, including PDK1, AKT1, TSC1/TSC2, and mTOR (Davis et al., 2014).
As a gatekeeper, TP53 is the most mutated gene across all cancer types. TP53 is the most frequently mutated gene in chRCC (33%) (Davis et al., 2014), but not in ccRCC (2%) or pRCC (4%). However, unlike chRCC, ccRCC may find an alternative way to interfere with p53 function through loss of VHL. VHL has been shown to interact with p53, enhancing the stability and activation of p53 (Roe et al., 2006).
Additional mechanisms activating the PI3K/AKT pathway in RCC
Activation of the PI3K/AKT pathway is not always initiated from extracellular signals and transmembrane receptors. Multiple other mechanisms of AKT activation or inhibition have been identified in RCC, contributing to aberrant PI3K/AKT pathway activation. We have shown that glucose deprivation induces a unique form of AKT phosphorylation and activation in multiple cell lines, including 786-0, RXF393, ACHN, and other RCC cells (Gao et al., 2014). AKT is selectively phosphorylated on Thr308 through enhanced protein complex formation with PDK1 and 78-kDa glucose-regulated protein under glucose-deprivation conditions, which likely represents a novel AKT activation mechanism in RCC, as well as in other cancers when metabolic stress is present (Dawood et al., 2014; Gao et al., 2014).
MicroRNA is emerging as a new class of important regulators of the PI3K/AKT pathway. miR-182-5p has been shown to be a negative regulator of AKT, and downregulation of this microRNA results in AKT activation and subsequent RCC proliferation (Xu et al., 2014). miR-122 has been shown to be a positive regulator of the PI3K/AKT signaling pathway, promoting proliferation, invasion, and migration of RCC cells (Lian et al., 2013).
Targeting the PI3K/AKT pathway in RCC
The treatment of ccRCC is currently leading the development of targeted therapy. Multiple targeted drugs have been approved by the US Food and Drug Administration for ccRCC. The first class of approved targeted drugs for metastatic RCC was anti-angiogenesis drugs that target elevated VEGF or VEGF receptors (VEGFR), on the basis of the rationale that ccRCC is a highly vascularized cancer owing to activated VEGF signaling resulting from HIF upregulation. This class of drugs includes the small-molecule inhibitors sorafenib (approved in 2005), sunitinib (2006), pazopanib (2009), and axitinib (2010), which target VEGFR and other RTKs, and a monoclonal antibody, bevacizumab (2009), which targets VEGF. Newer inhibitors (such as brivanib, cabozantinib, cediranib, dovitinib, foretinib, lenvatinib, linifanib, nintedanib, regorafenib, tivozanib, vandetanib, and aflibercept) for VEGFR and additional targets (such as PDGFR, FGFR, c-kit, and MET) and monoclonal antibodies (ramucirumab and rilotumumab) are being developed and tested in clinical trials for ccRCC (Dutcher, 2013; Randall et al., 2014). Although some studies (Huang et al., 2010) suggested that anti-angiogenesis drugs such as sunitinib primarily target tumor endothelial cells instead of cancer cells, the studies also showed that the drugs inhibited RTKs in cancer cells. The role of the PI3K/AKT pathway in cancer treated with anti-angiogenesis drugs has not been elucidated. Considering that these drugs inhibit multiple other RTKs in addition to VEGFR (Jonasch et al., 2014), it is likely that these drugs may target the PI3K/AKT pathway in cancer cells, contributing to tumor inhibition.
The second class of approved targeted drugs for metastatic RCC includes temsirolimus (approved in 2007) and everolimus (2009) (Figlin et al., 2013; Jonasch et al., 2014). These drugs target mTOR, a component of the PI3K/AKT pathway, on the basis of the rationale that mTOR activation resulting from PI3K/AKT and other pathway activation is important for ccRCC tumorigenesis and progression. Furthermore, mTOR is a key regulator of cell metabolism, whereas RCC is primarily a cancer of metabolism dysregulation (Linehan et al., 2010).
Currently available targeted drugs for metastatic and advanced ccRCC have shown some degree of efficacy; however, even for the subpopulation of patients that demonstrate responses to these drugs, the responses are short and patients almost always show disease progression within a year (Jonasch et al., 2012). Two-thirds of patients diagnosed with ccRCC are also diagnosed with metastatic RCC, for which the 5-year overall survival rate is only 20% (Jonasch et al., 2014). Thus, identification of new drug targets, development of new drugs and in particular rational drug combinations for RCC are still urgently needed. As described above, the PI3K/AKT pathway components are frequently targeted in RCC, suggesting that targeting the pathway, either alone or in combination with other drugs, holds great potential in RCC (Brugge et al., 2007; Fruman and Rommel, 2014).
The currently approved mTOR inhibitors, temsirolimus and everolimus, are both rapalog allosteric mTOR inhibitors, which result in only partial inhibition of mTORC1 activation toward some substrates (e.g., p70S6K) but not others (e.g., eIF4E-binding proteins) (Fruman and Rommel, 2014). This might be part of the reason that rapalogs provided only modest survival benefits for patients with ccRCC (Randall et al., 2014). A new class of mTOR inhibitors that directly target mTOR kinase activity, including AZD8055, MLN0128, and OSI-027, are in clinical development and clinical trials for the treatment of ccRCC (Cho, 2013). These agents target both mTORC1 and mTOCR2 conferring greater efficacy for the PI3K/AKT/mTOR pathway inhibition.
PI3K inhibitors have been tested in multiple cancer types including RCC. However, no drug has reached clinical use. Numerous pan or isoform specific PI3K inhibitors are in clinical development or clinical trials in RCC including GDC0941, XL147, BKM120, NVP-BYL719 (p110α specific), SAR260301 (p110β specific), and TGR-1202 (p110β specific) (Cho, 2013; Fruman and Rommel, 2014). PI3K and mTOR are structurally related. Many dual inhibitors for both PI3K and mTOR (such as NVP-BEZ235, GDC-0980 and XL765) are being investigated, which may overcome the limitations of single kinase inhibition resulting from feedback loops (O'Reilly et al., 2006; Cho, 2013; Figlin et al., 2013; Fruman and Rommel, 2014).
Allosteric AKT inhibitors (such as MK2206) and kinase inhibitors (such as GDC0068 and AZD5663) are being tested in ccRCC clinical trials (Cho, 2013; Fruman and Rommel, 2014). Different AKT isoforms mediate critical non-redundant functions in cancer pathophysiology. For example, AKT1 has been shown to be a major contributor to tumor initiation, whereas AKT2 appears to primarily promote tumor metastasis (Dillon et al., 2009; Endersby et al., 2011). In ccRCC cells, HIF2α expression is dependent on AKT2 but not on other isoforms, whereas HIF1α expression is dependent on AKT3 (Toschi et al., 2008). Therefore, isoform-specific AKT inhibition is also a promising approach in ccRCC.
In addition to treatments targeting angiogenesis and the PI3K/AKT/mTOR pathway, other novel treatment approaches are being explored and tested in RCC. Immunotherapy is an emerging promising approach. Monoclonal antibodies (such as ipilimumab and tremelimumab) against the immune checkpoint inhibitors cytotoxic T lymphocyte antigen 4 or programmed cell death 1 have shown antitumor activity and are being tested in multiple clinical trials for the treatment of RCC (Dutcher, 2013; Jonasch et al., 2014). Peptide vaccines have also been in development for certain types of RCC (Jonasch et al., 2014). Considering that one-third of all VHL mutations in ccRCC are missense point mutations, generating a full-length unstable protein with residual functionality (Lee et al., 2009; Rechsteiner et al., 2011), we proposed that destabilized VHL could be refunctionalized by modulating the proteostasis of missense point mutated VHL (Ding et al., 2012), and we provided evidence that the proteasome inhibitors bortezomib and carfilzomib, which are currently in clinical use, stabilize VHL-R167Q, the most common hereditary mutation, and increase its ability to downregulate HIF2α (Ding et al., 2014). The variety of novel approaches, together with the development of the next generation of inhibitors for angiogenesis and the PI3K/AKT/mTOR pathway, are expected to make an impact on ccRCC patient care.
Concluding remarks
The treatment of metastatic RCC has changed dramatically from traditional chemotherapies or radiotherapies to targeted cancer therapy, aiming to fulfill the promise of personalized molecular medicine (Jonasch et al., 2012). The efficient implementation of this new generation of cancer treatments is dependent on a clear understanding of protein function and signaling networks in cancer cells, especially the druggable oncogenic pathways such as the PI3K/AKT pathway. The latest large-scale genomic and proteomic studies of RCC have provided a molecular basis of the disease, which is likely to lead to rapid development of new targeted drugs for this deadly cancer.
Acknowledgements
The work is supported by the grant from The University of Texas MD Anderson Cancer Center Kidney Cancer Multidisciplinary Research Program to ZD, the NCI CCSG grant (No. P30 CA016672) and the GDAC grant (No. 5U24CA143883).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agoulnik IU, Hodgson MC, Bowden WA, Ittmann MM. INPP4B: the new kid on the PI3K block. Oncotarget. 2011;2:321–328. doi: 10.18632/oncotarget.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbani R, Ng PKS, Werner HMJ, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang J-Y, Yoshihara K, Li J, Ling S, Seviour EG, Ram PT, Minna JD, Diao L, Tong P, Heymach JV, Hill SM, Dondelinger F, Städler N, Byers LA, Meric-Bernstam F, Weinstein JN, Broom BM, Verhaak RGW, Liang H, Mukherjee S, Lu Y, Mills GB. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B α. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Celeste Simon M, Rafii S, Pandolfi PP. PML inhibits HIF-1α translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat. Rev. Urol. 2014;11:517–525. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBα/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol. Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Brugge J, Hung M-C, Mills GB. A new mutational AKTivation in the PI3K Pathway. Cancer Cell. 2007;12:104–107. doi: 10.1016/j.ccr.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Cai S-L, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian Y-W, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai M-HT, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JQ, Godwin AK, Bellacosa A, Taguchi T, Franke TF, Hamilton TC, Tsichlis PN, Testa JR. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc. Natl. Acad. Sci. USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, Testa JR. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LWT, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, Ju Z, Cantley LC, Scherer SE, Liang H, Lu KH, Broaddus RR, Mills GB. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, Lydia WT, Yu S, Zhang D, Li J, Ng, Patrick KS, Panupinthu N, Mitra S, Ju Z, Yu Q, Liang H, Hawke, David H, Lu Y, Broaddus, Russell R, Mills, Gordon B. Naturally Occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26:479–494. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D. Novel targeting of phosphatidylinositol 3-kinase and mammalian target of rapamycin in renal cell carcinoma. Cancer J. 2013;19:311–315. doi: 10.1097/PPO.0b013e31829d5cea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM. The AKT genes and their roles in various disorders. Am. J. Med. Genet A. 2013;161:2931–2937. doi: 10.1002/ajmg.a.36101. [DOI] [PubMed] [Google Scholar]
- Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, Hacker KE, Bhanot G, Gordenin DA, Chu A, Gunaratne PH, Biehl M, Seth S, Kaipparettu BA, Bristow CA, Donehower LA, Wallen EM, Smith AB, Tickoo SK, Tamboli P, Reuter V, Schmidt LS, Hsieh JJ, Choueiri TK, Hakimi AA, Cancer Genome Atlas Research, N. Chin L, Meyerson M, Kucherlapati R, Park WY, Robertson AG, Laird PW, Henske EP, Kwiatkowski DJ, Park PJ, Morgan M, Shuch B, Muzny D, Wheeler DA, Linehan WM, Gibbs RA, Rathmell WK, Creighton CJ. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood M, Mills GB, Ding Z. Shrewd AKT regulation to survive. Oncoscience. 2014;1:113–114. doi: 10.18632/oncoscience.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009;69:5057–5064. doi: 10.1158/0008-5472.CAN-08-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, German P, Bai S, Feng Z, Gao M, Si W, Sobieski MM, Stephan CC, Mills GB, Jonasch E. Agents that stabilize mutated von Hippel-Lindau (VHL) protein: results of a high-throughput screen to identify compounds that modulate VHL proteostasis. J. Biomol. Screen. 2012;17:572–580. doi: 10.1177/1087057112436557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, German P, Bai S, Reddy AS, Liu XD, Sun M, Zhou L, Chen X, Zhao X, Wu C, Zhang S, Mills GB, Jonasch E. Genetic and pharmacological strategies to refunctionalize the von Hippel Lindau R167Q mutant protein. Cancer Res. 2014;74:3127–3136. doi: 10.1158/0008-5472.CAN-13-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher JP. Recent developments in the treatment of renal cell carcinoma. Ther. Adv. Urol. 2013;5:338–353. doi: 10.1177/1756287213505672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersby R, Zhu X, Hay N, Ellison DW, Baker SJ. Nonredundant functions for Akt isoforms in astrocyte growth and gliomagenesis in an orthotopic transplantation model. Cancer Res. 2011;71:4106–4116. doi: 10.1158/0008-5472.CAN-10-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Figlin RA, Kaufmann I, Brechbiel J. Targeting PI3K and mTORC2 in metastatic renal cell carcinoma: New strategies for overcoming resistance to VEGFR and mTORC1 inhibitors. Int. J. Cancer. 2013;133:788–796. doi: 10.1002/ijc.28023. [DOI] [PubMed] [Google Scholar]
- Fisher R, Horswell S, Rowan A, Salm M, de Bruin E, Gulati S, McGranahan N, Stares M, Gerlinger M, Varela I, Crockford A, Favero F, Quidville V, Andre F, Navas C, Gronroos E, Nicol D, Hazell S, Hrouda D, O'Brien T, Matthews N, Phillimore B, Begum S, Rabinowitz A, Biggs J, Bates P, McDonald N, Stamp G, Spencer-Dene B, Hsieh J. Development of synchronous VHL syndrome tumors reveals contingencies and constraints to tumor evolution. Genome Biol. 2014;15:433. doi: 10.1186/s13059-014-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew IJ, Moch H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu. Rev. Pathol. 2015;10:263–289. doi: 10.1146/annurev-pathol-012414-040306. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:1–19. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liang J, Lu Y, Guo H, German P, Bai S, Jonasch E, Yang X, Mills GB, Ding Z. Site-specific activation of AKT protects cells from death induced by glucose deprivation. Oncogene. 2014;33:745–755. doi: 10.1038/onc.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, Martinez P, Phillimore B, Begum S, Rabinowitz A, Spencer-Dene B, Gulati S, Bates PA, Stamp G, Pickering L, Gore M, Nicol DL, Hazell S, Futreal PA, Stewart A, Swanton C. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, Pandolfi PP, Cantley LC. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi C, James P, Zanin S, Pinna LA, Ruzzene M. Differential phosphorylation of Akt1 and Akt2 by protein kinase CK2 may account for isoform specific functions. BBA-Mol. Cell Res. 2014;1843:1865–1874. doi: 10.1016/j.bbamcr.2014.04.020. [DOI] [PubMed] [Google Scholar]
- Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated mTOR mutations are Hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gao M, Lu Y, Liang J, Lorenzi PL, Bai S, Hawke DH, Li J, Dogruluk T, Scott KL, Jonasch E, Mills GB, Ding Z. Coordinate phosphorylation of multiple residues on single AKT1 and AKT2 molecules. Oncogene. 2014;33:3463–3472. doi: 10.1038/onc.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haar EV, Lee S.-i., Bandhakavi S, Griffin TJ, Kim D-H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Hers I, Vincent EE, Tavaré JM. Akt signalling in health and disease. Cell. Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Ding Y, Li Y, Luo W-M, Zhang Z-F, Snider J, VandenBeldt K, Qian C-N, Teh BT. Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res. 2010;70:1053–1062. doi: 10.1158/0008-5472.CAN-09-3722. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan K-L. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring P, Dela Vega T, Kenski DM, Bowman KK, Lorenzo M, Li H, Wu J, Modrusan Z, Stinson J, Eby M, Yue P, Kaminker JS, de Sauvage FJ, Backer JM, Seshagiri S. Somatic mutations in p85α promote tumorigenesis through Class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasch E, Futreal PA, Davis IJ, Bailey ST, Kim WY, Brugarolas J, Giaccia AJ, Kurban G, Pause A, Frydman J, Zurita AJ, Rini BI, Sharma P, Atkins MB, Walker CL, Rathmell WK. State of the science: an update on renal cell carcinoma. Mol. Cancer Res. 2012;10:859–880. doi: 10.1158/1541-7786.MCR-12-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. Br. Med. J. 2014 doi: 10.1136/bmj.g4797. doi: http://dx.doi.org/10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Hickey MM, Sanford CA, McGuire CG, Cowey CL, Simon MC, Rathmell WK. VHL Type 2B gene mutation moderates HIF dosage in vitro and in vivo. Oncogene. 2009;28:1694–1705. doi: 10.1038/onc.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, Yang J-Y, Broom BM, Verhaak RGW, Kane DW, Wakefield C, Weinstein JN, Mills GB, Liang H. TCPA: a resource for cancer functional proteomics data. Nat. Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JH, Wang WH, Wang JQ, Zhang YH, Li Y. MicroRNA-122 promotes proliferation, invasion and migration of renal cell carcinoma cells through the PI3K/Akt signaling pathway. Asian Pac. J. Cancer Prev. 2013;14:5017–5021. doi: 10.7314/apjcp.2013.14.9.5017. [DOI] [PubMed] [Google Scholar]
- Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, Guo W, Scherer SE, Carter H, Westin SN, Dyer MD, Verhaak RG, Zhang F, Karchin R, Liu CG, Lu KH, Broaddus RR, Scott KL, Hennessy BT, Mills GB. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat. Rev. Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Shvartsman MB, Lee AY, Shao JM, Murray MM, Kladney RD, Fan D, Krajewski S, Chiang GG, Mills GB, Arbeit JM. Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res. 2010;70:3287–3298. doi: 10.1158/0008-5472.CAN-09-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan K, Coppola D, Challa S, Fang B, Chen YA, Zhu W, Lopez AS, Koomen J, Engelman RW, Rivera C, Muraoka-Cook RS, Cheng JQ, Schonbrunn E, Sebti SM, Earp HS, Mahajan NP. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS One. 2010;5:e9646. doi: 10.1371/journal.pone.0009646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Fang X, Lu Y, Lapushin R, Bast RC, Jr., Mills GB. Inhibition of growth-factor-induced phosphorylation and activation of protein kinase B/Akt by atypical protein kinase C in breast cancer cells. Biochem. J. 352 Pt. 2000;2:475–482. [PMC free article] [PubMed] [Google Scholar]
- Murph M, Smith D, Hennessy B, Lu Y, Joy C, Coombes K, Mills G. Individualized Molecular Medicine: Linking Functional Proteomics to Select Therapeutics Targeting the PI3K Pathway for Specific Patients. In: Coukos G, Berchuck A, Ozols R, editors. Ovarian Cancer. Springer; New York: 2008. pp. 183–195. [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Mukeria A, Holcatova I, Schmidt LS, Toro JR, Karami S, Hung R, Gerard GF, Linehan WM, Merino M, Zbar B, Boffetta P, Brennan P, Rothman N, Chow WH, Waldman FM, Moore LE. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin. Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DJ, Hajduch E, Kular G, Hundal HS. Ceramide disables 3-phosphoinositide binding to the pleckstrin homology domain of protein kinase B (PKB)/Akt by a PKCζ-dependent mechanism. Mol.Cell. Biol. 2003;23:7794–7808. doi: 10.1128/MCB.23.21.7794-7808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall JM, Millard F, Kurzrock R. Molecular aberrations, targeted therapy, and renal cell carcinoma: current state-of-the-art. Cancer Metastasis Rev. 2014;33:1109–1124. doi: 10.1007/s10555-014-9533-1. [DOI] [PubMed] [Google Scholar]
- Rechsteiner MP, von Teichman A, Nowicka A, Sulser T, Schraml P, Moch H. VHL gene mutations and their effects on hypoxia inducible factor HIFα: identification of potential driver and passenger mutations. Cancer Res. 2011;71:5500–5511. doi: 10.1158/0008-5472.CAN-11-0757. [DOI] [PubMed] [Google Scholar]
- Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol. Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ruderman NB, Kapeller R, White MF, Cantley LC. Activation of phosphatidylinositol 3-kinase by insulin. Proc. Natl. Acad. Sci. USA. 1990;87:1411–1415. doi: 10.1073/pnas.87.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat. Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung M-C, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat. Commun. 2014;5:4846, 1–10. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Wang G, Paciga JE, Feldman RI, Yuan Z-Q, Ma X-L, Shelley SA, Jove R, Tsichlis PN, Nicosia SV, Cheng JQ. AKT1/PKBα kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 Cells. Am. J. Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia inducible factors 1 α and 2 α on mTORC1 and mTORC2. J. Biol. Chem. 2008;283:34495–34499. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay F, Brule S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, Marette A. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc. Natl. Acad. Sci. USA. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twardowski PW, Mack PC, Lara PN., Jr. Papillary renal cell carcinoma: current progress and future directions. Clin. Genitourin. Cancer. 2014;12:74–79. doi: 10.1016/j.clgc.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang Z, Wang X, Lin Y, Mao Y, Chen H, Luo J, Liu B, Zheng X, Xie L. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol. Cancer. 2014;13:109. doi: 10.1186/1476-4598-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]