Abstract
Introduction and Objectives
The zinc-finger transcription factor Krϋppel-like factor 2 (KLF2) transduces blood flow into molecular signals responsible for a wide range of responses within the vasculature. KLF2 maintains a healthy, quiescent endothelial phenotype. Previous studies report a range of phenotypes following morpholino antisense oligonucleotide-induced klf2a knockdown in zebrafish. Targeted genome editing is an increasingly applied method for functional assessment of candidate genes. We therefore generated a stable klf2a mutant zebrafish and characterised its cardiovascular and haematopoietic development.
Methods and Results
Using Transcription Activator-Like Effector Nucleases (TALEN) we generated a klf2a mutant (klf2a sh317) with a 14bp deletion leading to a premature stop codon in exon 2. Western blotting confirmed loss of wild type Klf2a protein and the presence of a truncated protein in klf2a sh317 mutants. Homozygous klf2a sh317 mutants exhibit no defects in vascular patterning, survive to adulthood and are fertile, without displaying previously described morphant phenotypes such as high-output cardiac failure, reduced haematopoetic stem cell (HSC) development or impaired formation of the 5th accessory aortic arch. Homozygous klf2a sh317 mutation did not reduce angiogenesis in zebrafish with homozygous mutations in von Hippel Lindau (vhl), a form of angiogenesis that is dependent on blood flow. We examined expression of three klf family members in wildtype and klf2a sh317 zebrafish. We detected vascular expression of klf2b (but not klf4a or biklf/klf4b/klf17) in wildtypes but found no differences in expression that might account for the lack of phenotype in klf2a sh317 mutants. klf2b morpholino knockdown did not affect heart rate or impair formation of the 5th accessory aortic arch in either wildtypes or klf2a sh317 mutants.
Conclusions
The klf2a sh317 mutation produces a truncated Klf2a protein but, unlike morpholino induced klf2a knockdown, does not affect cardiovascular development.
Introduction
KLF2 belongs to the KLF zinc finger transcription factor family, characterised by three tandem C2H2 zinc fingers that serve as a DNA binding motif [1]. Since its characterisation in mice [1] and humans [2] it is clear that mammalian KLF2 plays important roles in vascular development and homeostasis [3]. KLF2 modulates expression of genes critical in regulating vascular tone [4, 5], thrombosis [6], inflammation [3, 4] barrier function [7] and antioxidative properties of endothelium [8]. Within the vascular wall, KLF2 is expressed exclusively in endothelium [3, 5, 9]. Endothelial KLF2 expression is induced by blood flow via mechanotransduction [3, 10], as well as via non flow-dependent mechanisms such as inflammatory cytokines or statins [4, 6, 11]. Homozygous Klf2 knockout mice die between E12.5–14.5 from intra-embryonic and intra-amniotic haemorrhaging associated with defective blood vessel morphology. Impaired vascular smooth muscle cell and pericyte migration resulting in failure to organize into a compact tunica media has been reported [12, 13], although while another group confirmed the stage of lethality (E11.5–13.5) and abdominal bleeding, they but did not detect vessel wall abnormalities [14]. Conditional Klf2 deletion in murine embryonic tissues confirms that endothelial Klf2 deletion is responsible for embryonic mortality [15].
The zebrafish (Danio rerio) is an increasingly popular model to study vascular development [16]. Zebrafish possess two KLF2 paralogs, klf2a and klf2b [17] and klf2a expression in embryonic zebrafish vasculature is (like mammalian KLF2) regulated by blood flow [3].
Assessment of gene function in zebrafish has often used morpholino antisense oligonucleotide-mediated gene knockdown, and several studies have reported the effect of klf2a knockdown on zebrafish cardiovascular and haematopoietic development. klf2a knockdown has been reported to cause high-output cardiac failure with pericardial oedema and high aortic blood flow velocity attributed to loss of smooth muscle tone [15]. klf2a knockdown induced failure to form the 5th accessory aortic arch (AA5x) although with otherwise preserved vascular patterning [18]. In relation to haematopoietic development klf2a knockdown impaired maintenance of haematopoietic stem cell (HSC) gene expression via a blood flow dependent signalling cascade involving nitric oxide (NO) [19].
Morpholino oligonucleotides (MO) remain commonly used in zebrafish reverse genetic studies. However, their reliability has been called into question [20]. The advantages of MOs in terms of cost and speed are counterbalanced by their temporary and incomplete action and potential for off-target effects and toxicity [21]. The development of targeted genome editing to create stable, heritable mutations in genes of interest avoids the disadvantages of morpholino use. Several techniques are available, including Zinc Finger Nucleases (ZFN) [22], Transcription Activator-like Effector Nucleases (TALEN) [23] and most recently Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated systems (CRISPR/Cas9) [24]. These induce site-specific double stranded DNA breaks that are repaired by error-prone non-homologous end joining DNA repair resulting in mutations that can disrupt the reading frame.
Targeted mutation has been proposed to be a more reliable way of assessing gene function [20]. Recent publications have shown that several published morphants phenotypes are not reproduced in mutants [25]. Very recently, Rossi et al. found that the severe vascular defect induced by egfl7 morpholino knockdown was not seen in egfl7 mutants, due to a compensatory upregulation of Emilin 2 and 3 not seen in morphants [26].
The existence of mechanisms that compensate for the effects of loss-of-function mutations would explain the low rate of overt phenotypes seen in the Sanger zebrafish mutagenesis project [27]. However, as zebrafish were first established as a model for forward mutagenesis screens [28–30], clearly not all mutations can be so “rescued”. There is therefore a pressing need to rapidly establish which mutations can be compensated for, and which cannot.
We used TALENs to generate a stable klf2a mutant line (named klf2a sh317). This mutation produces an aberrant mRNA transcript that is predicted to produce a significantly truncated klf2a protein without the essential DNA binding motifs. However, homozygous klf2a sh317 mutants exhibit no phenotypic differences compared to wildtypes (WT) and did not reproduce previously described klf2a morphant phenotypes, nor did they show any effect in miR-126 levels. We examined the expression patterns of three evolutionary closest klf family members; klf2b, klf4a and biklf/klf4b/klf17 in the search for a gene that might compensate for the mutation in klf2a. Although we detected klf2b expression in the vasculature, which has not been described previously, we found no differences in expression of these genes between klf2a sh317 mutants and wildtypes, and klf2b knockdown did not reveal any phenotype in klf2a sh317 mutants.
Materials and Methods
Zebrafish Husbandry
Zebrafish were raised in the Bateson Centre aquaria and fed Artemia nauplii. Zebrafish were kept on a constant 14 hour on /10 hour off light cycle at 28°C. Studies performed on zebrafish conformed to Directive 2010/63/EU of the European Parliament and Home Office regulations under Home Office project licences 40/3434 held by TJAC and 40/3082 held by FVE.
Fish Strains
hey2/gridlock mutants [31] were kindly supplied by Randall Peterson. vhl hu2117 [32] was obtained from Hubrecht Institute in Utrecht, Netherlands and was crossed with Tg(fli1:eGFP) [33] kindly supplied by Brant Weinstein. Tg(kdrl:HRAS-mCherry) [34] was kindly supplied by Arndt Siekmann and Tg(flk1:EGFP-nls) was kindly supplied by Markus Affolter. klf2a sh317 mutant line was generated in Nacre wild type (WT) strain and was crossed with Tg(kdrl:HRAS-mCherry;flk1:EGFP-nls) and with vhl hu2117 +/−;Tg(fli1:eGFP).
Morpholino Studies
Morpholino oligonucleotides (GENE-TOOLS, OR, USA) were diluted with ultra-pure (miliQ) water and phenol red (SIGMA) and 2ng of each were injected into one-cell stage embryos. Morpholino sequences: tnnt2 morpholino 5′ CATGTTTGCTCTGACTTGACACGCA 3′ as published [35], klf2b morpholino 5′ AGTGTCAAATACTTACATCCTCCCA 3′, control morpholino 5′ CCTCTTACCTCAGTTACAATTTATA 3′ (GENE TOOLS stock control).
Drug Treatments
In order to reversibly block cardiac contraction and stop blood flow embryos were treated with the anaesthetic tricaine [2.53mM] or the myosin ATPase inhibitor 2,3-butanedione 2-monoxime (BDM) [15mM] (both SIGMA) from 46hpf until 65hpf as previously published [18, 36].
RNA Extraction, Reverse Transcription and Polymerase Chain Reaction
Total RNA was extracted from pools of 30 embryos or unfertilised eggs using TRIzol (LIFE TECHNOLOGIES) according to manufacturer`s instructions. Total RNA was reversely transcribed into complementary DNA using the VERSO cDNA Synthesis Kit (THERMO SCIENTIFIC). Primers for amplification of klf2a, klf2b, and gapdh were: klf2a F 5`GGATCCATGGCTTTGAGTGGAACG 3`, klf2a R 5`GAATTCCTACATATGACGTTTCAT 3`, klf2b F 5'-GCCTTCGATTTCAACGTTTGC-3', klf2b R 5'-ATCCTCGTCATCCTTACGGC-3', gapdh F 5`AGGCTTCTCACAAACGAGGA, gapdh R 5`GCCATCAGGTCACATACACG 3`.
Quantitative PCR (qPCR) of zebrafish mature miR-126-5p was performed according to the manufacturer’s instructions using the TaqMan MicroRNA Assay. Total RNA from 72hpf zebrafish embryos was purified using Trizol. cDNA was prepared using the TaqMan microRNA Reverse Transcription kit (4366596, APPLIED BIOSYSTEMS) and qPCR performed using miR-126-5p and U6 TaqMan Assays (4427975 and 4427975, APPLIED BIOSYSTEMS). The primers used for reverse transcription and qPCR were supplied with each assay. The qPCR primer is a stem-loop oligonucleotide containing the sequence of the mature miR-126-5p (CAUUAUUACUUUUGGUACGCG). qPCR was performed in duplicate using an ABI 7900HT Fast Real-Time PCR System. Expression of miR-126-5p was normalized to U6 for relative quantification. Individual points represent relative quantification of miR-126-5p in a pool of embryos relative to mean expression in the wild-type group. Summary data is displayed as mean ± standard error of the mean (SEM). Statistical comparison was made using a two-tailed Student’s t-test on log-transformed data.
Measurement of Cardiovascular Performance
Heart Rate
Embryos were kept in individual dishes and individually removed from the 28°C incubator immediately prior to measurement to minimize the effect of environmental temperature on heart rate [37]. Heart rates were measured directly by eye under a stereomicroscope for 30 seconds.
Blood flow velocity
Particle image velocimetry (PIV) analysis using erythrocytes as tracer particles was used to measure aortic blood velocity in the mid aorta immediately above the cloaca. Zebrafish embryos were imaged at 10x magnification and at 300 frames/second using a high speed camera (OLYMPUS IX81) and Video Savant 4.0 digital video recording software (IO INDUSTRIES). 400 images (frames) in.tiff format per each embryo were recorded. Images were analysed using ImageJ (version 1.45s public domain software http://imagej.nih.gov/ij) to obtain a kymograph with lines representing movements of individual erythrocytes in real time. Kymographs were uploaded to custom made software CORRELATOR which calculated instantaneous erythrocyte velocities throughout the cardiac cycle. Knowing the ratios of pixel/μm (0.8pixel/μm), pixel/frame (1:1) and the rate of imaging (300frames/second), velocity (v) was calculated as follows: v = distance/time [μm/sec]. Average erythrocyte velocity per cardiac cycle was calculated by averaging all instantaneous velocities measured during the cardiac cycle. Additionally, cardiac cycles from all embryos of the same group were put in phase, and average instantaneous erythrocyte velocities for each frame were calculated and plotted as a single velocity curve.
Whole-Mount In Situ Hybridization (WISH)
WISH was performed according to previously published protocols [38, 39]. Embryos were permeabilized with proteinase K (ROCHE) [15μg/ml] for following times depending on the developmental stage: 0min (24hpf), 15min (36hpf), 40min (48hpf), 80min (72hpf), 100min (4dpf) and 150min (5dpf). Maleic acid buffer was used instead of phosphate buffered saline for day 2 and day 3 washes. Expression was detected with digoxigenin (ROCHE) labelled riboprobes and resolved using BM Purple (ROCHE). EST clone IMAGp998A0510285Q (IMAGENES) containing full-length klf2a cDNA sequence was digested with XBaI and EcoRI (both NEB) resulting in a 1144bp fragment which was inserted into Bluescript KS expression plasmid and subsequently used for klf2a antisense riboprobe synthesis (EcoRI (NEB) and T7 RNA polymerase (ROCHE)). Total embryonic RNA at 48hpf was used as a template for klf2b, klf4a and biklf/klf4b/klf17 riboprobes synthesis. The following primers were designed to amplify approx. 1000bp cDNA fragments: klf2b F 5`CGTGGACATGGCTTTACCTT 3`, klf2b R 5`ATGGGAGCTTTTGGTGTACG 3`, klf4a F 5`TTGATAGCATGGCACTGAGC 3`, klf4a R 5`CCTGCGGAAATCCAGAATAA 3`, biklf/klf4b/klf17 F 5`ACCCCGGACATGAATTATCA 3`, biklf/klf4b/klf17 R 5`TGTCCGGTGTGTTTCCTGTA 3`. T3 promoter sequence 5`CATTAACCCTCACTAAAGGGAA 3`was added to 5`end of each of the F primers and T7 promoter sequence 5`TAATACGACTCACTATAGGG 3`was added to 5`end of each of the R primers. RT-PCR and subsequent PCR amplification of corresponding cDNA was performed in a single step using Superscript III One-Step RT-PCR kit (INVITROGEN) following the manufacturer`s instructions. Antisense riboprobes were synthesized using T7 RNA polymerase (ROCHE). The remaining riboprobes used in this study have been kindly donated by other research groups: dll4 [40] runx1 [41] and cmyb [42].
Microscopy
Visual assessment of embryos, monitoring of WISH staining and heart rate counting were performed using a Leica S6E stereo microscope (LEICA MICROSYSTEMS). Images of live embryos and WISH images were taken using a Leica M165 FC fluorescent stereo microscope with Leica DFC310 FX camera (both by LEICA MICROSYSTEMS). Fluorescence microscopy was performed using UltraVIEW Vox spinning disc confocal microscope and Volocity v5.3.2 imaging software (both by PERKIN ELMER). Zebrafish embryos were anaesthetised with Tricaine and mounted on a cover slip in 1% low-melting point agarose prior to imaging. Images were taken in 2μm slices across the region of interest and digitally processed by focal plane merging (Z stacking). Merged images were further analysed using ImageJ.
Western Blotting
Total protein was extracted from genotyped zebrafish embryos at 5dpf (4 embryos per group) using RIPA lysis buffer (SIGMA-ALDRICH) with added Halt Protease and Phosphatase inhibitor cocktail (THERMO SCIENTIFIC). Embryos were homogenised using an Eppendorf pestle (SIGMA-ALDRICH) and left on ice for 30min. Lysate was centrifuged at 14000rpm for 10min at 4°C and the supernatant was separated and protein amounts quantified using a Bradford assay. Protein samples were then mixed with 1M dithiothreitol (DTT) (SIGMA-ALDRICH) and NuPAGE LDS Sample Buffer (LIFE TECHNOLOGIES) heated to 95°C for 5min and loaded onto a NuPAGE NOVEX 4–12% Bis-Tris gel (LIFE TECHNOLOGIES). After the electrophoretic separation, proteins were transferred to Immobilon-P PVDF transfer membranes (MILLIPORE) using the Xcell II Blot Module (LIFE TECHNOLOGIES). After blocking with 5% non-fat milk for 1 hour at RT, transfer membranes were incubated with rabbit anti-mouse polyclonal Klf2 antibody (raised against a polypeptide containing AAs 70–270 of mouse wildtype Klf2a protein) (1:500) (MILLIPORE, Cat.# 09–820) or with a rabbit anti-human β-actin antibody (1:10000) (CELL SIGNALLING, Cat.# 4967) overnight. Membranes were washed in TBSTw (Tris buffered saline plus Tween 20: 136.8mM NaCl, 24.8mM Tris, 0.1% Tween 20 (vol/vol) (all from SIGMA), pH 7.6) for 3x5min on a rocker and incubated with a secondary antibody (goat anti-rabbit polyclonal antibody with conjugated horseradish peroxidase (HRP), 1:1000) (DAKO) for 45min on a rocker. After thorough washing of the membranes, immobilised proteins were detected by the EZ-ECL Chemiluminiscence Detection Kit (BIOLOGICAL INDUSTRIES).
klf2a TALEN Mutagenesis
We followed a published protocol for targeted TALEN mutagenesis [23]. klf2a genomic sequence was submitted to the Old TALEN Targeter software at https://tale-nt.cac.cornell.edu/node/add/talen-old. A target site at the 5`end of exon 2 5`TCAACCCATCACCACCTCCACCG/AT[ACACCACCAGCCTACTGGC]AGAGCTTCTGCAGTCTG 3`(19bp spacer sequence between target sequences for L and R TALEN subunits is annotated with square brackets) containing the XcmI (NEB) restriction enzyme site 5`CCANNNNNNNNNTGG 3`was chosen for the mutagenesis. Primers flanking the target site were designed amplifying a 281bp PCR product; klf2a TAL XcmI F 5`CAGGCGACTACAGAATGCAA 3`and klf2a TAL XcmI R 5`GCCCTCTTGTTTGACTTTGG 3`. Genomic DNA extraction was performed using REDExtract-N-Amp™ Tissue PCR Kit (SIGMA-ALDRICH). XcmI (NEB) restriction enzyme digest (3 hours, 37°C) results in generation of 180bp and 101bp fragments. Successful mutagenesis is indicated by the loss of the restriction site and the presence of an undigested 281bp band after the XcmI (NEB) digest. Following the assembly and capped mRNA synthesis, 1.5ng of capped mRNA coding for the klf2a TALEN construct was injected per embryo at one cell stage.
Statistical Analysis
F1 klf2a mutant studies were blinded to whether embryos were homozygous mutants, heterozygotes or wildtype (after imaging and quantification embryos were euthanized and genotyped). Statistical analysis was carried out using GraphPad Prism v5.04 software. Quantitative data are presented as mean ± standard error of the mean (SEM). Statistical comparisons between two groups were made by Student’s t-test. Statistical significance is indicated as: ns = non-significant, * = p<0.05, ** = p<0.01, *** = p<0.001, **** = p<0.0001.
Results
The Expression of klf2a in Developing Zebrafish Embryos
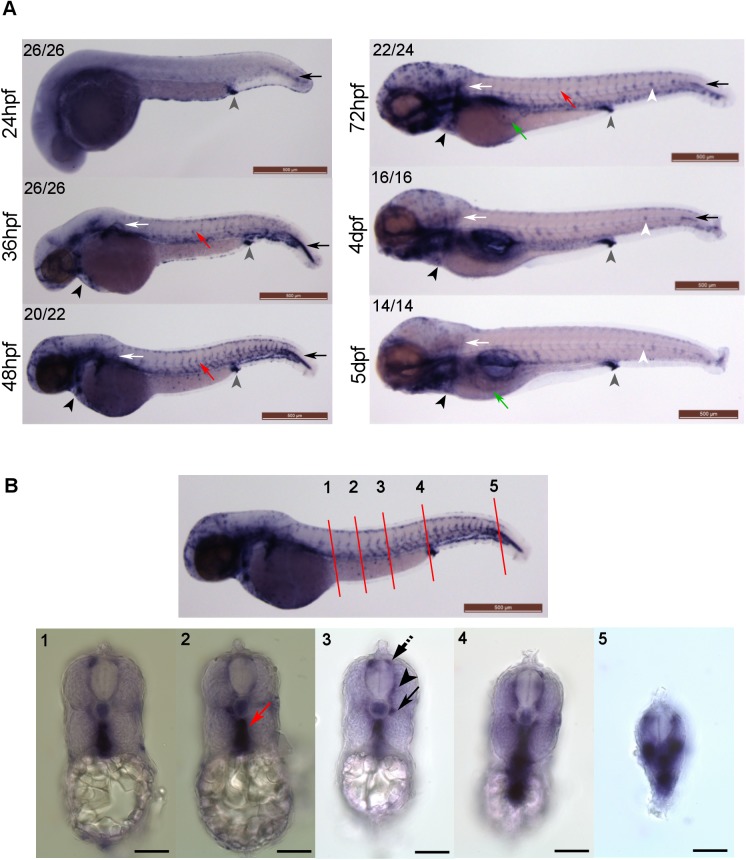
We analysed klf2a expression during zebrafish embryonic development daily from 1 to 5 days post fertilisation (dpf) in wildtype embryos using WISH (Fig 1A). Prior to onset of blood flow (24-26hpf) we could detect only non-vascular klf2a expression notably in the cloaca (Fig 1A, grey arrowheads) and in cells abutting the most posterior notochord (Fig 1A, black arrows). After onset of blood flow, klf2a was expressed in the developing vasculature, in particular at 48hpf (Fig 1A, red arrow). Cross sections at 48hpf confirmed vascular expression of klf2a (Fig 1B).These observations confirm previously published data on klf2a expression in early embryonic stages [17]. We next extended our studies to later stages of development to 5dpf and found klf2a expression in the trunk vasculature, intersegmental vessels and cardiac outflow tract (Fig 1A, red arrows and black arrowheads respectively). klf2a mRNA in the cardiac outflow tract was detectable up to 5dpf, whereas klf2a expression in the trunk vasculature and intersegmental vessels was only detectable up to 72hpf (Fig 1A). From 72hpf onwards, klf2a was expressed in the subintestinal veins that subsequently form the hepatic vein at 5dpf (Fig 1A, green arrows). We also detected klf2a expression in the neuromasts of the lateral line from 72hpf onwards (Fig 1A, white arrowheads).
Fig 1. klf2a expression patterns in developing zebrafish embryos.
(A) klf2a expression patterns were examined using whole-mount in situ hybridisation. Grey arrowheads indicate cloaca, black arrows indicate cells lateral to the most posterior notochord, white arrows indicate pectoral fin, red arrows indicate trunk vasculature, black arrowheads indicate the cardiac outflow tract, white arrowheads indicate neuromasts and green arrows indicate subintestinal veins (3dpf) or hepatic portal vein (5dpf). Numbers in the top left corners indicate number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm. (B) Cross sections of a 48hpf wildtype embryo showing klf2a expression in dorsal aorta (red arrow), parachordal vessel (black arrow), intersegmental vessel (ISV) (black arrowhead) and dorsal longitudinal anastomotic vessel (DLAV) (dotted black arrow). Anatomical positions of sections are indicated by the red lines and numbers on the top panel figure with corresponding cross sections images in the bottom panel. Scale bar = 500μm (top panel) and 100μm (bottom panel).
We confirmed that klf2a expression in the zebrafish vasculature is dependent on blood flow using three distinct approaches. These were by preventing cardiac contraction by morpholino antisense knockdown of cardiac troponin T2; preventing aortic blood flow to the trunk due to the aortic occlusion caused by homozygous mutation in hey2/gridlock; and finally by chemically-induced cessation of established blood flow between 46-65hpf by incubation in tricaine. When we examined vascular klf2a expression, this was reduced in all three models of hindered flow, in keeping with previous studies using tnnt2 knockdown [3] (S1 Fig).
Generation of a Stable klf2a Mutant Line
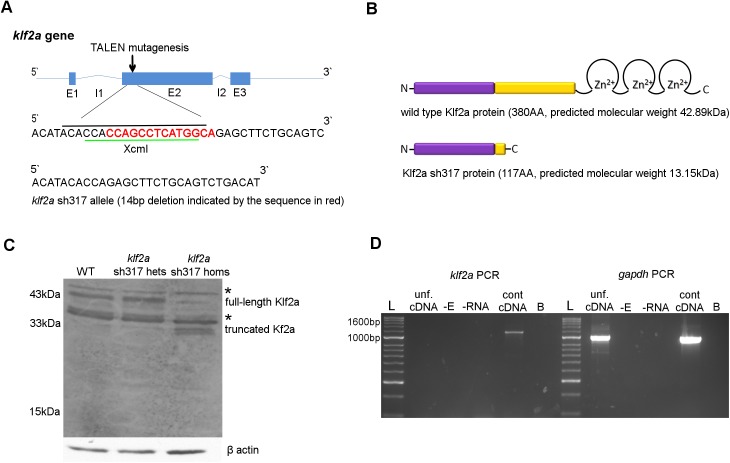
To examine the role of klf2a on cardiovascular and haematopoietic development we generated a klf2a mutant zebrafish using TALEN. The site for mutagenesis was chosen at the 5`end of exon 2 (Fig 2A). Mutations that produce a premature stop codon in this region are predicted to result in translation of a truncated Klf2a protein lacking the 3 tandem zinc fingers DNA binding motif located at the C terminus of the wildtype Klf2a protein (Fig 2B), rendering it unlikely to function as a transcription factor. We raised TALEN injected embryos and sequenced their genomic DNA and identified a founder carrying an allele with a 14 base pair (bp) deletion that disrupted the reading frame at the target site and generated a premature stop codon 14bp downstream. This allele was named klf2a sh317 and is predicted to generate a truncated Klf2a protein with molecular weight of 13.15 kilodalton (kDa) (Fig 2B). All mutant fish from the F1, F2 and F3 generation used in subsequent experiments were genotyped and the klf2a sh317 mutation confirmed by sequencing.
Fig 2. Generation of a stable klf2a sh317 mutant line.
(A) Schematic structure of wildtype klf2a gene with genomic sequence at the TALEN mutagenesis site. XcmI restriction site is indicated by the green line and the 19bp spacer for the TALEN structure of choice is indicated by the black line. The 14bp deletion identified in the klf2a sh317 allele is indicated by the sequence in red. (B) Schematic structure of Klf2a wildtype and Klf2a sh317 mutant proteins with amino acid (AA) lengths and predicted molecular weights in kilodaltons (kDa) Klf2a transactivation domain is in purple and transrepression domain is in yellow colour. (C) Substantial reduction of the 43kDa band representing the full-length Klf2a protein can be seen in a western blot of whole-embryo protein samples extracted from the F3 generation of klf2a sh317 mutant embryos at 5dpf, compared to wildtype embryos and heterozygous klf2a sh317 carriers. Additionally, two bands running at approximately 33kDa can be seen in the klf2a sh317 mutant, possibly representing truncated Klf2a. Asterixes indicate bands of approx. molecular weights of 38kDa and 47kDa consistent with expression of Klf4a and Biklf/Klf4b/Klf17 respectively, present in all three lanes in equal intensity. β actin was used as a loading control. (D) klf2a coding sequence was amplified from the control cDNA originating from 48hpf wildtype embryos giving an expected 1155bp PCR product. No band was detected in the sample with cDNA from unfertilised embryos (unf. cDNA) indicating the absence of maternal klf2a mRNA in unfertilised embryos. No 1674bp band was detected in unfertilised cDNA or control cDNA confirming absence of genomic DNA contamination. gapdh was used as a positive control. A band of expected size was detected in both the unfertilised eggs cDNA and control cDNA lanes indicating maternal gapdh mRNA in unfertilised zebrafish eggs. Abbreviations: -E: negative control with no reverse transcriptase added during reverse transcription (RT). -RNA: negative control with no RNA added during RT. B: blank, no template added to the PCR reaction. Abbreviations: L: Hyperladder II (NEB).
To determine the effect of the klf2a sh317 mutation on the mRNA transcript, we extracted whole embryonic RNA from 30 pooled embryos from the F3 generation of klf2a sh317 mutants. klf2a coding sequence was reversely transcribed, PCR amplified, cloned into an expression plasmid and transformed into bacterial cells. 23 separate colonies were sequenced. All confirmed the expected klf2a sh317 sequence without any additional transcripts (S2 Fig). We did not therefore find any evidence that the klf2a sh317 mutation induces alternative splicing.
Targeted mutation can induce gene loss-of-function by a number of mechanisms. As well as production of a truncated or non-functional protein, the mRNA can be targeted for nonsense mediated decay (NMD) [43]. We examined wither the klf2a sh317 mutation induced NMD by performing in situ hybridisation for klf2a in wildtypes and homozygous klf2a sh317 mutants, but found no difference in mRNA expression (S3 Fig). This, together with the fact that we could amplify and clone the expected mRNA transcript by rt-PCR (S2 Fig) strongly suggests that the klf2a sh317 mutation does not induce nonsense-mediated decay.
We next attempted to examine the effect of the klf2a sh317 mutation on Klf2a protein expression and size by western blotting in wildtype and klf2a sh317 mutant embryos (Fig 2C). Although there are no monoclonal antibodies available against zebrafish Klf2a we were able to obtain and use an anti-mouse polyclonal Klf2a antibody previously shown to bind to zebrafish Klf2a [19]. As might be expected from a polyclonal antibody raised against a largely conserved region of several Klf genes this blot shows multiple bands consistent with the presence of Klf2a, in addition to other bands which are consistent with Klf4a and Biklf/Klf4b/Klf17 expression (predicted molecular weights 38kDa and 47kDa respectively; indicated by asterixes in Fig 2C). However, a band likely to represent wildtype Klf2a protein with the correct size of 43kDa was detected in either wildtype or heterozygous klf2a sh317 embryos (Fig 2C). This band was greatly reduced in homozygous klf2a sh31 mutants, with the residual band migrating with a similar molecular weight likely to reflect the expression of Klf2b (predicted molecular weight 41.2kDa). Additionally, two bands of approximately 31-33kDa were detected in klf2a sh317 homozygous mutants, probably representing truncated Klf2a protein (Fig 2C). Although larger that the predicted molecular mass of Klf2a sh317 protein, the truncated protein would be expected to be very acidic (ph ~4, versus ~7.8 for full-length protein) and contain a significant proportion of proline residues, both features that contribute to aberrant behaviour on SDS-PAGE.
To determine the presence and potential contribution of maternal klf2a mRNA we analysed klf2a expression by RT-PCR in unfertilised zebrafish embryos. We found no evidence of maternal klf2a mRNA in unfertilised zebrafish eggs (Fig 2D).
Taken together, these data strongly suggest that the klf2a sh317 mutation induces a significant truncation of the klf2a protein that is highly likely to perturb its function as a transcription factor.
Characterisation of the klf2a sh317 Mutant Line
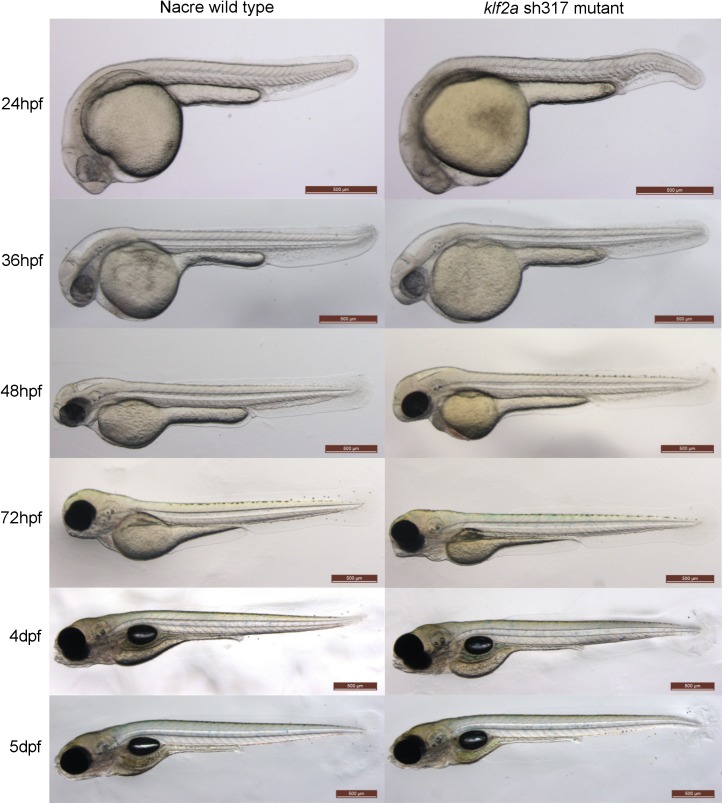
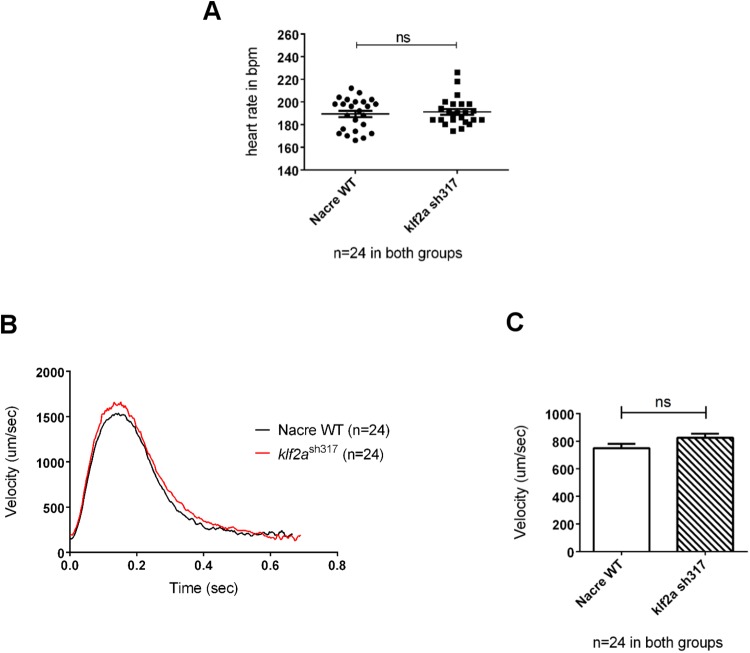
Homozygous klf2a sh317 mutants displayed no morphological abnormalities compared to wildtypes by at least 5dpf (Fig 3). In contrast to previously described klf2a morphants [15], no pericardial oedema was observed at any stage. klf2a sh317 mutants showed no difference in heart rate or blood flow velocity in the dorsal aorta at 48hpf compared to wildtypes, in contrast to the high-output cardiac failure described in klf2a morphants [15] (Fig 4). Furthermore, we raised homozygous klf2a sh317 mutants to adulthood and both these (F2 generation) and incrossed offspring (F3) were viable and fertile, making late-onset cardiac failure or major cardiovascular defects unlikely and excluding maternal effects that compensate for the mutation when homozygous mutants are derived from heterozygous females.
Fig 3. Comparison of the morphology of wildtype and klf2a sh317 mutant embryos.
There are no obvious morphological differences between wildtype and klf2a sh317 mutant embryos up to 5dpf and also beyond up to adulthood (not shown). Scale bar = 500μm.
Fig 4. Comparison of heart rates and blood flow velocities of wildtype and klf2a sh317 mutant embryos at 48hpf (A) No significant differences in heart rates were observed. (B) Instantaneous blood flow velocities throughout a single cardiac cycle in wildtype and klf2a sh317 mutants. (C) Average instantaneous blood flow velocities per cardiac cycle (n = 24, unpaired t-test).
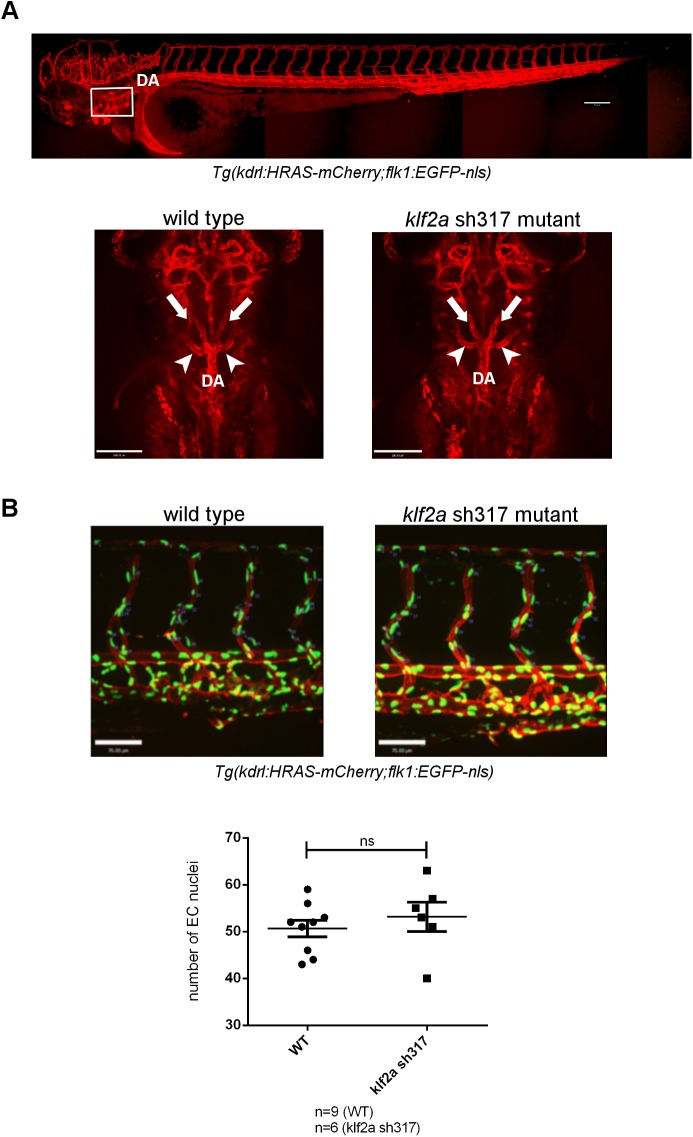
Next we examined the vascular anatomy of klf2a sh317 mutant embryos. We crossed klf2a sh317 mutants with the double transgenic line Tg(kdrl:HRAS-mCherry;flk1:EGFP-nls) that differentially labels endothelial cell cytoplasm and nuclei. We found no differences in vascular patterning in klf2a sh317 mutants (Fig 5A). Specifically, we examined the trunk vasculature and found no abnormal formation of the axial or intersegmental vessels, nor any difference in endothelial cell number in these regions (Fig 5B and 5C).Formation of the 5th accessory aortic arch (AA5x) has been shown to be impaired either in klf2a morphants or in the absence of blood flow [18]. We confirmed that in our hands, formation of the AA5x is blood flow dependent (S4 Fig) but formation of the AA5x was unaffected in klf2a sh317 mutants (Fig 5A).
Fig 5. The vascular anatomy of homozygous klf2a sh317 mutants.
(A) Formation of the 5th accessory aortic arch (AA5x) connecting the 5th and 6th aortic arch is not affected in homozygous klf2a sh317 mutants. Top panel figure shows vascular anatomy of a zebrafish embryo at 3dpf. White rectangle indicates the location of aortic arches. DA denotes dorsal aorta. Bottom panel figures represent dorsal views on aortic arches. White arrows indicate lateral dorsal aortae. White arrowheads indicate AA5x vessels. Scale bar = 200μm. (B) Quantification of endothelial cell nuclei in 4 ISVs closest to the cloaca and corresponding DLAV in WT and klf2a sh317 mutants at 3dpf. Scale bar = 70μm.
We recently showed that the excessive hypoxic signalling-driven trunk vessel angiogenesis in homozygous vhl mutants (vhl hu2117) is blood flow dependent [44]. Since klf2a is a major endothelial flow responsive transcription factor, we determined the effect of the klf2a sh317 mutation on angiogenesis in vhl hu2117 mutants. Heterozygotes for klf2a sh317 and vhl hu2117 in the Tg(fli1:eGFP) background were incrossed. Embryos homozygous for both klf2a sh317 and vhl hu2117 still display aberrant intersegmental vessel development (Fig 6) indicating that the klf2a sh317 mutation does not alter this process.
Fig 6. Vascular development in vhl hu2117 mutants is unaffected by the klf2a sh317 mutation.
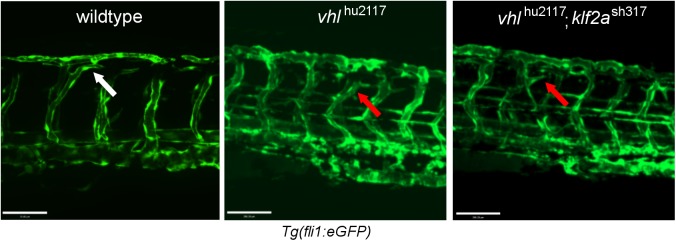
Wildtype embryos exhibit angiogenesis typical for this developmental stage– 3dpf (left figure, white arrow). Homozygous vhl hu2117 embryos show enlargement of vessels (ISVs and DLAV) with increased tortuosity and looping of the DLAV (middle figure, right arrow). The same vascular phenotype was be observed in vhl hu2117 embryos in a homozygous klf2a sh317 background (right figure, red arrow). Confocal images of Tg(fli1:eGFP) zebrafish embryos at 3dpf.. Scale bar = 70μm.
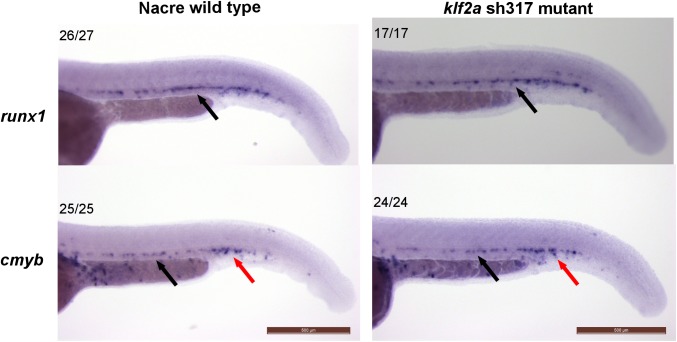
Since maintenance of haematopoetic stem cell (HSC) gene expression is impaired in klf2a morphants, or in the absence of blood flow [19] we examined this in klf2a sh317 mutants. While we confirmed that runx1 and cmyb expression was reduced in the absence of blood flow (S5 Fig), expression of both genes was unaffected in homozygous klf2a sh317 mutants (Fig 7).
Fig 7. Expression of HSC markers runx1 and cmyb is unaffected in homozygous klf2a sh317 mutants at 36hpf.
Expression patterns of HSC markers runx1 (top panel) and cmyb (bottom panel) do not differ between the wildtype and klf2a sh317 mutants at 36hpf. Black arrows indicate the aorta-gonad-mesonephros (AGM) region and red arrows indicate caudal haematopoietic tissue (CHT). Numbers in the top left corners indicate number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
Since homozygous klf2a sh317 mutants (unlike klf2a morphants) displayed no cardiovascular phenotype, we sought to determine whether this could be explained by redundancy between other klf family members. We examined expression of the three genes phylogenetically closest to klf2a, namely the paralog klf2b and both KLF4 orthologs klf4a and biklf/klf4b/klf17, to assess whether these might be compensating for the klf2a sh317 mutation. In addition to the close molecular phylogeny to KLF2, KLF4 has been identified as another endothelial blood flow dependent transcription factor in human and mouse [45, 46]. Vascular expression of klf2b, klf4a or biklf/klf4b/klf17 in wildtype zebrafish embryos has not been described [17, 47].
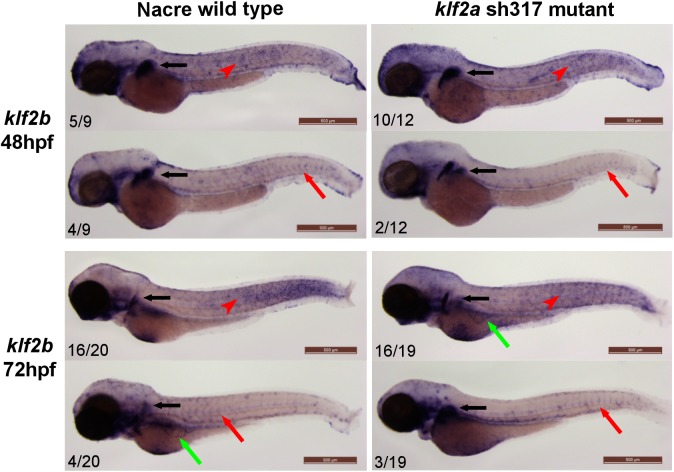
At 48hpf klf2b mRNA was detected in the cleithrum and the mesenchyme of the pectoral fin bud (Fig 8, black arrow) and in the epidermal cells (Fig 8, red arrowhead) in keeping with previously published data [17, 48]. Additionally, klf2b mRNA was detected in the ISVs of some wildtype and klf2a sh317 mutant embryos (Fig 8, red arrows). At 72hpf klf2b expression persisted in the pectoral fin and epidermis. As in 48hpf embryos, a small proportion of both wildtype and klf2a sh317 mutants at 72hpf exhibited vascular expression of klf2b in the ISVs and the subintestinal veins (Fig 8, red arrows). No differences in klf2b expression were apparent between wildtype and klf2a sh317 mutants.
Fig 8. klf2b expression patterns do not differ between wildtype and klf2a sh317 mutants.
klf2b mRNA is detectable in the developing pectoral fin bud (black arrows). Most of the embryos examined exhibit klf2b mRNA presence on the surface of the embryos representing epidermal cells as described before [17, 48] (red arrowheads). A small proportion of both wildtype and klf2a sh317 mutant embryos show vascular staining in the ISVs (red arrows) and/or in the subintestinal veins (green arrows). There were no differences in staining patterns and especially in the level of vascular staining observed between wildtype and klf2a sh317 mutant embryos. Figures in bottom left corner of each image indicate the number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
We detected klf4a expression in the epidermis (Fig 9, black arrows) and in the pectoral fin (Fig 9, red arrowheads) of 48hpf wildtype embryos in keeping with previously published data [47]. klf4a expression in klf2a sh317 mutants was not different to wildtype and no vascular expression of klf4a was detected (Fig 9). Similarly, in situ hybridisation for biklf/klf4b/klf17 at 48hpf confirmed previously published data [17] showing expression in the neuromasts of the lateral line organ (Fig 9, black arrowheads) and in the hatching gland (Fig 9, red arrows). Again, no differences in biklf/klf4b/klf17 expression were observed between wildytpe and klf2a sh317 mutants and no vascular biklf/klf4b/klf17 expression was detected (Fig 9).
Fig 9. klf4a and biklf/klf4b/klf17 expression patterns do not differ between wildtype and klf2a sh317 mutants at 48hpf.
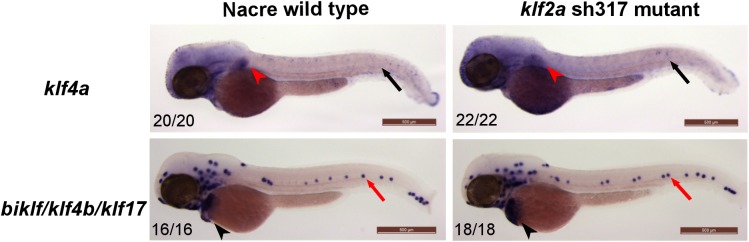
klf4a expression was detected in epidermis (black arrows) and pectoral fins (red arrowheads) and biklf/klf4b/klf17 expression was detected in neuromasts of lateral line organ (red arrows) and in hatching glands (black arrowheads) of wildtype embryos and klf2a sh317 mutants in keeping with previously published data [17, 47] No vascular expression of klf4a or biklf/klf4b/klf17 could be detected in any of the wildtype embryos or klf2a sh317 mutants examined. Figures in bottom left corner of each image indicate the number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
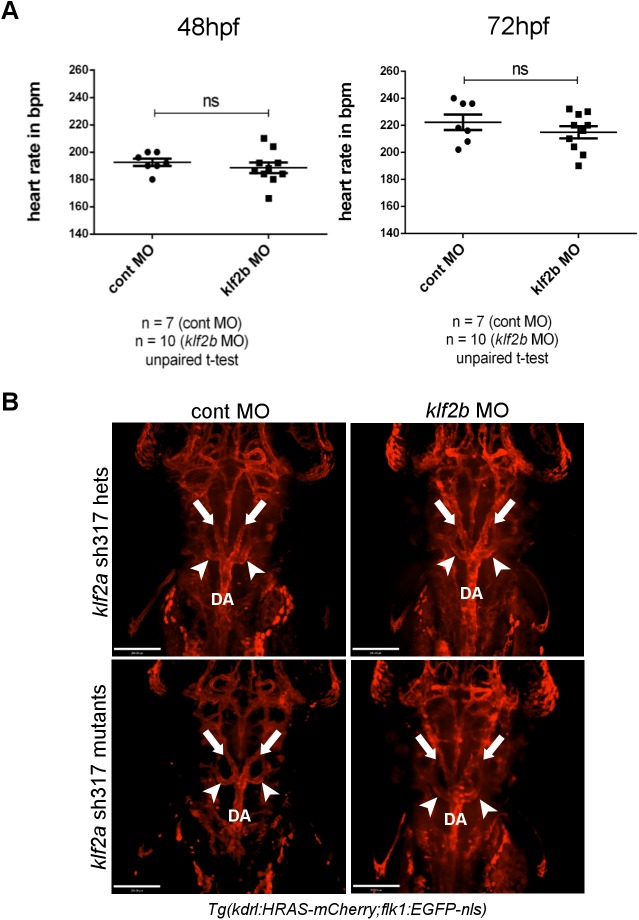
The above detailed experiments found no evidence of compensatory upregulation of klf2b, klf4a, or biklf/klf4b/klf17 in klf2a sh317 mutants. However, even without evidence of upregulation, normal levels of expression of these genes could conceivable compensate for loss-of-function of klf2a. Since only klf2b was seen to be expressed in the vasculature, we examined whether morpholino knockdown of klf2b induced cardiovascular phenotypes in klf2a sh317 mutants. Morpholino-induced klf2b knockdown had no effect on either heart rate or AA5x formation in either heterozygous or homozygous klf2a sh317 mutants, strongly suggesting that redundancy between klf2a and klf2b does not explain the lack of a cardiovascular phenotype in klf2a sh317 mutants (Fig 10).
Fig 10. klf2b knockdown does not alter heart rate or AA5x formation in klf2a sh317 mutants.
(A) klf2b morpholino injection did not affect the heart rate of klf2a sh317 mutants at either 48 or 72hpf. (B) AA5x vessel formation (white arrowheads) is intact in klf2b morphants in either a homozygous or heterozygous klf2a sh317 mutant background. White arrows indicate lateral dorsal aortae. DA indicates dorsal aorta. Scale bar = 200μm.
microRNA 126 (miR-126) has been shown to be upregulated by klf2a and morpholino-induced klf2a knockdown resulted in reduced miR-126 expression levels in zebrafish [18]. To attempt to further explore the reason for the lack of phenotype in klf2a sh317 mutants, we quantified miR-126 levels in 72hpf klf2a sh317 mutants compared with wildtypes. We found no reduction in miR-126 levels in klf2a sh317 mutants compared to wild types (Fig 11). This suggests that although the mutation clearly induces a significant alteration in the klf2a transcript in association with a truncated Klf2a protein, the downstream functions of klf2a appear not to be perturbed.
Fig 11. Comparison of miR-126 expression levels in klf2a sh317 mutants with wildtypes at 72hpf.
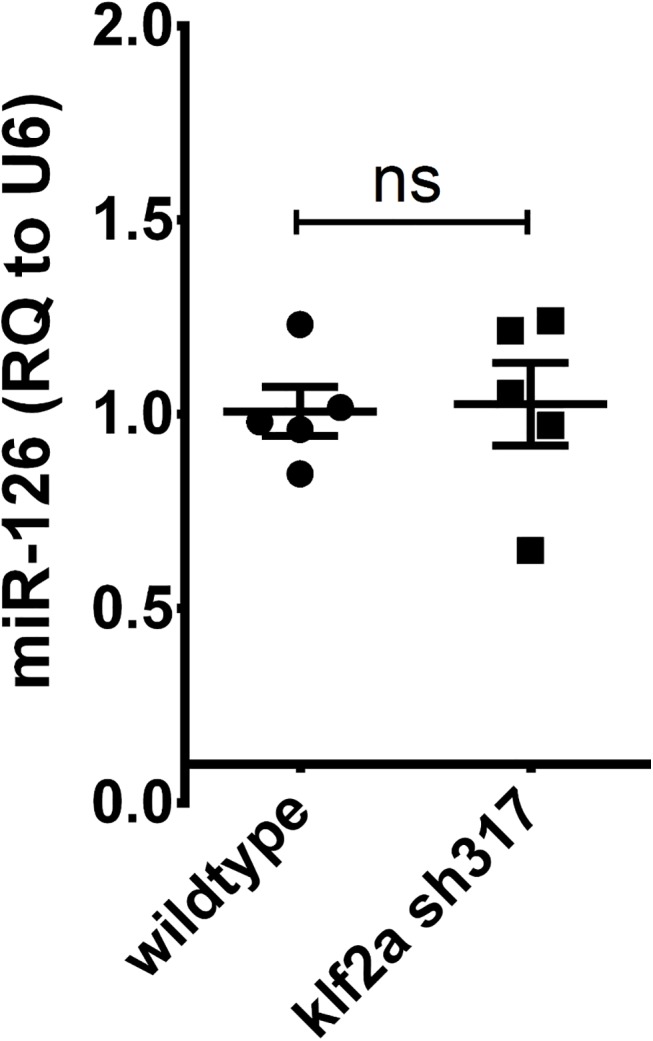
Expression levels of miR-126 do not differ between klf2a sh317 and wildtypes at the observed time point. Expression of miR-126 was normalized to U6 for relative quantification. Individual points represent relative quantification of miR-126 in a pool of embryos relative to the mean expression in the wild-type group. Summary data is displayed as mean ± SEM.
Discussion
In this study we describe for the first time the generation and characterisation of a stable klf2a mutant zebrafish. We find that, in contrast to several studies describing a range of cardiovascular phenotypes in klf2a morphant zebrafish, homozygous klf2a sh317 mutant embryos display no discernible cardiovascular abnormalities, survive into adulthood and are fertile. There are several possible explanations for the discrepancy between the morphant and klf2a sh317 mutant phenotype, a phenomenon that is increasingly being recognised as targeted mutants are generated for genes previously studied using morpholinos [25].
It is possible that some or all of the published klf2a morpholino studies are describing off-target or toxic effects of the reagent used, rather than specific effects of klf2a knockdown. This is perhaps particularly possible in the case of klf2a, expression of which is regulated by blood flow. Non-specific effects that reduce cardiac output would indirectly reduce klf2a expression and confound interpretation of the phenotype. This could explain the findings that klf2a morpholino knockdown impairs AA5x [18] and HSC formation [19], both of which are impaired in the absence of blood flow. It is harder to ascribe the phenotype of high-output heart failure [15] to off-target effects or toxicity since this is a phenotype not commonly seen in morpholino studies, although the pericardial oedema also reported in this study is commonly induced by high-dose morpholino injections. However, high-output heart failure was not reported in the later klf2a morpholino studies, suggesting that it was not observed by these groups.
Conversely, it is possible that the klf2a sh317 mutation does not induce loss-of-function and this accounts for the lack of a mutant phenotype. Despite concerns about the reliability of morpholinos, and the increasing preference for using stable mutants, it remains challenging to confirm that a targeted mutation truly induces loss-of-function. The deletion in the genomic DNA sequence of klf2a sh317 mutants leads to a frame shift resulting in a premature stop codon that would remove the DNA binding motifs necessary for klf2a to function as a transcription factor. Sequencing the cDNA from klf2a sh317 mutants found no evidence that the deletion leads to an alternatively spliced transcript that could remain functional. We were fortunate to obtain an antibody with at least some degree of cross-reactivity against zebrafish Klf2a, since a lack of suitable antibodies prevents assessment of the effect of targeted mutation on protein expression in the case of most genes. Western blotting with this antibody confirmed loss of wildtype Klf2a in klf2a sh317 mutants. However, we did not detect a band of the predicted size of the mutant transcript (13kDa) but instead detected a novel band of around 33kDa. Although it is possible that this represents the predicted mutant protein with additional post-translational modifications, it is also possible that some mechanism has allowed the ribosome to produce a truncated yet functional Klf2a protein in the face of the klf2a sh317 mutation.
After initial submission of this paper, Rossi et al. published a description of a compensatory mechanism that rescues the effect of egfl7 mutants but not of egfl7 morpholino knockdown, via upregulation of Emilins [49]. Since the klf2a sh317 mutation seems highly likely to perturb klf2a function, yet does not affect miR-126 levels, we consider it most likely that a similar mechanism exists for klf2a. We have shown that this compensation is not mediated by klf2b, but it is entirely possible that other genes may rescue the effect of the the klf2a sh317 mutation. Since forward mutagenesis screens in zebrafish have been highly successful, not least for cardiovascular phenotypes [50], such mechanisms appear not to compensate for many genes. However, the existence of such mechanisms may explain the observation that in a large-scale zebrafish mutagenesis study only 6% of disruptive mutations in protein-coding genes induced an observable phenotype [27].
In summary, our work shows that even in the face of clear evidence of a potentially disruptive mutation induced in a gene of interest, it is currently very difficult to be certain that this leads to loss-of-function, and hence to be confident about the role of the gene in embryonic development.
Supporting Information
(PDF)
klf2a is expressed in zebrafish embryonic vasculature of a WT embryo at 48hpf. Cessation of blood flow in the trunk vasculature by an occlusion of proximal aorta in the gridlock mutants (indicated by a green arrow) results in a complete loss of klf2a vascular expression distally to the occlusion. Blockage of embryonic heart contractions by tnnt2 MO results in significantly decreased klf2a vascular expression. Pharmacological inhibition of heart contractions by tricaine from 32 to 48hpf results in significantly decreased klf2a vascular expression. Interestingly tricaine also reduces klf2a expression in the heart region and in the cells lateral to the most posterior notochord. Red arrows indicate trunk vasculature, black arrows indicate the cells lateral to most posterior notochord, white arrows indicate pectoral fins, black arrowheads indicate the cardiac outflow tract and white arrowheads indicate cloaca. Numbers in top right corners indicate the number of embryos with similar staining pattern out of all embryos examined. klf2a riboprobe used. Scale bar = 500μm.
(TIF)
(A) klf2a wildtype and klf2a sh317 coding sequences. Exons are colour coded in alternating black and blue colours. Nucleotides within the 19bp spacer between the klf2a TALEN L and R subunit binding sites are underlined. Targeted TALEN-induced mutagenesis occurs around this region. Codons coding for amino acids that bind the zinc atoms within the 3 tandem C2H2 zinc fingers at the C terminus of wildtype Klf2a protein are highlighted in red. 14bp deletion in the klf2a sh317 coding sequence causes disruption of the reading frame. (B) Primary protein structures of Klf2a WT and Klf2a sh317 proteins are aligned. Cysteine (C) and histidine (H) amino acids that bind zinc atoms within the 3 tandem C2H2 zinc fingers at the C terminus of wildtype Klf2a protein are highlighted in red. (C) Sequencing of reversely transcribed klf2a sh317 mRNA confirmed the presence of expected sequence in 23/23 sequencing reactions without any additional transcripts detected. Sequence highlighted in blue (ACACCA) indicates the area of targeted mutagenesis in klf2a gene.
(TIF)
No differences in klf2a WISH staining patterns were detected in klf2a sh317 mutant embryos when compared to wildtype counterparts at examined time points indicating the absence of nonsense-mediated mRNA decay (NMD) in klf2a sh317 mutants. Grey arrowheads indicate cloaca, black arrows indicate cells lateral to the most posterior notochord, white arrows indicate pectoral fin, red arrows indicate trunk vasculature, black arrowheads indicate the cardiac outflow tract, white arrowheads indicate neuromasts and green arrows indicate subintestinal veins. Numbers in the bottom left corners indicate number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
(TIF)
(A) Vascular anatomy of a zebrafish embryo at 3dpf. White rectangle indicates the location of aortic arches. DA denotes dorsal aorta. Scale bar = 200μm. (B) Formation of AA5x vessels in unaffected in the WT embryos (untreated controls) as indicated by the white arrowheads. White arrows point at lateral dorsal aortae. AA5x vessel is missing in embryos treated with tricaine (middle panel) and with BDM (right figure) which both prevent blood flow (green arrowheads). Formation of lateral dorsal aortae is unaffected (white arrows). These findings confirm the previously published data [18]. Scale bar = 70μm.
(TIF)
Expression patterns of HSC markers runx1 (top panel) and cmyb (bottom panel) is significantly diminished in the tnnt2 morphants which lack blood flow confirming previously published data [19]. Black arrows indicate the aorta-gonad-mesonephros (AGM) region and red arrows indicate caudal haematopoietic tissue (CHT). Numbers in the top left corners indicate number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
(TIF)
Acknowledgments
The authors would like to thank Eleanor Markham for technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Medical Research Council (www.mrc.ac.uk) Centre Grant (G0700091) provided a clinical training fellowship to PN. This work was supported by British Heart Foundation (www.bhf.org.uk) project grants 09/287/28051 and 12/12/29433 awarded to TJAC. FvE was supported by European Commission (http://cordis.europa.eu/result/rcn/140792_en.html) as part of its Seventh Framework Programme (EC Grant Agreement HEALTH-F4-2010-242048). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15(11):5957–65. Epub 1995/11/01.: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wani MA, Conkright MD, Jeffries S, Hughes MJ, Lingrel JB. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Kruppel-like factor. Genomics. 1999;60(1):78–86. Epub 1999/08/25. 10.1006/geno.1999.5888 S0888-7543(99)95888-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 3. Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, et al. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116(1):49–58. Epub 2005/12/13. 10.1172/JCI24787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199(10):1305–15. Epub 2004/05/12. 10.1084/jem.20031132 jem.20031132 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dekker RJ, van Thienen JV, Rohlena J, de Jager SC, Elderkamp YW, Seppen J, et al. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am J Pathol. 2005;167(2):609–18. Epub 2005/07/29. doi: S0002-9440(10)63002-7 [pii] 10.1016/S0002-9440(10)63002-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin Z, Kumar A, SenBanerjee S, Staniszewski K, Parmar K, Vaughan DE, et al. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ Res. 2005;96(5):e48–57. Epub 2005/02/19. doi: 01.RES.0000159707.05637.a1 [pii] 10.1161/01.RES.0000159707.05637.a1 . [DOI] [PubMed] [Google Scholar]
- 7. Lin Z, Natesan V, Shi H, Dong F, Kawanami D, Mahabeleshwar GH, et al. Kruppel-like factor 2 regulates endothelial barrier function. Arterioscler Thromb Vasc Biol. 2010;30(10):1952–9. Epub 2010/07/24. doi: ATVBAHA.110.211474 [pii] 10.1161/ATVBAHA.110.211474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fledderus JO, Boon RA, Volger OL, Hurttila H, Yla-Herttuala S, Pannekoek H, et al. KLF2 primes the antioxidant transcription factor Nrf2 for activation in endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28(7):1339–46. Epub 2008/05/10. 10.1161/ATVBAHA.108.165811 ATVBAHA.108.165811 [pii]. . [DOI] [PubMed] [Google Scholar]
- 9. Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2). Blood. 2002;100(5):1689–98. Epub 2002/08/15. 10.1182/blood-2002-01-0046 . [DOI] [PubMed] [Google Scholar]
- 10. Huddleson JP, Srinivasan S, Ahmad N, Lingrel JB. Fluid shear stress induces endothelial KLF2 gene expression through a defined promoter region. Biol Chem. 2004;385(8):723–9. Epub 2004/09/29. 10.1515/BC.2004.088 . [DOI] [PubMed] [Google Scholar]
- 11. Parmar KM, Nambudiri V, Dai G, Larman HB, Gimbrone MA Jr., Garcia-Cardena G. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J Biol Chem. 2005;280(29):26714–9. Epub 2005/05/10. doi: C500144200 [pii] 10.1074/jbc.C500144200 . [DOI] [PubMed] [Google Scholar]
- 12. Wu J, Bohanan CS, Neumann JC, Lingrel JB. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. J Biol Chem. 2008;283(7):3942–50. Epub 2007/12/08. doi: M707882200 [pii] 10.1074/jbc.M707882200 . [DOI] [PubMed] [Google Scholar]
- 13. Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11(22):2996–3006. Epub 1997/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wani MA, Means RT Jr., Lingrel JB. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998;7(4):229–38. Epub 1998/12/22. . [DOI] [PubMed] [Google Scholar]
- 15. Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11(6):845–57. Epub 2006/12/05. doi: S1534-5807(06)00399-6 [pii] 10.1016/j.devcel.2006.09.006 . [DOI] [PubMed] [Google Scholar]
- 16. Quaife NM, Watson O, Chico TJ. Zebrafish: an emerging model of vascular development and remodelling. Curr Opin Pharmacol. 2012;12(5):608–14. Epub 2012/08/04. 10.1016/j.coph.2012.06.009 S1471-4892(12)00107-5 [pii]. . [DOI] [PubMed] [Google Scholar]
- 17. Oates AC, Pratt SJ, Vail B, Yan Y, Ho RK, Johnson SL, et al. The zebrafish klf gene family. Blood. 2001;98(6):1792–801. Epub 2001/09/06. . [DOI] [PubMed] [Google Scholar]
- 18. Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464(7292):1196–200. Epub 2010/04/07. doi: nature08889 [pii] 10.1038/nature08889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Zhang P, Wei Y, Gao Y, Patient R, Liu F. A blood flow-dependent klf2a-NO signaling cascade is required for stabilization of hematopoietic stem cell programming in zebrafish embryos. Blood. 2011;118(15):4102–10. Epub 2011/08/19. 10.1182/blood-2011-05-353235 blood-2011-05-353235 [pii]. . [DOI] [PubMed] [Google Scholar]
- 20. Schulte-Merker S, Stainier DY. Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development. 2014;141(16):3103–4. Epub 2014/08/08. 10.1242/dev.112003 141/16/3103 [pii]. . [DOI] [PubMed] [Google Scholar]
- 21. Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135(10):1735–43. Epub 2008/04/12. 10.1242/dev.001115 dev.001115 [pii]. . [DOI] [PubMed] [Google Scholar]
- 22. Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods. 2011;8(1):67–9. Epub 2010/12/15. 10.1038/nmeth.1542 nmeth.1542 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82 Epub 2011/04/16. 10.1093/nar/gkr218 gkr218 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–9. Epub 2013/01/31. 10.1038/nbt.2501 nbt.2501 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, et al. Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev Cell. 2015;32(1):97–108. Epub 2014/12/24. 10.1016/j.devcel.2014.11.018 S1534-5807(14)00735-7 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015. Epub 2015/07/15. 10.1038/nature14580 nature14580 [pii]. . [DOI] [PubMed] [Google Scholar]
- 27. Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496(7446):494–7. Epub 2013/04/19. 10.1038/nature11992 nature11992 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. Epub 1996/12/01. . [DOI] [PubMed] [Google Scholar]
- 29. Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. Epub 1996/12/01. . [DOI] [PubMed] [Google Scholar]
- 30. Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. Epub 1996/12/01. . [DOI] [PubMed] [Google Scholar]
- 31. Weinstein BM, Stemple DL, Driever W, Fishman MC. Gridlock, a localized heritable vascular patterning defect in the zebrafish. Nat Med. 1995;1(11):1143–7. Epub 1995/11/01. . [DOI] [PubMed] [Google Scholar]
- 32. van Rooijen E, Voest EE, Logister I, Korving J, Schwerte T, Schulte-Merker S, et al. Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood. 2009;113(25):6449–60. Epub 2009/03/24. 10.1182/blood-2008-07-167890 blood-2008-07-167890 [pii]. . [DOI] [PubMed] [Google Scholar]
- 33. Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248(2):307–18. Epub 2002/08/09. doi: S0012160602907116 [pii]. . [DOI] [PubMed] [Google Scholar]
- 34. Hogan BM, Bos FL, Bussmann J, Witte M, Chi NC, Duckers HJ, et al. Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat Genet. 2009;41(4):396–8. Epub 2009/03/17. 10.1038/ng.321 ng.321 [pii]. . [DOI] [PubMed] [Google Scholar]
- 35. Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31(1):106–10. Epub 2002/04/23. 10.1038/ng875 ng875 [pii]. . [DOI] [PubMed] [Google Scholar]
- 36. Serluca FC, Drummond IA, Fishman MC. Endothelial signaling in kidney morphogenesis: a role for hemodynamic forces. Curr Biol. 2002;12(6):492–7. Epub 2002/03/23. doi: S0960982202006942 [pii]. . [DOI] [PubMed] [Google Scholar]
- 37. Barrionuevo WR, Burggren WW. O2 consumption and heart rate in developing zebrafish (Danio rerio): influence of temperature and ambient O2. Am J Physiol. 1999;276(2 Pt 2):R505–13. Epub 1999/02/10. . [DOI] [PubMed] [Google Scholar]
- 38. Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. Epub 2008/01/15. 10.1038/nprot.2007.514 nprot.2007.514 [pii]. . [DOI] [PubMed] [Google Scholar]
- 39. Wilkinson RN. Hedgehog and TGF-beta family signalling in zebrafish blood and endothelial development: University of Oxford; 2008. [Google Scholar]
- 40. Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134(5):839–44. Epub 2007/01/26. doi: dev.003244 [pii] 10.1242/dev.003244 . [DOI] [PubMed] [Google Scholar]
- 41. Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129(8):2015–30. Epub 2002/04/06. . [DOI] [PubMed] [Google Scholar]
- 42. Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW 3rd, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197(2):248–69. Epub 1998/06/19. doi: S0012-1606(98)98887-X [pii] 10.1006/dbio.1998.8887 . [DOI] [PubMed] [Google Scholar]
- 43. Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. Epub 2007/03/14. 10.1146/annurev.biochem.76.050106.093909 . [DOI] [PubMed] [Google Scholar]
- 44. Watson O, Novodvorsky P, Gray C, Rothman AM, Lawrie A, Crossman DC, et al. Blood flow suppresses vascular Notch signalling via dll4 and is required for angiogenesis in response to hypoxic signalling. Cardiovasc Res. 2013;100(2):252–61. Epub 2013/07/03. 10.1093/cvr/cvt170 cvt170 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2001;98(16):8955–60. Epub 2001/08/02. 10.1073/pnas.171259298 98/16/8955 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, et al. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282(18):13769–79. Epub 2007/03/07. doi: M700078200 [pii] 10.1074/jbc.M700078200 . [DOI] [PubMed] [Google Scholar]
- 47. Li IC, Chan CT, Lu YF, Wu YT, Chen YC, Li GB, et al. Zebrafish kruppel-like factor 4a represses intestinal cell proliferation and promotes differentiation of intestinal cell lineages. PLoS One. 2011;6(6):e20974 Epub 2011/06/21. 10.1371/journal.pone.0020974 PONE-D-10-05091 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Expression of the zebrafish genome during embryogenesis [Internet]. ZFIN. 2001. [Google Scholar]
- 49. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–3. 10.1038/nature14580 . [DOI] [PubMed] [Google Scholar]
- 50. Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–92. Epub 1996/12/01. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
klf2a is expressed in zebrafish embryonic vasculature of a WT embryo at 48hpf. Cessation of blood flow in the trunk vasculature by an occlusion of proximal aorta in the gridlock mutants (indicated by a green arrow) results in a complete loss of klf2a vascular expression distally to the occlusion. Blockage of embryonic heart contractions by tnnt2 MO results in significantly decreased klf2a vascular expression. Pharmacological inhibition of heart contractions by tricaine from 32 to 48hpf results in significantly decreased klf2a vascular expression. Interestingly tricaine also reduces klf2a expression in the heart region and in the cells lateral to the most posterior notochord. Red arrows indicate trunk vasculature, black arrows indicate the cells lateral to most posterior notochord, white arrows indicate pectoral fins, black arrowheads indicate the cardiac outflow tract and white arrowheads indicate cloaca. Numbers in top right corners indicate the number of embryos with similar staining pattern out of all embryos examined. klf2a riboprobe used. Scale bar = 500μm.
(TIF)
(A) klf2a wildtype and klf2a sh317 coding sequences. Exons are colour coded in alternating black and blue colours. Nucleotides within the 19bp spacer between the klf2a TALEN L and R subunit binding sites are underlined. Targeted TALEN-induced mutagenesis occurs around this region. Codons coding for amino acids that bind the zinc atoms within the 3 tandem C2H2 zinc fingers at the C terminus of wildtype Klf2a protein are highlighted in red. 14bp deletion in the klf2a sh317 coding sequence causes disruption of the reading frame. (B) Primary protein structures of Klf2a WT and Klf2a sh317 proteins are aligned. Cysteine (C) and histidine (H) amino acids that bind zinc atoms within the 3 tandem C2H2 zinc fingers at the C terminus of wildtype Klf2a protein are highlighted in red. (C) Sequencing of reversely transcribed klf2a sh317 mRNA confirmed the presence of expected sequence in 23/23 sequencing reactions without any additional transcripts detected. Sequence highlighted in blue (ACACCA) indicates the area of targeted mutagenesis in klf2a gene.
(TIF)
No differences in klf2a WISH staining patterns were detected in klf2a sh317 mutant embryos when compared to wildtype counterparts at examined time points indicating the absence of nonsense-mediated mRNA decay (NMD) in klf2a sh317 mutants. Grey arrowheads indicate cloaca, black arrows indicate cells lateral to the most posterior notochord, white arrows indicate pectoral fin, red arrows indicate trunk vasculature, black arrowheads indicate the cardiac outflow tract, white arrowheads indicate neuromasts and green arrows indicate subintestinal veins. Numbers in the bottom left corners indicate number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
(TIF)
(A) Vascular anatomy of a zebrafish embryo at 3dpf. White rectangle indicates the location of aortic arches. DA denotes dorsal aorta. Scale bar = 200μm. (B) Formation of AA5x vessels in unaffected in the WT embryos (untreated controls) as indicated by the white arrowheads. White arrows point at lateral dorsal aortae. AA5x vessel is missing in embryos treated with tricaine (middle panel) and with BDM (right figure) which both prevent blood flow (green arrowheads). Formation of lateral dorsal aortae is unaffected (white arrows). These findings confirm the previously published data [18]. Scale bar = 70μm.
(TIF)
Expression patterns of HSC markers runx1 (top panel) and cmyb (bottom panel) is significantly diminished in the tnnt2 morphants which lack blood flow confirming previously published data [19]. Black arrows indicate the aorta-gonad-mesonephros (AGM) region and red arrows indicate caudal haematopoietic tissue (CHT). Numbers in the top left corners indicate number of embryos with similar staining patterns out of total number of embryos examined. Scale bar = 500μm.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.