Abstract
Aims
To investigate if the single nucleotide polymorphisms rs3753841, rs1015213 and rs11024102, recently implicated in the development of acute primary angle closure or primary angle closure glaucoma, are associated with ocular biometric characteristics of British adults in the European Prospective Investigation of Cancer-Norfolk eye study.
Methods
Genotyping data on rs1015213 (between PCMTD1 and ST18), rs11024102 (at PLEKHA7) and rs3753841 (at COL11A1) were available on 3268 participants. Direct genotypic data was available for rs1015213 and rs3753841. Data was imputed for rs11024102. Ocular biometric data was available on 1137 participants who attended the third European Prospective Investigation of Cancer health examination and 988 (87%) of these participants had no previous cataract surgery either eye. Axial length (AL), anterior chamber depth (ACD) and corneal keratometry were measured by using the Zeiss IOLMaster.
Results
Presence of at least one A allele (AG or AA genotype) for rs1015213 was associated with a shallower ACD (−0.07 mm, 95% CI −0.01 to −0.14 mm, p=0.028) after adjusting for age and sex (both p≤0.001). There was no association with AL or corneal keratometry for rs1015213 genotypes. AL, ACD and keratometry were not associated with rs3753841 or rs11024102 genotypes including after adjusting for age and sex.
Conclusions
This study suggests that primary angle closure glaucoma susceptibility at the PCMTD1-ST18 locus may be partly explained by an association between rs1015213 and ACD in European populations. This effect is equivalent to almost 20% of the SD of the mean ACD of phakic individuals in this cohort. We were not able to identify any association between rs3753841 or rs11024102 and ocular biometry.
INTRODUCTION
Glaucoma is the leading cause of irreversible blindness worldwide.1 Although primary open angle glaucoma is more common, accounting for approximately three quarters of primary glaucoma cases, primary angle closure glaucoma (PACG) is more severe and the numbers of those blind from each type are almost equal.1 Recently three novel susceptibility loci for PACG have been identified by a genome-wide association study (GWAS) with replication experiments.2 These effect loci are rs11024102 G at PLEKHA7, rs3753841 G at COL11A1 and rs1015213 A, which is located between PCMTD1 and ST18 on chromosome 8q.2 The mechanism by which these three loci influence PACG risk is unknown. PACG has a well recognised ocular phenotype with associated biometric characteristics including shallow anterior chamber, short axial length (AL), small corneal radius of curvature, and a thicker, anteriorly positioned crystalline lens.3–9 Consequently we hypothesised that these PACG effect loci may be associated with ocular biometric characteristics and therefore PACG susceptibility. To investigate this, we performed a genotype–phenotype correlation study of single nucleotide polymorphisms (SNPs) recently implicated in the development of PACG and ocular biometry in participants of the European Prospective Investigation of Cancer (EPIC)-Norfolk Eye Study.
METHODS
EPIC is a pan-European study that started in 1989 with the primary aim of investigating the relationship between diet and cancer risk.10 The EPIC-Norfolk cohort was broadened to include additional endpoints and exposures. The third health examination was conducted between 2006 and 2011 with the objective of investigating various physical and ocular characteristics of participants now aged 48–91 years. The third health examination was reviewed and approved by the East Norfolk and Waverney National Health Service Research Governance Committee (2005EC07L) and the Norfolk Research Ethics Committee (05/Q0101/191). All participants gave written informed consent and the study was performed in accordance with the principles of the Declaration of Helsinki.
All participants underwent a detailed health examination performed by trained nurses following standard operating procedures. Ocular biometry was measured by non-contact partial coherence interferometry using the Zeiss IOLMaster Optical Biometer (IOLMaster, Carl Zeiss Meditech Ltd., Welwyn Garden City, UK). Five measurements of AL and anterior chamber depth (ACD, defined as corneal epithelium to anterior crystalline lens surface) and three measurements of central keratometry were made to allow calculation of mean values. Investigations were performed on both eyes of participants, and the mean values of both eyes used for analyses as consistent with previous studies.11–13 Participants who had undergone previous cataract surgery (pseudophakic participants) were excluded from analyses for ACD and corneal keratometry.
Genotyping data was available for rs3753841 and rs1015213 using the Infinium HumanHap300 SNP chip (Illumina, San Diego, California, USA), containing 317503 tagging SNPs derived from phase I of the International HapMap project.14 Genotyping was performed at the Wellcome Trust Sanger Institute (Hinxton, Cambridgeshire, UK) following manufacturer’s instructions (http://www.illumina.com). No direct genotyping data were available for rs11024102, and this was imputed on the basis of HapMap using IMPUTE v2 (https://mathgen.stats.ox.ac.uk/impute/impute.html). The imputation quality score was 0.986 and the imputed most likely genotypes were used.
Statistical analysis was performed using SPSS V.20. SNPs were tested for deviation from the Hardy-Weinberg equilibrium using the χ2 test. Mean values between groups were compared by analysis of variance. Each SNP was assessed for association under an additive genetic model with each continuous biometric parameter (dependent variables) after adjusting for age and sex in linear regression models. An additive genetic model was used for SNP rs1015213, rs3753841 and rs11024102 (GG, AG and AA genotypes coded as 0, 1 or 2, respectively), and an additional dominant genetic model for SNP rs1015213 (due to AA genotype frequency of <1%); thus GG coded as 0, AG and AA combined coded as 1 for analysis). A significance level of p≤0.05 was used. Bonferroni correction was not indicated as each of the tested SNPs were previously validated effect alleles for PACG.2
RESULTS
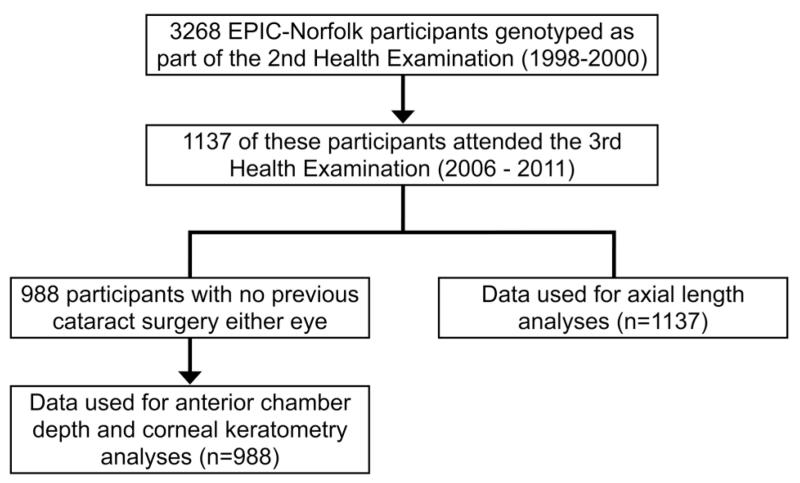
Genotypic data were available on 3268 EPIC-Norfolk participants of which 1137 participants had undergone ocular biometric measurements as part of the third health examination. The frequencies of all three SNPs were in Hardy-Weinberg equilibrium. Almost 100% of participants were of Caucasian ethnicity. Of the 1137 participants, 988 participants were phakic in both eyes (see figure 1; 107 bilateral previous cataract surgery and 42 unilateral cataract surgery either eye). Comparison of data between all participants (n=1137) and those phakic both eyes (n=988) showed the former to be older with deeper ACDs (table 1). Evaluations of ocular biometric data showed AL, ACD and corneal keratometry values were all significantly associated with age and sex. These factors were therefore included in multivariable linear regression models investigating possible effects of rs1015213, rs3753841 and rs11024102 genotype on ocular biometric parameters. Table 2 shows the biometric data for participants by SNP genotype. Participants who were phakic in both eyes were significantly younger with shallower anterior chambers than in the overall cohort of phakic and pseudophakic participants. There was no association between age or sex and genotype for rs1015213, rs3753841 or rs11024102.
Figure 1.
Flow chart showing data sources for analyses.
Table 1.
Demographic and biometric differences between data for all participants (used for axial length (AL) analyses); and those phakic both eyes (used for anterior chamber depth (ACD) and corneal keratometry (K) analyses)
| All participants (n=1137) | Phakic both eyes (n=988) | p Value | |
|---|---|---|---|
| Age, years | 69.7 (69.2 to 70.1) | 68.6 (68.1 to 69.1) | 0.001 |
| Sex, n=male | 494/1137 (43.4%) | 437/988 (44.2%) | 0.69 |
| AL, mm | 23.54 (23.47 to 23.61) | 23.51 (23.43 to 23.58) | 0.50 |
| ACD, mm | 3.15 (3.12 to 3.17) | 3.06 (3.03 to 3.08) | <0.001 |
| K, dioptres | 43.63 (43.53 to 43.72) | 43.64 (43.54 to 43.74) | 0.84 |
| rs1015213, n (GG/AG/AA/NN) | 951/173/10/3 | 825/152/8/3 | 1.0 |
| rs3753841, n (GG/AG/AA/NN) | 192/543/394/8 | 163/478/341/6 | 0.98 |
| rs11024102, n (GG/AG/AA/NN) | 89/462/545/41 | 72/408/473/35 | 0.97 |
Means with 95% CI are shown. Genotype data numbers: n=GG/AG/AA/missing (NN) with χ2 test result comparing between all participants and phakic both eyes groups.
Table 2.
Biometric data (with 95% CI) for rs1015213, rs375384 and rs11024102 SNPs
| SNP | AL (mm) | ACD (mm) | K (dioptres) |
|---|---|---|---|
| rs1015213 | |||
| GG | 23.55 (23.47 to 23.62) | 3.07 (3.04 to 3.10) | 43.65 (43.54 to 43.76) |
| AG and AA* | 23.51 (23.34 to 23.69) | 3.00 (2.94 to 3.06) | 43.61 (43.36 to 43.85) |
| p Value | 0.74 | 0.046 | 0.76 |
| rs3753841 | |||
| GG | 23.45 (23.29 to 23.61) | 3.04 (2.97 to 3.10) | 43.59 (43.35 to 43.82) |
| AG | 23.57 (23.46 to 23.67) | 3.06 (3.02 to 3.09) | 43.60 (43.47 to 43.74) |
| AA | 23.56 (23.44 to 23.67) | 3.07 (3.03 to 3.11) | 43.70 (43.53 to 43.88) |
| p Value | 0.50 | 0.69 | 0.61 |
| rs11024102 | |||
| GG | 23.45 (23.21 to 23.69) | 3.02 (2.94 to 3.10) | 43.69 (43.25 to 44.13) |
| AG | 23.56 (23.45 to 23.67) | 3.06 (3.03 to 3.10) | 43.58 (43.43 to 43.72) |
| AA | 23.54 (23.44 to 23.65) | 3.06 (3.02 to 3.08) | 43.68 (43.54 to 43.82) |
| p Value | 0.75 | 0.69 | 0.61 |
Comparisons of means between GG, AG and AA genotypes for rs3753841 and rs11024102; and
between GG, and AG and AA combined for rs1015213 (due the AA genotype frequency <1%).
AL, axial length; ACD, anterior chamber depth; K, mean corneal keratometry; SNP, single nucleotide polymorphism.
For rs1015213, linear regression using an additive genetic model considering wild type (GG), heterozygous (AG) and homozygous variants (AA) showed mean ACD was associated with rs1015213 (−0.07 mm, 95% CI <0.00 to −0.13, p=0.037) after adjusting for age and sex (both p≤0.001). Comparing means between GG, and AG and AA combined (due to AA genotype frequency of <1%) showed mean ACD for rs1015213 AG and AA (3.00 mm, 95% CI 2.94 to 3.06) was significantly lower than GG (3.07 mm, 95% CI 3.04 to 3.10, p=0.046) (see table 2). Univariable linear regression estimated this ACD difference to be −0.07 mm (95% CI <0.00 to −0.13 mm, p=0.046). The magnitude of this was unchanged after adjusting for age and sex (−0.07 mm, 95% CI −0.01 to −0.14 mm, p=0.028), (see table 3). No association with AL and corneal keratometry were found in similar models.
Table 3.
Regression coefficients and p values for linear regression models with predictors: age (per decade), sex and SNP for dependent variables
| Age (per decade) |
Sex |
SNP |
||||
|---|---|---|---|---|---|---|
| SNP | Regression coefficient (95% CI) | p Value | Regression coefficient (95% CI) | p Value | Regression coefficient (95% CI) | p Value |
| rs1015213 | ||||||
| AL (mm) | −0.14 (−0.05 to −0.23) | 0.002 | −0.47 (−0.32 to −0.60) | <0.001 | −0.07 (−0.25 to 0.12) | 0.32 |
| ACD (mm) | – | – | – | – | −0.07 (<0.00 to −0.13) | 0.046 |
| ACD (mm) | −0.06 (−0.03 to −0.09) | <0.001 | −0.09 (−0.04 to −0.13) | 0.001 | −0.07 (−0.01 to −0.14) | 0.028 |
| K (dioptres) | 0.15 (0.02 to 0.28) | 0.028 | 0.42 (0.22 to 0.62) | <0.001 | −0.02 (−0.29 to 0.24) | 0.87 |
| rs3753841 | ||||||
| AL (mm) | −0.14 (−0.06 to −0.23) | 0.002 | −0.45 (−0.31 to −0.59) | <0.001 | 0.03 (−0.07 to 0.13) | 0.56 |
| ACD (mm) | −0.06 (−0.02 to −0.09) | 0.001 | −0.08 (−0.03 to −0.13) | 0.001 | 0.01 (−0.02 to 0.05) | 0.54 |
| K (dioptres) | 0.16 (0.02 to 0.29) | 0.022 | 0.43 (0.24 to 0.63) | <0.001 | 0.08 (−0.06 to 0.22) | 0.28 |
| rs11024102 | ||||||
| AL (mm) | −0.13 (−0.04 to −0.22) | 0.005 | −0.47(−0.33 to −0.61) | <0.001 | 0.00 (−0.11 to 0.11) | 0.95 |
| ACD (mm) | −0.06 (−0.02 to −0.09) | 0.001 | −0.08 (−0.03 to −0.12) | 0.003 | 0.00 (−0.04 to 0.04) | 0.89 |
| K (dioptres) | 0.15 (0.02 to 0.29) | 0.025 | 0.44 (0.24 to 0.64) | <0.001 | 0.05 (−0.10 to 0.21) | 0.50 |
Regressions were performed with GG, AG and AA genotypes coded as 0, 1, 2; with the exception of rs1015213 where GG was coded as 0, and AG and AA combined coded (as 1 due to the AA genotype frequency of <1%, see text).
AL, axial length; ACD, anterior chamber depth; K, corneal keratometry.
No biometric factors, including ACD, were significantly associated with rs3753841 or rs11024102 genotype in comparisons of means or in regression models including after adjusting for age and sex (table 3).
DISCUSSION
Angle closure results from a combination of predisposing anterior segment anatomy, unfavourable physiological behaviour15 and precipitating environmental factors.16 Recognised ocular biometric risk factors include a short AL, shallow anterior chamber, hyperopia, small radius of corneal curvature and a thick crystalline lens.3–9 Of these, ACD is believed to be the primary risk factor for angle closure glaucoma.17 We hypothesised that the three effect loci recently reported by Vithana et al2 may be associated with ocular biometric characteristics that predispose towards angle closure development. We performed a genotype–phenotype correlation study in a cohort of participants of the EPIC Norfolk eye study and found the presence of at least one A allele (AG or AA) for rs1015213 to be associated with a 0.07 mm shallower ACD compared with wild type homozygotes after adjusting for the effects of age and sex. This is equivalent to almost 20% of the SD of the mean ACD of phakic participants in our cohort and the entire difference in mean ACD between male and female participants. As females are almost twice as likely as males to have an occludable anterior chamber angle,18,19 and ACD is associated with the likelihood of an occludable anterior chamber angle (OR 9.1 per 1 mm shallower anterior chamber),19 then the implied risk of at least one A allele for rs1015213 may be consistent with up to a two times higher likelihood of an occludable anterior chamber angle. We are not aware of any publications describing the association between rs1015213 genotype and ACD.
Vithana et al2 performed their GWAS using genotyping data from five sites in Asia to identify effect loci, and then performed replication experiments using genotyping data from six independent collections sites (two in China, and one site each in the UK, Singapore, India and Saudi Arabia). Review of the supplementary data by Vithana et al2 shows rs1015213 reached significance as a PACG effect locus in the replication experiment for both the Caucasian cohorts, but only some of the Asian cohorts. This is presumably due to the low minor allele frequency for rs1015213 of 1–3% in for example, Chinese and Vietnamese subjects, but could suggest inter-racial differences in effect loci for PACG. rs1015213 is located between PCMTD1 and ST18 on chromosome 8q. Little is known on the function of PCMTD1 or ST18, the latter encodes for suppression of tumorigenecity 18 which is thought to modulate inflammation and apoptosis in fibroblasts20 and has been reported as a breast cancer tumour suppressor gene.21 PCMTD1 is expressed in the human foetal eye, and in human and mouse retina.22 We found no association between ocular biometric characteristics and rs11024102 at PLEKHA7, which is an adherens junction protein and has been identified in several tissues including mammalian cornea.23 We also found no association between ocular biometric characteristics and rs3753841 at COL11A1. This is interesting considering mutations in COL11A1 are found in Stickler syndrome type 224 which is associated with axial myopia, while a mutation in COL11A2 has been reported in a family with non-ocular Stickler syndrome.25 COL11A1 is also a non-housekeeping gene expressed in human trabecular meshwork.26,27
Our study has a number of limitations. Of the 3268 participants with genotyping data, ocular biometry was only available on approximately a third and for some genotypes (eg, rs1015213 AA) numbers were very low. Hence our analysis for rs1015213 considered an additive (GG/AG/AA) and a dominant model (GG vs AG/AA), with both of these showing a significant association with ACD after adjusting for age and sex (additive model: p=0.037, dominant model: p=0.028). Another limitation is genotyping data were not available on all those with eye data, as genotyping was performed following the second EPIC health examination for the primary purpose of an obesity GWAS, while ocular biometric data were recorded at the third health examination. Consequently, we performed a sensitivity analysis that showed obesity status was not related to any ocular biometric characteristic. Also direct genotyping data were not available for rs11024102 requiring us to impute data for these analyses. Finally, it is also possible our finding of an association between rs1015213 and ACD could be due to chance.
Our study suggests that PACG susceptibility locus rs1015213 (between PCMTD1 and ST18) may exert at least part of its risk effect through an association with ACD in European populations. The magnitude of this effect is equivalent to almost 20% of the SD of the mean ACD of phakic individuals in this cohort. Further genotype–phenotype correlation studies are required in other populations to confirm this finding.
Acknowledgements
The authors thank Ananth Viswanathan and Valentina Cipriani for their advice on this manuscript.
Funding Supported by Grant G0401527 from the Medical Research Council, UK and Grant 262 from Research into Ageing, UK. PJF was also supported by the Richard Desmond Charitable Trust (via Fight for Sight, grant 1956). ACD and PJF were supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Competing interests None.
Ethics approval Waverney NHS Research Governance Committee (2005EC07L) and the Norfolk Research Ethics Committee (05/Q0101/191).
Provenance and review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–7. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vithana EN, Khor C-C, Qiao C, et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat Genet. 2012;44:1142–6. doi: 10.1038/ng.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsbirk P-H. Anatomical risk factors in primary angle-closure glaucoma. A ten year follow up survey based on limbal and axial anterior chamber depths in a high risk population. Int Ophthalmol. 1992;16:265–72. doi: 10.1007/BF00917973. [DOI] [PubMed] [Google Scholar]
- 4.Lowe RF. A history of primary angle closure glaucoma. Surv Ophthalmol. 1995;40:163–70. doi: 10.1016/s0039-6257(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 5.Alsbirk P-H. Anterior chamber depth in Greenland Eskimos. I. A population study of variation with age and sex. Acta Ophthalmol. 1974;52:551–64. doi: 10.1111/j.1755-3768.1974.tb01766.x. [DOI] [PubMed] [Google Scholar]
- 6.Alsbirk P-H. Corneal diameter in Greenland Eskimos. Anthropometric and genetic studies with special reference to primary angle-closure glaucoma. Acta Ophthalmol. 1975;53:635–46. doi: 10.1111/j.1755-3768.1975.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 7.Lowe RF. Aetiology of the anatomical basis for primary angle-closure glaucoma. Biometrical comparisons between normal eyes and eyes with primary angle-closure glaucoma. Br J Ophthalmol. 1970;54:161–9. doi: 10.1136/bjo.54.3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsbirk P-H. Limbal and axial chamber depth variations. A population study in Eskimos. Acta Ophthalmol. 1986;64:593–600. doi: 10.1111/j.1755-3768.1986.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 9.Salmon JF. Predisposing factors for chronic angle-closure glaucoma. Prog Retin Eye Res. 1999;18:121–32. doi: 10.1016/s1350-9462(98)00007-x. [DOI] [PubMed] [Google Scholar]
- 10.Riboli E. Nutrition and cancer: background and rationale of the European Prospective Investigation into Cancer and Nutrition (EPIC) Ann Oncol. 1992;3:783–91. doi: 10.1093/oxfordjournals.annonc.a058097. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, Dimasi DP, Hysi PG, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6:e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solouki AM, Verhoeven VJM, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandhu MS, Waterworth DM, Debenham SL, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet Elsevier. 2008;371:483–91. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quigley HA, Friedman DS, Congdon NG. Possible mechanisms of primary angle-closure and malignant glaucoma. J Glaucoma. 2003;12:167–80. doi: 10.1097/00061198-200304000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Subak-Sharpe I, Low S, Nolan W, et al. Pharmacological and environmental factors in primary angle-closure glaucoma. Br Med Bull. 2010;93:125–43. doi: 10.1093/bmb/ldp042. [DOI] [PubMed] [Google Scholar]
- 17.He M, Wang D, Zheng Y, et al. Heritability of anterior chamber depth as an intermediate phenotype of angle-closure in Chinese: the Guangzhou Twin Eye Study. Investigat Ophthalmol Vis Sci. 2008;49:81–6. doi: 10.1167/iovs.07-1052. [DOI] [PubMed] [Google Scholar]
- 18.Lavanya R, Wong TY, Friedman DS, et al. Determinants of angle closure in older Singaporeans. Arch Ophthalmol. 2008;126:686–91. doi: 10.1001/archopht.126.5.686. [DOI] [PubMed] [Google Scholar]
- 19.Casson RJ, Marshall D, Newland HS, et al. Risk factors for early angle-closure disease in a Burmese population: the Meiktila Eye Study. Eye (Lond) 2009;23:933–9. doi: 10.1038/eye.2008.102. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Siqueira MF, Behl Y, et al. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J. 2008;22:3956–67. doi: 10.1096/fj.08-111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jandrig B, Seitz S, Hinzmann B, et al. ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2. Oncogene. 2004;23:9295–302. doi: 10.1038/sj.onc.1208131. [DOI] [PubMed] [Google Scholar]
- 22.NEIBank [accessed 21 Jan 2013]; http://www.neibank.nei.nih.gov.
- 23.Pulimeno P, Bauer C, Stutz J, et al. PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PLoS ONE. 2010;5:e12207. doi: 10.1371/journal.pone.0012207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards AJ, Yates JR, Williams R, et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum Mol Genet. 1996;5:1339–43. doi: 10.1093/hmg/5.9.1339. [DOI] [PubMed] [Google Scholar]
- 25.Vikkula M, Mariman EC, Lui VC, et al. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell. 1995;80:431–7. doi: 10.1016/0092-8674(95)90493-x. [DOI] [PubMed] [Google Scholar]
- 26.Michael I, Shmoish M, Walton DS, et al. Interactions between trabecular meshwork cells and lens epithelial cells: a possible mechanism in infantile aphakic glaucoma. Investigat Ophthalmol Vis Sci. 2008;49:3981–7. doi: 10.1167/iovs.08-1674. [DOI] [PubMed] [Google Scholar]
- 27.Paylakhi SH, Yazdani S, April C, et al. Non-housekeeping genes expressed in human trabecular meshwork cell cultures. Mol Vis. 2012;18:241–54. [PMC free article] [PubMed] [Google Scholar]