SUMMARY
Analysis of de novo CNVs (dnCNVs) from the full Simons Simplex Collection (SSC) (N = 2,591 families) replicates prior findings of strong association with autism spectrum disorders (ASDs) and confirms six risk loci (1q21.1, 3q29, 7q11.23, 16p11.2, 15q11.2-13, and 22q11.2). The addition of published CNV data from the Autism Genome Project (AGP) and exome sequencing data from the SSC and the Autism Sequencing Consortium (ASC) shows that genes within small de novo deletions, but not within large dnCNVs, significantly overlap the high-effect risk genes identified by sequencing. Alternatively, large dnCNVs are found likely to contain multiple modest-effect risk genes. Overall, we find strong evidence that de novo mutations are associated with ASD apart from the risk for intellectual disability. Extending the transmission and de novo association test (TADA) to include small de novo deletions reveals 71 ASD risk loci, including 6 CNV regions (noted above) and 65 risk genes (FDR ≤ 0.1).
INTRODUCTION
Autism Spectrum Disorder (ASD) is characterized by impairments in social communication and restricted or repetitive behavior or interests. Until recently, the genetic etiology of ASD has remained obscure. Over the last decade, however, a key role for de novo germline mutation has been established definitively. Such mutations have led to the discovery of dozens of ASD risk loci and genes (De Rubeis et al., 2014; Dong et al., 2014; Iossifov et al., 2012, 2014; Neale et al., 2012; O’Roak et al., 2012; Sanders et al., 2012), as well as yielding important insights into the genomic architecture and biological mechanisms underlying social disability (Chang et al., 2015; Parikshak et al., 2013; Pinto et al., 2014; Willsey et al., 2013). The Simons Simplex Collection (SSC), a cohort of simplex ASD families designed to facilitate the discovery de novo variation (Fischbach and Lord, 2010), has played a central role in this progress. Analysis of the SSC has demonstrated an excess of rare de novo mutations in probands versus unaffected siblings across a wide range of mutation types, from copy number variants (CNVs) (Levy et al., 2011; Sanders et al., 2011), to small insertion/deletions (indels) (Dong et al., 2014), and single nucleotide variants (SNVs) (Iossifov et al., 2012, 2014; O’Roak et al., 2012; Sanders et al., 2012). Moreover, the cohort has helped lay a foundation for the creation of rigorous statistical frameworks to evaluate the association of de novo mutations (He et al., 2013; Liu et al., 2014; Sanders et al., 2011, 2012). In combination with exome analyses of additional large ASD cohorts (De Rubeis et al., 2014; Liu et al., 2013; Neale et al., 2012), these frameworks have dramatically accelerated gene discovery in ASD.
Previous reports of approximately 1,000 SSC families (Levy et al., 2011; Sanders et al., 2011) replicated the association between ASD and de novo CNVs (dnCNVs) (Itsara et al., 2010; Marshall et al., 2008; Sebat et al., 2007) and the role of CNVs at 16p11.2 in ASD (Kumar et al., 2008; Marshall et al., 2008; Weiss et al., 2008). By developing methods to assess the genome-wide significance of recurrent de novo events, we identified novel risk loci, including duplications at 7q11.23 (Sanders et al., 2011). The current study extends these analyses to the entire SSC cohort (N = 10,220 individuals in 2,591 families). We replicate our prior findings in the newly analyzed SSC cohort; refine the estimates of locus heterogeneity for dnCNVs in ASD to between 93 and 246 distinct loci; confirm the genome-wide significance of four CNV loci (Table 1); and revisit earlier findings of an increased mutation burden in females (Figure 2) and genotype-phenotype correlations (Figure 3). In addition, we combine dnCNV findings from the Autism Genome Project (AGP) (Pinto et al., 2014) with the SSC data in an omnibus analysis of large-scale dnCNVs that yields four additional ASD risk loci with a false discovery rate (FDR) ≤ 0.1 (Table 2).
Table 1.
Regions with Multiple dnCNVs in the SSC (FDR ≤ 0.1)
| Band | Location (hg19) | dnCNVs (del/dup) | RefSeq Genes | Genesa | p Value (Corrected) | q Value (FDR) |
|---|---|---|---|---|---|---|
| 1q21.1 | chr1:146,467,203-147,858,208 | 5 (0/5) | 13 | – | 0.00008 | 0.00002 |
| 3q29 | chr3:195,747,398-197,346,971 | 3 (3/0) | 21 | – | 0.14 | 0.05 |
| 7q11.23 | chr7:72,773,570-74,144,177 | 4 (0/4) | 22 | – | 0.004 | 0.001 |
| 7q11.23 | chr7:73,978,801-74,144,177b | 5 (0/5) | 2 | GTF2I, GTF2IRD1 | 0.00008 | 0.00002 |
| 7q11.23 | chr7:74,455,447-74,488,775 | 3 (1/2) | 1 | WBSCR16 | 0.31 | 0.06 |
| 15q11.2-13.1 | chr15:23,683,783-28,471,141 | 5 (0/5) | 13 | – | 0.00008 | 0.00002 |
| 15q12 | chr15:26,971,834-27,548,820c | 6 (0/6) | 3 | GABRB3, GABRA5, GABRG3 | 1 × 10−6 | 6 × 10−7 |
| 15q13.2-13.3 | chr15:31,245,880-32,515,849 | 4 (2/2) | 7 | – | 0.01 | 0.002 |
| 16p11.2 | chr16:29,655,864-30,195,048 | 13 (8/5) | 27 | – | <1 × 10−10 | <1 × 10−10 |
| 16q23.3 | chr16:82,660,399-83,830,215d | 3 (3/0) | 1 | CDH13 | 0.13 | 0.05 |
| 22q11.21 | chr22:18,886,915-21,052,014 | 4 (2/2) | 36 | – | 0.31 | 0.06 |
Where ≤3 genes are present, they are listed to clarify the genomic location.
This is the region of intersection between an atypical dnCNV and the Williams-Beuren Syndrome locus (see Figure S5).
This is the region of intersection between an atypical dnCNV and the 15q11.2-13.1 locus (see Figure 6F).
Three de novo deletions overlap at least one exon of this gene.
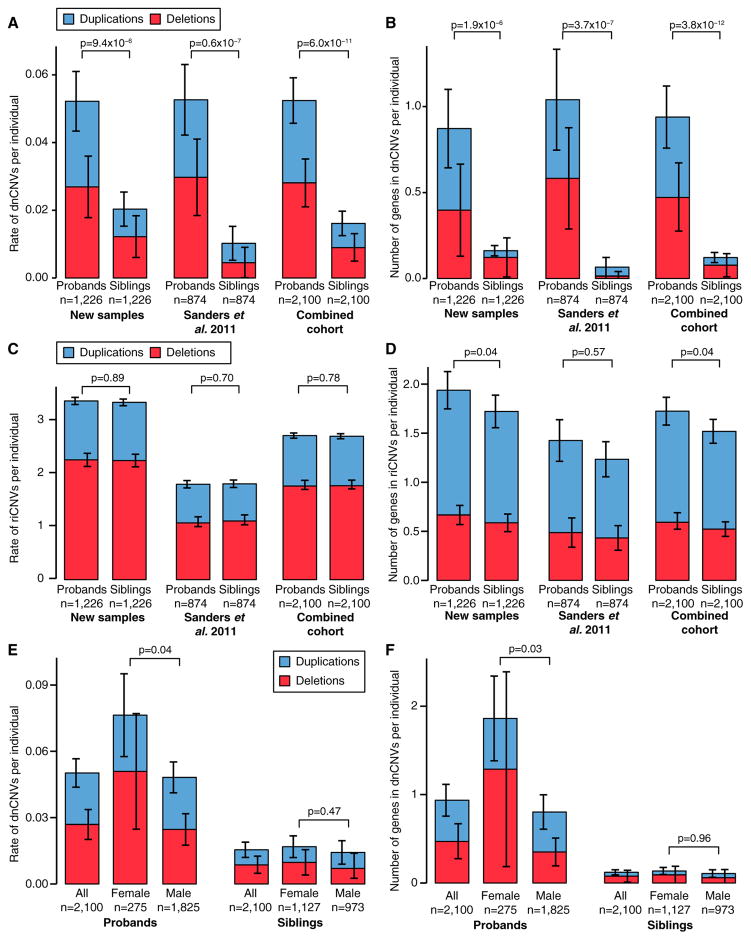
Figure 2. CNV Burden in the SSC.
(A) The rate of dnCNVs per individual in probands and family-matched sibling controls for deletions (red) and duplications (blue) are compared for new families (n = 1,226; left), previously published families (n = 874; middle), and the combination of these two cohorts (n = 2,100; right).
(B) The analysis presented in (A) is repeated except the number of genes within dnCNVs per individual is displayed rather than the rate of dnCNVs per individual. (C and D) The analyses presented in (A) and (B) are repeated using riCNVs instead of dnCNVs.
(E) The rate of dnCNVs per individual is shown for probands (left three bars) and siblings (right three bars). Within each group, the rate of dnCNVs is shown for all individuals (left), females (middle), and males (right). No statistical comparison was made between probands and siblings for this analysis.
(F) The analysis in (E) is repeated except the number of genes within dnCNVs per individual is displayed rather than the rate of dnCNVs per individual.
Statistical significance was calculated using a one-sided sign test for (A), a one-sided paired Wilcoxon ranked-sum test (WRST) for (B)–(D), and a two-sided unpaired WRST for (E) and (F). Whiskers show the 95% confidence intervals throughout (A)–(F).
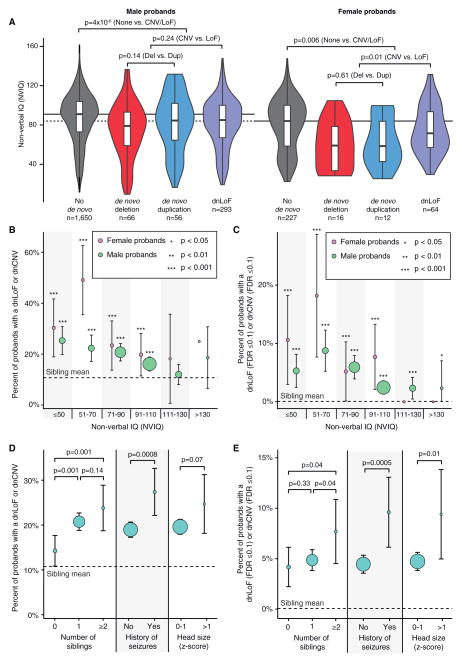
Figure 3. Genotype-Phenotype Correlations in the SSC.
(A) The violin plot shows the distribution of non-verbal IQ (NVIQ) in male probands (left four violins) and female probands (right four violins). Each sex is split into four sets: probands with no dnCNVs or dnLoF mutations (gray), probands with a de novo deletion (red), probands with a de novo duplication (blue), and probands with a dnLoF (purple). Individuals with multiple de novo events in more than one category were included in all of the corresponding distributions. The overlaid boxplot shows the median and interquartile range (IQR). The horizontal black lines show the median for the probands with no dnCNVs or dnLoFs in each sex; the dashed line extends this estimate for females to the y axis. Statistical significance was calculated using a one-sided WRST; violin plots of deletions and duplications together and deletions, duplications, and LoF together are not shown.
(B) The percent of probands with a dnLoF or dnCNV (y axis) is shown for male (green) and female (pink) probands binned by NVIQ (x axis). p values reflect the difference in de novo rate compared with siblings (horizontal dashed line at 10.7%) using a one-sided Fisher’s exact test; the whiskers show the 95% confidence intervals. The size of the circles represents the number of individuals in each group ranging from 4 to 694.
(C) The analysis in (B) is repeated considering only de novo mutations at loci with an FDR ≤ 0.1.
(D) The percent of probands with a dnLoF or dnCNV (y axis) is shown for three phenotypic factors. p values reflect the difference in de novo rate between groups of probands using a one-sided Fisher’s exact test; the whiskers show the 95% confidence intervals. The size of the circles represents the number of individuals in each group ranging from 170 to 2,177. The head size Z score is for the genetic deviation (Chaste et al., 2013).
(E) The analysis in (D) is repeated considering only de novo mutations at loci with an FDR ≤ 0.1.
Table 2.
Regions with Multiple dnCNVs in the SSC and AGP (FDR ≤ 0.1)
| Band | Location (hg19) | dnCNVs (del/dup) | RefSeq Genes | Genesa | p Value (Corrected) | q Value (FDR) |
|---|---|---|---|---|---|---|
| 1q21.1 | chr1:146,467,203-147,801,691 | 9 (1/8) | 13 | – | 6 × 10−9 | 2 × 10−9 |
| 2p16.3 | chr2:50,145,643-51,259,674b | 8 (7/1) | 1 | NRXN1 | 1 × 10−7 | 4 × 10−8 |
| 3q29 | chr3:195,747,398-196,191,434 | 4 (4/0) | 7 | – | 0.07 | 0.02 |
| 7q11.23 | chr7:72,773,570-74,144,177 | 5 (1/4) | 22 | – | 0.005 | 0.0008 |
| 7q11.23 | chr7:72,773,570-73,158,061c | 6 (1/5) | 10 | – | 0.0002 | 0.00003 |
| 7q11.23 | chr7:73,978,801-74,144,177c | 6 (1/5) | 2 | GTF2I, GTF2IRD1 | 0.0002 | 0.00003 |
| 15q11.2-13.1 | chr15:23,683,783-28,446,765 | 10 (0/10) | 13 | – | <1 × 10−10 | <1 × 10−10 |
| 15q12 | chr15:26,971,834-27,548,820d | 11 (0/11) | 3 | GABRA5, GABRB3, GABRG3 | <1 × 10−10 | <1 × 10−10 |
| 15q13.2-13.3 | chr15:30,943,512-32,515,849 | 5 (3/2) | 7 | – | 0.005 | 0.0008 |
| 16p11.2 | chr16:29,655,864-30,195,048 | 19 (12/7) | 27 | – | <1 × 10−10 | <1 × 10−10 |
| 22q11.21 | chr22:18,889,490-21,463,730 | 8 (4/4) | 45 | – | 1 × 10−7 | 4 × 10−8 |
| 22q13.33 | chr22:51,123,505-51,174,548 | 4 (4/0) | 1 | SHANK3 | 0.07 | 0.02 |
Where ≤ genes are present they are listed to clarify the genomic location.
Eight dnCNVs overlap at least one exon of this gene.
These are the regions of intersection between two atypical dnCNVs and the Williams-Beuren Syndrome locus (see Figure S5).
This is the region of intersection between an atypical dnCNV and the 15q11.2-13.1 locus (see Figure 6F).
Recent collaborative efforts have applied exome sequencing technology to the entire SSC cohort (Iossifov et al., 2014) identifying 27 ASD associated genes (FDR of ≤ 0.1). In parallel, 33 ASD risk genes (FDR ≤ 0.1) were identified in the Autism Sequencing Consortium (ASC) cohort (De Rubeis et al., 2014) with 12 genes identified in both analyses (Table S6), in part due to the inclusion of 825 SSC trios in the ASC. Importantly, these approaches to gene discovery that are agnostic to hypothesized biological mechanism have enabled a series of similarly agnostic systems biological analyses of ASD. These have reliably pointed to the contribution of chromatin modification and synaptic functioning and provided insights into the neuroanatomical and developmental dimensions of ASD pathology, highlighting in particular the contribution of mid-fetal cortical projection neurons and striatal medium spiny neurons (Chang et al., 2015; Cotney et al., 2015; Parikshak et al., 2013; Pinto et al., 2014; Sugathan et al., 2014; Willsey et al., 2013; Xu et al., 2014).
Completion of genotyping and exome sequencing of the SSC now allows for a combined analysis of CNV, indel, and SNV data to assess further the contribution of rare and de novo variation in these simplex families (Figure 2). We find that small de novo deletions often contain a single ASD gene of high effect that overlaps with de novo loss of function (dnLoF; nonsense, canonical splice site, and frameshift indels; also called “likely gene disrupting” [LGD]) mutations (Figure 5). In contrast, large dnCNVs do not show similar evidence and are likely to contain multiple genes of low effect. Moreover, expanding the TADA methodology (He et al., 2013), we combine evidence from de novo small deletions, indels, and SNVs to provide a unified statistical quantification of ASD association that, in combination with published data from the ASC and AGP, identifies 65 ASD risk genes (FDR ≤ 0.1) (Table 4). These 65 genes form a network of protein-protein interactions composed of two sub-networks that are enriched for genes that encode either chromatin regulators or synaptic proteins (Figure 7). Of note, mutations in male and female probands are randomly distributed in these networks rather than clustering on sex-specific genes.
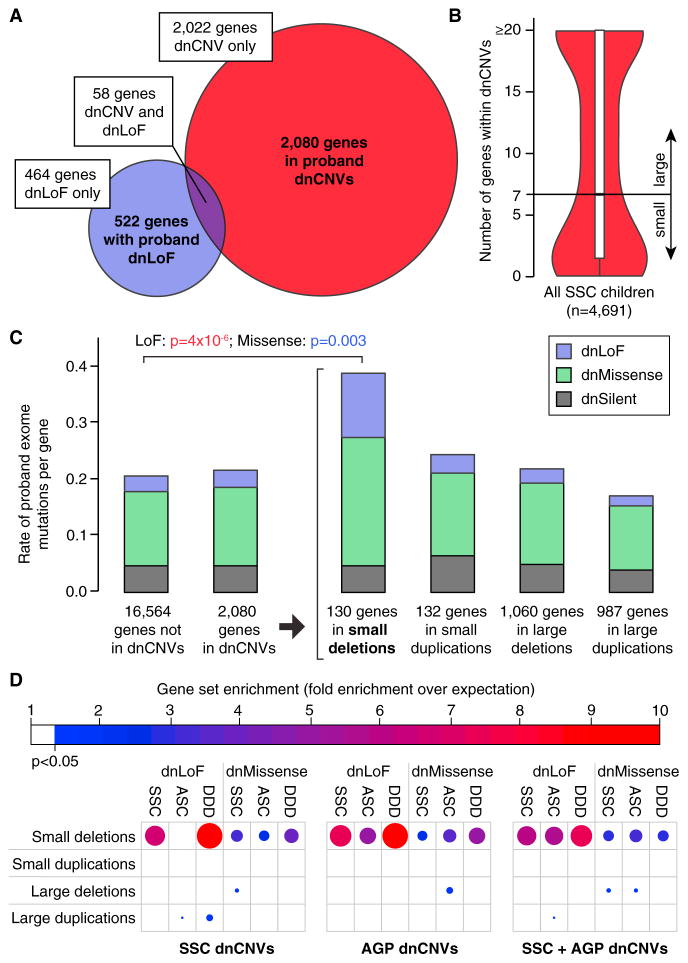
Figure 5. Small De Novo Deletions Are Enriched for Exome Mutations.
(A) 2,080 unique genes are identified within pro-band dnCNVs (red) and 522 unique genes have dnLoFs in probands (purple); 58 unique genes are observed in both these datasets.
(B) The median number of genes within validated dnCNVs in the SSC is seven; this threshold is used to distinguish small and large dnCNVs.
(C) The number of de novo mutations per gene observed with exome sequencing of the SSC and ASC are shown in different groups of genes based on dnCNV overlap. Mutation rates are normalized for gene mutability based on gene size and GC content. Exome mutations are divided into silent (gray), missense (green), and LoF (purple). No excess of exome mutations is observed in the 2,080 genes within dnCNV regions compared to the 16,564 genes outside of dnCNVs. Dividing the dnCNV regions by size (≤7 genes versus >7 genes) and type (deletion versus duplication) reveals strong enrichment for dnLoF (p = 4 × 10−6, Fisher Exact Test) and dnMissense (p = 0.003) in small de novo deletions only.
(D) The enrichment of genes within dnCNVs is shown by the size and shade of the circle (red and large = high degree of enrichment; blue and small = modest degree of enrichment); only results reaching nominal significance (hypergeometric test) are shown. Small de novo deletions show consistent enrichment for dnLoF and dnMissense mutations across three cohorts: SSC, Autism Sequencing Consortium (ASC), and Deciphering Developmental Disorders (DDD). This result is observed for dnCNVs detected in the SSC and Autism Genome Project (AGP) independently and in combination.
Table 4.
Integrating Small De Novo Deletions in TADA Identified 65 ASD Genes
| dnLoF Count | FDR ≤ 0.01 | 0.01 < FDR ≤ 0.05 | 0.05 < FDR ≤ 0.1 |
|---|---|---|---|
| ≥2 | ADNP, ANK2, ARID1B, ASH1L, CHD2, CHD8, CUL3, DSCAM, DYRK1A, GRIN2B, KATNAL2, KDM5B, KMT2C, NCKAP1, POGZ, SCN2A, SUV420H1, SYNGAP1, TBR1, TCF7L2, TNRC6B, WAC | BCL11A, FOXP1, GIGYF1, ILF2, KDM6B, PHF2, RANBP17, SPAST, WDFY3 | DIP2A, KMT2E |
| 1 | NRXN1, PTEN, SETD5, SHANK2, SHANK3, TRIP12 | DNMT3A, GABRB3, KAT2B, MFRP, MYT1L, P2RX5 | AKAP9, APH1A, CTTNBP2, ERBB2IP, ETFB, INTS6, IRF2BPL, MBD5, NAA15, NINL, OR52M1, PTK7, TRIO, USP45 |
| 0 | – | MIB1, SLC6A1, ZNF559 | ACHE, CAPN12, NLGN3 |
Genes with a small de novo deletion are in bold. FDR, false discovery rate.
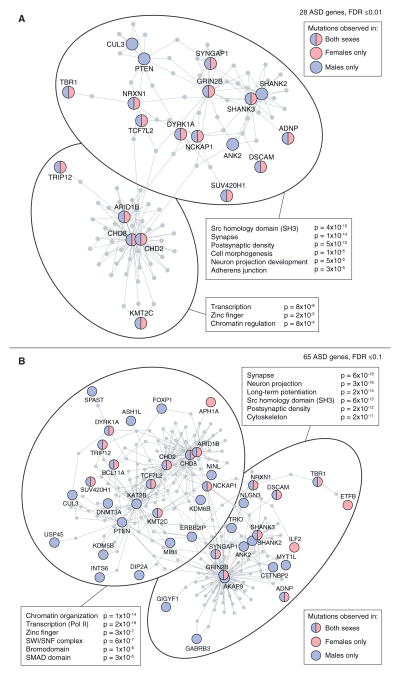
Figure 7. Protein-Protein Interaction Networks in ASD.
(A) 28 ASD genes identified with a TADA FDR ≤0.01 were submitted as seeds to form a DAPPLE PPI network (Rossin et al., 2011). The seed genes are shown as circles in red and/or blue based on the sex of the ASD cases in whom the mutations were identified; the distribution of male and female mutations in the network does not differ from expectation (p = 0.97). Protein-protein interactions are shown as gray lines (edges) and additional genes are pulled into the network to form indirect connections. The network has a clear distinction into two halves (shown by the large ovals). All seed and network genes in each oval were submitted to DAVID (Dennis et al., 2003) and the top gene ontology terms are shown with Benjamini Hochberg corrected p values.
(B) The analysis in (A) was repeated using all 65 ASD genes with an FDR ≤ 0.1 (Table 4).
RESULTS
SNP Genotyping, Sample Selection, and CNV Detection
High-quality SNP genotyping data were generated for 10,220 individuals in 2,591 families from the SSC using the Illumina Omni2.5, 1Mv3, or 1Mv1 arrays (Figure 1 and Table S1). Of these, 2,100 families were quartets, consisting of an affected proband, two unaffected parents, and at least one unaffected sibling, whereas 491 families were trios with no unaffected siblings. As described previously, CNVs were predicted using PennCNV (Wang et al., 2007), QuantiSNP (Colella et al., 2007), and GNOSIS and merged with CNVision (Sanders et al., 2011). To improve the specificity of dnCNV predictions, we developed a novel metric to estimate a per CNV p value (pCNV) based on the per SNP variability in Log R Ratio (LRR) and the number of SNPs consistent with a deletion/duplication based on B Allele Frequency (BAF).
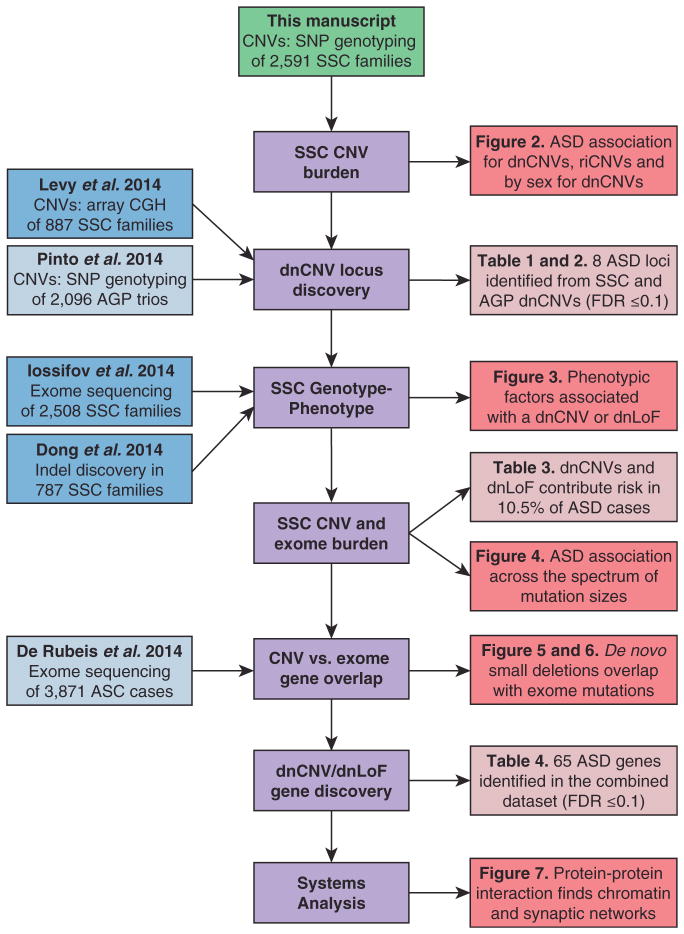
Figure 1. Overview of the Analysis.
This manuscript describes the analysis of CNVs predicted from SNP genotyping data in 2,591 families from the SSC (green). The analysis steps are shown in the middle of the flowchart (purple). Additional datasets from genomic analysis of the SSC (blue) and other ASD cohorts (light blue) are integrated to maximize power. The results of the analysis are shown in the figures (red) and tables (light red), along with the text of the manuscript.
This metric outperformed our prior approach of selecting the intersection of PennCNV and QuantiSNP calls (Sanders et al., 2011). Performance was assessed by rediscovery of validated dnCNVs detected on the 1Mv3 array in biological replicates run on the higher-resolution Omni2.5 array. A receiver operating characteristic (ROC) curve was used to compare the two approaches with an area under the curve (AUC) of 0.82 for the prior approach compared to 0.99 for the new pCNV metric (Figure S2). For dnCNVs detectable by microarray, a pCNV threshold of p ≤ 1 × 10−9 resulted in 80% sensitivity, similar to that obtained in our prior analysis, and increased specificity from 60% to 100%. We therefore elected to use this threshold, eliminating the need for blinded visual inspection prior to confirmation (Figure S3). Rare variation was defined as a population frequency ≤ 0.1% in either the Database of Genomic Variation (DGV) (MacDonald et al., 2014) or among all 5,382 SSC parents. This same population frequency threshold was used in the exome analyses (De Rubeis et al., 2014; Iossifov et al., 2014).
Overall, we detected 180 autosomal dnCNVs, of which 175 were validated by qPCR (97% confirmation rate); all validations were performed blinded to affected status. Nine dnCNVs (5.0%) at six loci were excluded due to germline mosaicism, based on a mosaic CNV in the parent or the same dnCNV in multiple siblings (Table S2). The ensuing analysis was performed on the remaining 166 validated dnCNVs, of which 110 were present in the probands of 2,100 quartet families, 34 were present in the siblings of the same 2,100 families, and 22 were found among 491 trios (Table S2).
De Novo CNVs Are Reproducibly Associated with ASD Risk
We first assessed whether our new CNV data replicated prior findings (Sanders et al., 2011). As observed previously (Levy et al., 2011; Sanders et al., 2011; Sebat et al., 2007), dnCNVs are more frequent in cases than controls (Figure 2). In the newly analyzed data (n = 1,226 quartets), we observed 64 dnCNVs in probands versus 25 dnCNVs in siblings (0.052 versus 0.020 dnCNVs per individual; ratio: 2.5, 95% CI: 1.6–4.1, p = 9.3 × 10−6, one-sided sign test). This increased burden is observed for both de novo deletions and duplications (p = 0.006 and p = 0.0007 respectively; Table S4). These results are consistent with our prior findings from a cohort of 874 quartet families (Sanders et al., 2011). The combined cohort of 2,100 quartet families shows 0.052 dnCNVs per proband compared to 0.016 dnCNVs per sibling (ratio: 3.1, 95% CI: 2.2–4.7, p = 6.0 × 10−11, one-sided sign test; Figure 2A).
As before (Sanders et al., 2011), a larger number of genes underlie dnCNVs in probands than siblings. In probands, 0.94 genes per individual were within the boundaries of a dnCNV compared with 0.12 genes per individual in siblings (ratio: 7.7, 95% CI: 4.2–17.1, p = 3.8 × 10−12, one-sided paired Wilcoxon signed-rank test (WSRT); Figure 2B). The difference between the individual-based and gene-based analyses is due to the larger size of dnCNVs in probands (median size 875 kbp in pro-bands versus 147 kbp in siblings) and higher density of genes in proband dnCNVs (p = 0.04, linear regression; Figure S4). Therefore, dnCNVs in probands are more frequent, larger, and more gene-rich than those observed in siblings.
Previously, we noted that more genes map within dnCNVs in female probands than male probands, consistent with a female protective effect (Sanders et al., 2011). This observation was replicated in an independent cohort of neurodevelopmental disorders (Jacquemont et al., 2014). In the current analysis, 1.9 genes are found within dnCNVs per proband among 275 female probands versus 0.8 genes within dnCNVs per proband among 1,825 male probands (ratio: 2.3, 95% CI: 1.0–4.6, p = 0.01, one-sided unpaired WRST; Figure 2F). We previously noted a trend toward a higher burden of dnCNVs in female probands (Sanders et al., 2011). With the increased power afforded by the current expanded dataset this trend is now significant; dnCNVs are observed at a rate of 0.076 in female probands compared to 0.049 in male probands (ratio: 1.6, 95% CI: 0.9–2.3, p = 0.04, one-sided Fisher’s exact test; Figure 2E). Of note, these findings are predominantly due to de novo deletions (ratio: 2.0, 95% CI: 1.0–3.5, p = 0.02, Fisher’s exact test; Figure 2E) rather than de novo duplications (ratio: 1.1, 95% CI: 0.3–1.9, p = 0.51, Fisher’s exact test). No difference in burden between the sexes was observed in unaffected siblings (Figure 2E; Table S4).
Rare Inherited CNVs Show Limited Evidence of ASD Association
We conducted similar analyses evaluating rare inherited autosomal CNVs (riCNVs) (Table S3). As before (Sanders et al., 2011), no significant excess of riCNVs was observed in probands with 5,713 riCNVs in 2,100 probands versus 5,687 riCNVs in 2,100 siblings (2.72 versus 2.71 per individual; ratio: 1.00, 95% CI: 0.97–1.04, p = 0.70, one-sided paired WRST; Figure 2C). A slight excess of genes was observed within proband riCNVs compared to sibling riCNVs (1.72 genes per proband versus 1.52 genes per sibling; ratio: 1.13, 95% CI: 1.04–1.24, p = 0.04, one-sided paired WRST; Figure 2D). These results were unchanged considering deletions or duplications separately (Table S4). The findings suggest that, overall, the contribution of riCNVs detectable by microarray must be small in simplex ASD. However, the observation of transmitted CNVs mapping to known ASD risk loci in some affected individuals supports the conclusion that a small subset of riCNVs does confer ASD risk.
Given the very modest risks imparted by riCNVs, we anticipated little difference in riCNV burden between the sexes, despite strong evidence for a female protective effect for de novo mutations. Accordingly, we observed no excess of riCNVs in female probands versus male probands (2.80 riCNVs/female versus 2.71 riCNVs/male; ratio: 1.03, 95% CI: 0.93–1.15, p = 0.21, one-sided paired WRST). Consistent with prior microarray analyses (Pinto et al., 2014; Sanders et al., 2011), but in contrast to analyses of small CNVs detected in exome data (Krumm et al., 2013; Krumm et al., 2015), we observe no excess of maternally inherited riCNVs in probands, with 2,813 riCNVs inherited from the father compared with 2,680 riCNVs inherited from the mother (p = 0.96, binomial distribution, one-sided). This difference may be the consequence of varying approaches and detection thresholds in CNV prediction. Similarly, we observed no increased burden of riCNVs in the mothers of SSC families compared to the fathers (5,350 in mothers versus 5,505 in fathers, p = 0.93, Binomial test, one-sided) using the detection thresholds described in a prior study that reported such a finding (Desachy et al., 2015).
Recurrent De Novo CNVs Identify Six ASD Risk Loci
The clustering of dnCNVs at a given locus in unrelated probands can be used to assess association when compared to the null expectation derived from dnCNVs in unaffected siblings (Sanders et al., 2011). Using this approach, we previously identified two loci with genome-wide significance in the SSC and predicted the discovery of two to three additional regions in the entire SSC. As anticipated, in the current study, a total of four loci reach genome-wide significance (p < 0.05; FDR ≤ 0.01): two previously identified (7q11.23 duplications and 16p11.2 [BP4-5] deletions and duplications) and two additional loci (1q21.1 duplications and 15q11.2-13.1 [BP2-3] duplications). Relaxing the detection threshold to an FDR ≤ 0.1 identifies three further loci: 3q29 deletions, 22q11.2 deletions and duplications, and deletions at Cad-herin 13 (CDH13) (Table 1). No locus met this threshold in the sibling data.
Integrating the SSC data with previously published dnCNVs identified among 2,096 trios from the AGP (Pinto et al., 2014) identifies two additional intervals reaching genome-wide significance: deletions at Neurexin 1 (NRXN1) and deletions and duplications at 22q11.2 (Table 2). Moreover, at an FDR threshold of ≤0.1, two further loci are identified: deletions at 3q29 and deletions at 22q13.33 that include the gene SHANK3. No further dnCNVs were reported in the AGP at the CDH13 locus, resulting in an FDR q value of 0.20 for the combined analysis. All eight loci (Table 2) have previously been implicated in ASD (Bucan et al., 2009; Kumar et al., 2008; Leblond et al., 2014; Marshall et al., 2008; Mefford et al., 2008; Moreno-De-Luca et al., 2013b; Sanders et al., 2011; Weiss et al., 2008).
More than 200 De Novo CNV Loci Carry ASD Risk
By comparing the burden of dnCNVs in siblings to that of probands, and considering the degree of dnCNV recurrence in probands, we previously estimated a total of 234 distinct dnCNV loci that mediate ASD risk. Repeating this calculation (Supplemental Experimental Procedures) in the entire SSC, we estimate a total of 93 ASD risk loci for dnCNVs, 61 loci for de novo deletions, and 37 loci for de novo duplications. In the combined SSC and AGP cohort, we estimate 246 total dnCNV risk loci, 181 loci for de novo deletions, and 168 loci de novo duplications.
The Presence of a De Novo Mutation Is Associated with a Reduction in IQ
Previously, we reported that the presence of a dnCNV was associated with a lower IQ in probands but that IQ was a weak predictor of de novo status (Sanders et al., 2011). Recent reports have described a similar reduction in IQ in the presence of a de novo LoF (dnLoF) mutation (Iossifov et al., 2014; Robinson et al., 2014; Samocha et al., 2014). Using the combined CNV and exome data in the SSC, we considered how sex and type of de novo mutation interact with non-verbal IQ (NVIQ).
For both males and females, we observe a reduction in NVIQ in the presence of either a dnLoF or dnCNV (8 point decrease in males, p = 4 × 10−6; 18 point decrease in females, p = 0.006; one-sided WRST; Figure 3A). In males there was no significant difference in the NVIQ between de novo deletions, duplications, or dnLoFs. In females, there was no difference in NVIQ between de novo deletions and duplications (p = 0.61); however, the median decrease in NVIQ was 12.5 points for dnLoF compared to 26 points for a dnCNV (p = 0.01; Figure 3A).
Overall, probands with an NVIQ below the proband median (89) had a 1.7-fold higher rate of de novo mutations compared to those with an NVIQ above the median (95% CI: 1.4–2.1; p = 1 × 10−7; one-sided WRST) and this effect was more pronounced in females (2.2-fold 95% CI: 1.3–3.8; p = 0.001) than males (1.6-fold 95% CI: 1.3–2.0; p = 3 × 10−5).
While a reduction in NVIQ is clearly associated with a dnCNV or dnLoF, we still observe a robust excess of de novo mutations in both male and female probands with an NVIQ between 91 and 110 compared to de novo mutations in siblings (p = 6 × 10−6 for males; p = 3 × 10−3 for females; WRST; Figure 3B). Furthermore, for the mutations with the highest confidence (FDR ≤0.1; Tables 2 and Table 4), we observe an excess burden in males even at an IQ above 130 (p = 0.04; WRST; Figure 5C). Therefore, despite the association between NVIQ and de novo status, a low NVIQ does not guarantee detecting a de novo mutation and a high NVIQ does not exclude an ASD-associated de novo mutation.
Phenotypic Features Associated with De Novo Mutations
Using the rich phenotypic data in the SSC, we tested whether other factors, along with sex and NVIQ, were associated with the presence of a dnCNV or dnLoF. The presence of an unaffected sibling increased the likelihood of observing a dnCNV or dnLoF (p = 0.001; WRST; Figures 3D and 3E) and this effect was amplified in the presence of multiple unaffected siblings. Similarly a history of seizures was associated with a higher de novo rate (p = 0.0008; WRST; Figures 3D and 3E); of note, this increase was observed equally for febrile and afebrile seizures (Figure S7). Similarly, a head circumference deviation of over 1 SD in either direction (Chaste et al., 2013) was associated with increased mutational burden. In contrast, we observed no difference in de novo burden in the presence of developmental regression or higher paternal or maternal age (Figure S7).
De Novo Mutations Contribute to ASD Risk in over 10% of Simplex Cases
The family structure of the SSC, and the availability of both SNP genotyping and exome data (Iossifov et al., 2014), offer an exceptional opportunity to explore the genomic architecture of ASD risk factors. By subtracting the rate of de novo mutations in siblings from the rate in probands, we can estimate the fraction of observed proband de novo mutations that contribute to ASD risk. In probands, we estimate that 70% (95% CI: 55%–80%) of dnCNVs and 46% (95% CI: 32%–56%) of dnLoF mutations carry ASD risk (Table 3). Both estimates are higher in females than males (Table 3).
Table 3.
Contribution of De Novo Mutations to ASD Risk
| Category of de novo mutation | Mutations per sample
|
Percent of mutations contributing to ASD risk (95% CI)
|
|||||
|---|---|---|---|---|---|---|---|
| Probands
|
Siblings
|
Probands
|
|||||
| All | Male | Female | All | All | Male | Female | |
| Deletions | 0.03 | 0.03 | 0.06 | 0.01 | 69.1% (43.2%–85.4%) | 64.6% (38.7%–83.2%) | 83.7% (66.2%–91.9%) |
|
| |||||||
| Duplications | 0.03 | 0.03 | 0.03 | 0.01 | 70.8% (48.3%–86.8%) | 70.8% (55.3%–88.6%) | 70.9% (0.0%–88.2%) |
|
| |||||||
| All CNVs | 0.06 | 0.05 | 0.09 | 0.02 | 69.9% (55.1%–79.6%) | 67.7% (49.2%–79.1%) | 79.7% (56.9%–88.5%) |
|
| |||||||
| Nonsense | 0.06 | 0.06 | 0.05 | 0.03 | 52.9% (33.8%–68.7%) | 53.9% (34.6%–66.4%) | 44.5% (0.0%–70.4%) |
|
| |||||||
| Splice Site | 0.02 | 0.02 | 0.06 | 0.01 | 52.4% (18.9%–73.7%) | 39.6% (0.0%–72.3%) | 80.8% (57%–91.6%) |
|
| |||||||
| Frameshift | 0.08 | 0.08 | 0.10 | 0.05 | 38.9% (14.8%–51.2%) | 37.4% (21.1%–52.9%) | 47.8% (4.9%–65.6%) |
|
| |||||||
| All LoF | 0.17 | 0.16 | 0.21 | 0.09 | 45.9% (31.8%–55.5%) | 43.9% (31.4%–53.8%) | 56.6% (41.1%–67.7%) |
|
| |||||||
| All LoFs and CNVs | 0.22 | 0.21 | 0.29 | 0.11 | 52.2% (45.2%–59.7%) | 50.0% (41.0%–58.9%) | 63.4% (49.4%–71.8%) |
| Category of de novo mutation | Percent of cohort with a mutation
|
Percent of cases with a mutation contributing to ASD risk (95% CI)
|
|||||
|---|---|---|---|---|---|---|---|
| Probands
|
Siblings
|
Probands
|
|||||
| All | Male | Female | All | All | Male | Female | |
| Deletions | 3.1% | 2.7% | 6.0% | 1.0% | 2.2% (1.1%–3.2%) | 1.8% (0.8%–2.5%) | 5.0% (2.3%–8.4%) |
|
| |||||||
| Duplications | 2.7% | 2.7% | 2.8% | 0.8% | 1.9% (1.2%–2.6%) | 1.9% (1.0%–3.0%) | 2.0% (0.2%–4.3%) |
|
| |||||||
| All CNVs | 5.8% | 5.3% | 8.7% | 1.7% | 4.1% (2.6–5.7%) | 3.6% (2.3%–4.9%) | 7.0% (3.2%–11.4%) |
|
| |||||||
| Nonsense | 5.9% | 6.0% | 5.0% | 2.8% | 3.1% (1.4–4.4%) | 3.2% (1.8%–4.9%) | 2.2% (0.0%–6.2%) |
|
| |||||||
| Splice Site | 2.4% | 1.9% | 6.0% | 1.1% | 1.3% (0.5–2.3%) | 0.7% (0.0%–1.5%) | 4.8% (1.5%–8.8%) |
|
| |||||||
| Frameshift | 7.8% | 7.7% | 8.7% | 4.8% | 3.0% (1.2–4.8%) | 2.9% (1.2%–4.4%) | 3.9% (0.2%–7.5%) |
|
| |||||||
| All LoF | 15.4% | 14.9% | 18.8% | 8.5% | 6.9% (4.9–8.9%) | 6.4% (3.9%–8.8%) | 10.3% (6.3%–16.2%) |
|
| |||||||
| All LoFs and CNVs | 20.6% | 19.7% | 26.6% | 10.1% | 10.5% (7.8–13.1%) | 9.6% (6.8%–12.0%) | 16.6% (11.4%–22.6%) |
By subtracting the percentage of siblings with ≥1 de novo mutation from the percentage of probands with ≥1 de novo mutation we can estimate the percentage of cases in whom a de novo mutation is contributing ASD risk. Based on this calculation, we estimate that 4% (95% CI: 3%–6%) of cases have a dnCNV mediating ASD risk and 7% (95% CI: 5%–9%) of cases have a dnLoF mediating ASD risk. In total, 11% (95% CI: 8%–13%) of simplex cases have a dnCNV and/or dnLoF mediating ASD risk (Table 3). In females, de novo mutations play a greater role in the ASD phenotype, contributing to ASD risk in 17% (95% CI: 11%–23%) of female probands and 10% (95% CI: 7%–12%) of male probands. Of note, these are conservative estimates for the overall contribution of de novo mutations to simplex ASD since they do not include very small CNVs, balanced structural variation, or variants discovered with sequencing aside from dnLoF.
ASD Risk Varies Based on the Size of the Mutation
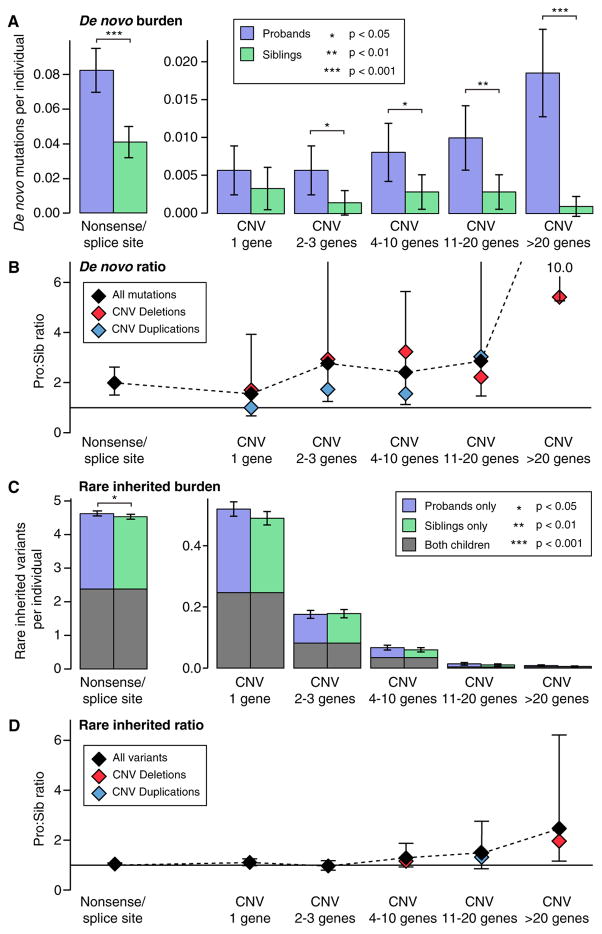
To further understand the genomic architecture of ASD in the SSC, we estimated the burden of both SNVs and CNVs in pro-bands compared to their unaffected siblings divided by variant size and mode of inheritance (Figure 4). Variants were divided into six bins based on size, the first bin was nonsense/splice site SNVs and the remaining five bins were CNVs covering: 1 gene, 2–3 genes, 4–10 genes, 11–20 genes, and >20 genes. For this analysis only, frameshift indels were excluded due to the absence of accurate population frequency data to identify rare inherited variants. Of note, we previously showed that both the burden and proband:sibling ratio of de novo frameshift indels is similar to that of de novo nonsense/splice site mutations (Dong et al., 2014).
Figure 4. Association of Genetic Factors with ASD across the Size Spectrum.
(A) The number of rare autosomal de novo mutations per individual are shown for dnLoF (nonsense and splice site only) in 1,911 SSC probands (purple) and 1,911 family-matched sibling controls (green) and for dnCNVs binned into five sizes by gene content in 2,100 SSC probands (purple) and 2,100 family-matched sibling controls (green). A significantly higher burden of de novo mutation is observed across the size range with the exception of “1 gene”; one-sided sign test; whiskers represent 95% confidence intervals.
(B) The proband:sibling ratio for each size of de novo mutation is shown by the black diamonds and the black dashed line; whiskers represent the 95% confidence interval estimated by bootstrapping. The ratio is also shown for deletions (red) and duplications (blue).
(C) The analysis shown in (A) is repeated for rare inherited variants in the same individuals. Significance was estimated using a one-sided paired Wilcoxon ranked-sum test with only rare inherited nonsense/splice site variants reaching nominal significance.
(D) The analysis in (C) is repeated for rare inherited variants in the same individuals.
De novo mutations were observed more frequently in pro-bands than siblings across the range of sizes. A similar proband: sibling ratio is observed for de novo nonsense/splice site and small de novo deletions, suggesting a similar fraction of these events mediate ASD risk. A higher ratio is observed for large dnCNVs. In contrast, no significant excess of rare inherited CNVs is observed at any size, though there is a trend toward an excess in probands for larger structural events (Figure 4).
Large De Novo CNVs and De Novo Point Mutations Target Different Genes
Multiple efforts have been made to fine map large multigenic dnCNVs in search of one or a small number of risk genes. Using dnCNV data from 4,687 probands in the SSC (this manuscript; Levy et al., 2011; Sanders et al., 2011) and the AGP (Pinto et al., 2014) alongside exome data from 3,982 probands in the SSC (Iossifov et al., 2014) and ASC (De Rubeis et al., 2014), we sought to address the broader question of whether, overall, the overlap between genes within dnCNVs and those altered by dnLoF mutations (Figure 5A) was greater than expected by chance, as would be predicted if large dnCNVs included only a small number of risk genes.
To address this question, we asked whether there was an overall excess of proband dnLoF, de novo missense (dnMissense), and de novo silent (dnSilent) mutations in the 1,794 unique genes within 119 large dnCNVs in probands. As there are very few large dnCNVs in siblings, we elected to use the number of proband dnLoF, dnMissense, and dnSilent mutations in the 16,564 genes not within any proband dnCNVs to calculate the null distribution. We observed no evidence of an excess of mutations from exome data in the genes within large dnCNVs, with 0.024 dnLoFs per gene within large dnCNVs compared with 0.031 dnLoFs per gene outside of dnCNVs (OR: 0.8; 95% CI: 0.5–1.0; p = 0.10, two-sided Fisher’s exact test; estimates corrected for gene mutability based on size and GC content; Figure 5C). Similarly, no difference was observed when we restricted the analysis of point mutations to dnMissense or dnSilent separately, when we considered various types of dnCNVs including large deletions, large duplications, and large dnCNVs with the strongest evidence of association with ASD based on recurrence (FDR ≤ 0.1), or when we considered the SSC cohort independently (Figures 5C and S8).
Small De Novo Deletions and De Novo LoFs Target a Common Set of Genes
In contrast to large dnCNVs, a similar analysis considering only small dnCNVs (N ≤ 7 genes) (Figure 5C) showed an excess of dnLoF and dnMissense mutations. The 130 unique genes within 73 small de novo deletions in probands contained 0.127 dnLoF per gene compared with 0.031 dnLoFs per gene outside of dnCNVs (OR: 4.4; 95% CI: 2.4–17.5; p = 3.8 × 10−6, two-sided Fisher’s exact test; estimates corrected for gene mutability; Figure 5C). A modest excess of dnMissense mutations (OR: 1.9; 95% CI: 1.2–2.9; p = 0.003), but not dnSilent mutations, was also observed (Figure 5C).
To demonstrate that this result was driven by ASD association, we repeated the analysis substituting in the sibling data. These were not used for the primary analysis due to the sparseness of de novo events in the siblings and the consequent reduction in power. There was no evidence of enrichment of sibling dnLoFs within proband small de novo deletions (p = 0.71, two-sided Fisher’s exact test), proband dnLoFs within sibling small de novo deletions (p = 0.24, two-sided Fisher’s exact test), or sibling dnLoFs within sibling small de novo deletions (p = 1.00, two-sided Fisher’s exact test).
To ensure that the overlap between exome mutations and small de novo deletions was not driven by a single dataset, we next divided the exome data by cohort (SSC versus ASC) and also considered de novo mutations identified by exome sequencing in the Deciphering Developmental Disorders study (Deciphering Developmental Disorders Study, 2015), an independent cohort of individuals with developmental disability. Similarly, we split the dnCNVs by cohort (SSC, AGP, and the combination of SSC and AGP). Enrichment of de novo point mutations was consistently observed within small de novo deletions, but not for the other three categories of dnCNV (Figure 5D). Of note, these analyses are robust to variation of the number of genes used to define small versus large dnCNVs between three and ten (Figure S9).
We also compared the genes within small de novo deletions to five datasets previously reported to intersect with ASD-associated genes. All five datasets showed enrichment in genes in small de novo deletions in probands, specifically: two datasets of genes that show evolutionary constraint (p = 0.04, p = 1 × 10−5, logistic regression) (Petrovski et al., 2013; Samocha et al., 2014), one dataset of genes that are targeted by the Fragile X protein FMRP in mouse brain (p = 4 × 10−8, logistic regression) (Darnell et al., 2011), and two datasets of genes that are bound by CHD8 using ChIP-seq (p = 0.04, p = 0.03, logistic regression) (Cotney et al., 2015; Sugathan et al., 2014). In contrast, none of these datasets were enriched in genes in small de novo deletions from the SSC siblings (p = 0.99, p = 0.46, p = 0.99, p = 0.99, p = 0.97, respectively; logistic regression).
The consistently strong enrichment of dnLoF mutations in small de novo deletions raises the possibility that these two classes of mutation target a common set of genes that mediate ASD risk. Based on this hypothesis, we would expect this enrichment to be the most dramatic for the genes with the strongest evidence of ASD association in the exome data. To test this hypothesis, we used the transmitted and de novo association (TADA) test to combine exome data from the SSC and ASC (Table S5). The model was built on the background of the published TADA analysis from the ASC, including the ASC rare inherited exome variants (De Rubeis et al., 2014). We elected not to include the rare inherited exome variants from the SSC, since these were not analyzed in a consistent manner to the ASC, their confirmation rate was not known, and their contribution to the TADA score is minimal. The TADA test generates an FDR q value for every gene based on the evidence from exome sequencing and the per gene mutability (He et al., 2013). A low q value represents strong evidence for ASD association, therefore if dnLoF and de novo small deletions target a common set of genes we would expect specific genes within the deletions to have lower than expected TADA scores.
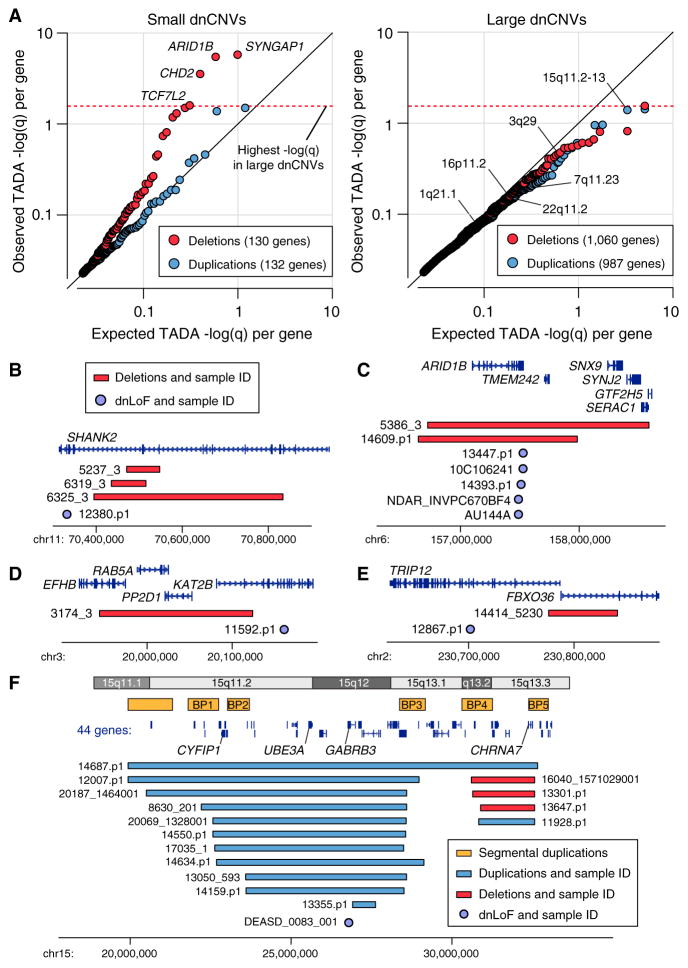
To assess the distribution of TADA q values in small de novo deletions, it was necessary to determine the expected q values under the null hypothesis (i.e., that small de novo deletions and exome variants do not target a common set of genes). A permutation test was used based on the mutability of genes within a dnCNV and the ability to detect these dnCNVs on a microarray (a factor of gene size including introns, number of SNPs, and the interaction of these two terms; Figures S10–S12). Comparing the expected TADA q values from permutated genes (median q value of 100 permutations) with the observed TADA q values showed that over half the genes within small de novo deletions showed evidence of overlap with the exome data (Figure 6A). In contrast, like the large dnCNVs, the q values of de novo small duplications match expectation closely (Figure 6A). Therefore, after correcting for the number of genes within a CNV, small de novo deletions are enriched for ASD risk genes identified by exome data, while small de novo duplications (Figure 6A) and large dnCNVs (Figure 6B) are not.
Figure 6. Small De Novo Deletions Intersect with ASD Genes.
(A) The TADA FDR q value is an assessment of ASD association based on de novo and inherited variants identified by exome sequencing in the context of estimates of gene mutability. A low TADA FDR q value (high −log(q)) represents stronger ASD association. Observed TADA −log(q) values are shown against expected TADA −log(q) values derived from permutation testing. Each point represents one gene within a proband dnCNV. The black line represents random sampling of the genome, with no increased overlap between genes in dnCNVs and the genes identified by exome sequencing in ASD. Small de novo deletions (red, on the left) deviate dramatically from this expectation while the other three categories show expected or slightly less than expected enrichment for ASD genes. The four genes with the strongest evidence for ASD association are labeled for the small de novo deletions (left). The individual genes with the highest −log(q) value (Table S6) within each of six large dnCNV loci with the strongest evidence for ASD association (Table 2) are indicated by the locus name (right).
(B) Three small de novo deletions and one dnLoF are observed in SHANK2.
(C) Two small de novo deletions and five dnLoF are observed in ARID1B.
(D) One small de novo deletion and one dnLoF are observed in KATB2.
(E) One small de novo deletion and one dnLoF are observed in TRIP12.
(F) Of the six large dnCNV loci with the strongest evidence for ASD association (Table 2) the 15q11.2-13 contains the gene with the lowest −log(q) value from the exome data: GABRB3.
The observed distribution of TADA q values would be consistent with a model in which a single gene within each de novo small deletion is responsible for all of the ASD risk from the dnCNV. In keeping with this model there are no examples of a small de novo deletion in which a dnLoF is observed in more than one gene, There are, in contrast, several examples of multiple small de novo deletions in unrelated individuals targeting the same gene (e.g., NRXN1) or multiple dnLoFs targeting the same gene (e.g., SYNGAP1, ARID1B; Figure 6C).
Integrating Small De Novo Deletions with Exome Data Reveals 65 ASD Risk Genes
Because small de novo deletions and exome mutations appear to target a common set of ASD risk genes, we integrated the evidence from the small de novo deletions into the TADA model to enhance gene discovery (Supplemental Experimental Procedures). Applying this model to the entire SSC cohort identified eight ASD genes (Table S6) in addition to the 27 identified previously at an FDR of ≤ 0.1 (Iossifov et al., 2014). Integrating the data from the recent ASC exome analysis (De Rubeis et al., 2014) and small de novo deletions from the AGP (Pinto et al., 2014) and a separate analysis of the SSC (Levy et al., 2011) identified 65 ASD genes (FDR ≤ 0.1; Table 4; Table S6). Of these, 27 had not previously met this threshold in the independent datasets (21 added by the combined TADA analysis, 6 by the inclusion of small de novo deletions).
At the more conservative threshold of FDR ≤0.01, we identify 28 ASD genes. Of these, 12 genes (ASH1L, KMT2C, NCKAP1, NRXN1, PTEN, SETD5, SHANK2, SHANK3, TCF7L2, TNRC6B, TRIP12, and WAC) had not previously met this threshold in independent genome-wide datasets (4 added by the combined TADA analysis, 8 by the inclusion of dnCNVs).
Several of the 65 ASD risk genes were previously described in the literature but had not met detection thresholds in the exome data. These include NRXN1, SHANK2 (Figure 6B), and SHANK3. While these genes have been previously considered definitive ASD risk loci (Berkel et al., 2010; Durand et al., 2007; Leblond et al., 2014; Pinto et al., 2010; Sanders et al., 2011), the current analysis allows a side-by-side comparison of the evidence in favor of association of these loci with the other genes identified in recent studies.
Many of the novel ASD risk genes were identified through the integration of the SSC and ASC datasets using the TADA methodology. This set of genes includes the ATPase gene Spastin (SPAST) that is associated with autosomal-dominant spastic paraplegia (Hazan et al., 1999) but was predicted to be associated with ASD by virtue of its relationship to other ASD genes in gene co-expression using the DAWN framework (Liu et al., 2014). The gene DNA-methyltransferase 3 alpha (DNMT3A), a key gene in the establishment of genomic imprinting in the embryo (Kaneda et al., 2004), is also identified by combining the SSC and ASC data. De novo heterozygous missense mutations in this gene have recently been associated with an overgrowth syndrome characterized by tall stature, distinctive facial appearance, and intellectual disability (Tatton-Brown et al., 2014). Three individuals in the SSC had a non-synonymous de novo mutation in DNTM3A: a boy with a NVIQ of 71 had a de novo frameshift mutation; a boy with an NVIQ of 82 had a de novo missense mutation (V665L); and a girl with an NVIQ of 49 also had a de novo missense mutation (P904L). Consistent with an overgrowth syndrome, all three individuals had heights and weights above the 95th percentile despite normal body mass index (BMI).
Finally, several novel genes are added to the list through the combination of small de novo deletions and dnLoFs. These include the p300/CBP-associated transcriptional regulator K(lysine) acetyltransferase 2B (KAT2B; Figure 6D) that has not previously been associated with neurodevelopmental abnormalities, the E3 ubiquitin ligase Thyroid hormone receptor interactor 12 (TRIP12; Figure 6E) in which a further dnLoF has been identified through targeted sequencing (O’Roak et al., 2014), and Methyl-CpG binding domain protein 5 (MBD5) that has previously been identified as the critical gene at the 2q23.1 locus through the observation of small de novo deletions and is associated with microcephaly, intellectual disability, neurodevelopmental abnormalities, and autistic features (Hodge et al., 2014; Talkowski et al., 2012).
ASD-Associated Genes Form a Protein-Protein Interaction Network with Two Subnetworks Enriched for Chromatin Regulating and Synaptic Genes
The integration of the small de novo deletions and exome data using the TADA metric resulted in a considerably expanded set of high confidence ASD genes. We therefore considered how this list of genes could inform our view of the etiology of ASD. Gene ontology analysis of the 65 genes with an FDR ≤0.1 using DAVID (Dennis et al., 2003) showed strong enrichment for chromatin regulation (3.2-fold over expectation; Benjamini Hochberg corrected (BHC) p = 0.0004), with eight genes contributing to this process (ARID1B, ASH1l, CHD8, DNMT3A, KMT2C, KMT2E, KDM5B, and SUV420H1); no other distinct processes showed significant enrichment.
We next considered whether there was evidence of protein-protein interaction (PPI) using DAPPLE (Rossin et al., 2011). A single network was derived from the 28 genes with an FDR ≤0.01 (p = 0.05 for direct interactions; p = 0.02 for indirect interactions). The resulting network shows a clear distinction into two subnetworks (Figure 7A); using DAVID, one subnetwork is enriched for synaptic/neuronal genes (9.5-fold enrichment; BHC p = 1 × 10−14), while the other subnetwork is enriched for chromatin regulator/transcription genes (6.6-fold enrichment; BHC p = 8 × 10−8). Of note, the gene Branched Chain Ketoacid Dehydrogenase Kinase (BCKDK) is drawn into the chromatin subnetwork by virtue of its interactions with both CHD8 and CHD2. This gene was previously associated with ASD through the observation of homozygous variants that disrupt BCKDK function in three consanguineous families (Novarino et al., 2012) resulting in markedly reduced plasma levels of branched chain amino acids. This result may indicate a functional relationship between ASD-associated chromatin regulators and a metabolic cause of ASD.
Repeating the DAPPLE analysis with all 65 genes (FDR ≤0.1) resulted in one large network (p = 0.02 for indirect interactions). The distinct divide between synaptic/neuronal and chromatin regulator/transcription subnetworks also persists (10.5-fold enrichment, BHC p = 6 × 10−18 and 14.5-fold enrichment, BHC p = 1 × 10−14, respectively; Figure 7B).
Mutations in Male and Female Cases Target a Common Set of Genes
One possible explanation for the increased male prevalence in ASD would be a set of genes in which mutations contribute ASD risk in males only. Our data do not support this hypothesis. In the 65 ASD genes (FDR ≤ 0.1), there were 109 dnLoF mutations or small de novo deletions in male probands compared to 33 in female probands. If the mutations targeted a common set of genes, irrespective of sex, we would expect 19 genes to include mutations from both sexes. The presence of male-specific risk genes would result in fewer than 19 genes. In contrast, we observed 20 genes with a mutation from both sexes (p = 0.97; permutation analysis). This result was not altered by restricting to genes in the chromatin PPI subnetwork (9 expected, 10 observed; p = 0.73) or in the synaptic PPI subnetwork (6 expected, 7 observed; p = 0.76; Figure 7B).
DISCUSSION
Analysis of CNVs detected in 2,591 families from the SSC highlights the key role of de novo mutations in the etiology of ASD. We replicate prior findings that dnCNVs are associated with ASD through the observation of an increased burden in pro-bands compared to unaffected sibling controls. Using the sibling distribution of dnCNVs to establish rigorous statistical thresholds, we previously identified two loci at genome-wide significance (deletions and duplications at 16p11.2 and duplications at 7q11.23) and, as predicted (Sanders et al., 2011), identified two further loci: 1q21.1 duplications and 15q11.2-13.1 duplications (Table 1). Including additional dnCNV data (Levy et al., 2011; Pinto et al., 2014) identifies an additional two loci: deletions at NRXN1 and deletions and duplications at 22q11.2. Relaxing the threshold to an FDR ≤ 0.1 identifies two more: 3q29 deletions and SHANK3 deletions, leading to a total of eight ASD risk loci from dnCNVs (Table 2); however, two of these loci involve only a single gene (NRXN1 and SHANK3) and consequently were included in the list of ASD risk genes, leading to a total of 6 loci and 65 genes.
The majority of these CNV loci are also associated with developmental delay (Coe et al., 2014) and, to a lesser extent, schizophrenia. The overlap with schizophrenia loci appears to be more selective; for example, 16p11.2 duplications are associated with schizophrenia, while 16p11.2 deletions are not (Szatkiewicz et al., 2014). These observations are consistent with a model in which CNVs contribute risk to a number of neuropsychiatric disorders (Moreno-De-Luca et al., 2013a; Stefansson et al., 2014); however, the extent of this risk varies between phenotypes for each locus (Moreno-De-Luca et al., 2014).
Our prior exome analysis demonstrated that the observation of even a small number of dnLoF mutations in the same gene among unrelated individuals could provide considerable statistical power to establish association (Sanders et al., 2012). The TADA test has extended this approach further and provides a framework to incorporate case-control data, transmitted variants, and missense mutations alongside dnLoF, resulting in a single metric of genome-wide association (He et al., 2013). Applying this model to 3,871 ASD cases and 9,937 controls in the ASC yielded 33 ASD risk genes (FDR ≤ 0.1) (De Rubeis et al., 2014). Here we present the latest iteration of this approach that incorporates structural variation. We use the TADA model to integrate data from small de novo deletions in 4,687 ASD cases and 2,100 matched sibling controls from the SSC and AGP (Pinto et al., 2014) alongside exome data of 5,563 ASD cases and 13,321 controls in the combination of the SSC (Iossifov et al., 2014) and ASC (De Rubeis et al., 2014) datasets. This comprehensive analysis identifies 65 ASD genes (FDR ≤ 0.1), including 28 at the more stringent threshold of FDR ≤ 0.01 (Table 4). The TADA framework provides a common standard of association that enables researchers to choose an association threshold tailored to their needs (Table S6).
These 65 ASD risk genes show enrichment for protein-protein interactions (PPIs) and coalesce into a PPI network with two distinct sub-networks corresponding to genes involved in chromatin regulation and the synapse (Figure 7), consistent with previous analyses (Chang et al., 2015; Li et al., 2014; Parikshak et al., 2013; Pinto et al., 2014). One explanation for how these two sets of genes both contribute to the ASD phenotype is that the genes involved in chromatin regulation may regulate the expression of the synaptic genes, a hypothesis supported by recent analyses of the regulatory targets of CHD8, an ASD gene and chromatin regulator, though these data do not establish clear directionality in this relationship (Cotney et al., 2015; Sugathan et al., 2014).
Our data also lend support to the female protective effect (FPE) hypothesis as a mechanism for the increased male prevalence in ASD through the observation of an increased burden of de novo mutations in female probands than male probands (Figures 2E and 2F). Further support of the FPE hypothesis comes from the observation that mutations in male and female pro-bands target a common set of genes (Figure 7), as opposed to an independent set of genes contributing male-specific risk.
The importance of large dnCNVs in human disorders, including disorders of childhood neurodevelopment, has been appreciated for over 30 years (Dobyns et al., 1983; Ewart et al., 1993; Ledbetter et al., 1981; Schmickel, 1986). Considerable effort has gone into identifying critical regions and critical genes that are responsible for the associated neurodevelopmental phenotypes. While there have been some notable successes in this endeavor, for example UBE3A in Angelman Syndrome (Matsuura et al., 1997), the majority of risk loci have been difficult to reduce to a single critical gene. By considering the intersection of de novo mutations in the genes targeted by dnCNVs, we were able to address the question of whether ASD risk is mediated by a single high-effect risk gene in either small or large dnCNVs.
Three strands of evidence support a role for such a critical gene in small de novo deletions. First, the ratio of mutations between probands and siblings is similar between dnLoF, which affect single genes, and small de novo deletions (Figure 4). Second, small de novo deletions in probands with ASD are greatly enriched for genes associated with ASD from exome analysis (Figures 5 and 6). Finally, we observe no instances of multiple genes associated with ASD (FDR ≤0.1) mapping within a single small de novo deletion.
In contrast, two strands of evidence support the hypothesis that multiple risk genes are present within large dnCNVs: First, NVIQ is inversely related to the number of genes within dnCNVs in probands (Figure S6). Second, many large CNVs contain sufficient numbers of genes that numerous ASD risk genes would be expected based solely on the fact that 800 genes are likely contributing to ASD risk (He et al., 2013; Sanders et al., 2012). Our data also support the hypothesis that genes within large dnCNVs carry modest individual effects. Notably, we find no evidence that significant risk genes identified via exome sequencing overlap with those mapping within large dnCNVs (Figure 6B). This finding is not altered by restricting the search to dnLoF mutations within large de novo deletions or dnMissense mutations in large de novo duplications (Figure 5C). Of course, there is considerable evidence that, cumulatively, large CNVs mediate large effects as has been noted throughout the literature (Itsara et al., 2010; Pinto et al., 2010, 2014; Sanders et al., 2011) and further supported by our observation here that large CNVs are rarely inherited, regardless of genomic location (Figures 2 and 4).
Given our approach, the current analysis does not preclude the presence of large effect non-coding mutations within these intervals. Moreover, if a single large-effect gene was present within a dnCNV in which a mutation led to a more severe phenotype, such as a structural brain abnormality or lethality, this would explain the observation of a paucity of dnLoF mutations mapping within CNVs in this ASD cohort (Figure 6B).
While we have used a threshold of seven genes to distinguish between small and large dnCNVs, our results are robust to varying the threshold between three and ten genes. A more general statement of this model is that dnCNV size is: (1) positively correlated to the number of ASD risk genes contained within; and (2) negatively correlated to the ASD risk mediated per gene. Of note, this model achieves a balance in the phenotypic contribution of de novo mutations across the spectrum of mutation sizes, so that a dnLoF in a single gene and large multigenic dnCNV can have a similar contribution to the ASD phenotype (Figure 3A).
Overall, through an integrative analysis of de novo mutations in ASD, we further clarify the genomic architecture of ASD, estimating that 50% of dnCNVs/dnLoFs mediate ASD risk, that more than 200 CNV risk loci and 800 risk genes are vulnerable to de novo mutation, and that de novo mutations contribute to the ASD phenotype in at least 11% of simplex ASD cases. We also provide further clarification of the relationship between de novo ASD risk mutations and intellectual disability. While we replicate the reported correlation of lower IQ with the presence of a de novo mutation, we find de novo mutations across the entire IQ distribution, with an excess burden of mutations in the highest confidence ASD risk genes, even in males with IQ above 130. In short, the data support the conclusion that large-effect de novo mutations contribute to ASD risk apart from intellectual disability.
Using a rigorous approach to assessing the significance of recurrent de novo mutations, we identify 71 independent ASD risk loci (FDR ≤ 0.1), composed of eight ASD risk loci (Table 2) and 65 ASD risk genes (Table 4) with NRXN1 and SHANK3 appearing in both lists. Finally, using a systems biological approach, we show that these 65 genes form a single network of protein-protein interactions that is enriched for genes involved in chromatin regulation and the synapse (Figure 7).
EXPERIMENTAL PROCEDURES
This study was overseen by the IRB at both Yale (HIC 0301024156) and UCSF (IRB: 14-14726 Ref: 146621). The data reported in this paper can be downloaded from SFARI Base (http://sfari.org/resources/sfari-base).
Genotyping
Illumina SNP genotyping data were generated for 10,220 individuals from 2,591 families from the SSC. Three types of genotyping array were used, with all family members run on the same array type: 333 families on the 1Mv1, 1,189 families on the 1Mv3, and 1,069 on the Omni2.5. A complete list of individuals is shown in Table S1.
Quality Control
All individuals included in the analysis passed stringent quality control measures, including genotypic identity within the family using PLINK (Purcell et al., 2007); sex check based on chromosome X heterozygosity and sex chromosome LRR (Figure S1); genotypes matching the exome data; genotyping rate ≥ 97%; and all members of the family passing the quality metrics of the CNV detection algorithms.
CNV Detection
Three algorithms were used to predict CNVs with default settings: PennCNV (Wang et al., 2007), QuantiSNPv2.3 (Colella et al., 2007), and GNOSIS (Sanders et al., 2011). The CNVs from the three algorithms were merged using CNVision (https://sourceforge.net/projects/cnvision/). Based on assessment of dnCNVs in technical replicates, a pCNV threshold of ≤1 × 10−9 was used for de novo prediction (Figure S2) and ≤1 × 10−4 for rare inherited prediction (Figure S3). Only CNVs with a population frequency ≤0.1% based on parental data and the Database of Genomic Variation (MacDonald et al., 2014) were included in the analysis. A complete list of de novo (Table S2) and rare inherited (Table S3) CNVs are included in the SOM.
Assessment of De Novo Recurrence
The significance of observing multiple dnCNVs at the same genomic loci was assessed using the method described previously (Sanders et al., 2011). The degree of overlap between dnCNVs in siblings was used to estimate the number of potential genomic locations a dnCNV occurred at, under the null distribution based on the unseen species model. Using this estimate, we predicted the likelihood of observing two or more dnCNVs in probands at a given location based on permutation testing.
Observed versus Expected TADA Values
To estimate the expected distribution of TADA values in small or large CNVs (Figure 6), we used a permutation test. For each observed dnCNV the same number of genes was selected at random, based on the mutation rate of those genes within CNVs (Figure S10–S12). The corresponding TADA FDR q values were obtained. This was repeated for all the dnCNVs in the probands. The TADA scores were then sorted from high to low. This process was repeated 100 times and the median of the highest score from 100 iterations was used as the expected value to compare with the highest score from the observed TADA values. This was repeated with the second highest, third highest, etc. to account for all the observed genes.
Supplementary Material
Highlights.
De novo copy number variants (dnCNV) are associated with Autism Spectrum Disorder/ASD
De novo mutations are associated with ASD in individuals with a high IQ
Small de novo deletions, but not large dnCNVs, contain one high-effect ASD risk gene
Identifies 6 ASD loci and 65 ASD genes, many of which target chromatin or the synapse
Acknowledgments
We are grateful to all the families participating in this research, including the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC), the Autism Sequencing Consortium (ASC), and the Autism Genome Project (AGP). This work was supported by grants from the Simons Foundation (SFARI #124827 to M.W.S. and SFARI #307705 to S.J.S.), the National Institute for Health/National Institute for Mental Health (MH100233 to J.D.B., MH100229 to M.J.D., MH100239 to M.W.S., MH057881 and MH100209 to B.D., DC009410 to D.M.M., MH074090 to D.H.L. and C.L.M., and MH071584-07 and MH19961-14 supporting D.M.M.), Donita B. Sullivan MD Research Professorship funds to D.M.M., the Howard Hughes Medical Institute (International Student Research Fellowship to S.J.S.), and the Canadian Institutes of Health Research (Doctoral Foreign Study Award to A.J.W.). C.L. receives royalties for the ADOS and ADI-R from Western Psychological Services; royalties related to this project are donated to charities. We would like to thank the SSC principal investigators (A.L. Beaudet, R. Bernier, J. Constantino, E.H. Cook, Jr, E. Fombonne, D. Geschwind, D.E. Grice, A. Klin, D.H. Led-better, C. Lord, C.L. Martin, D.M. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M.W. State, W. Stone, J.S. Sutcliffe, C.A. Walsh, and E. Wijsman) and the coordinators and staff at the SSC clinical sites; the SFARI staff, in particular M. Benedetti; the Rutgers University Cell and DNA repository for accessing biomaterials; the Yale Center of Genomic Analysis for SNP genotyping data; and I. Hart and X. Sanders for their unwavering support. D.H.L. acts as a consultant to Natera, Inc.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, 12 figures, and 6 tables and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2015.09.016.
AUTHOR CONTRIBUTIONS
Conceptualization, S.J.S., B.D., K.R., and M.W.S.; Methodology and Software, S.J.S., X.H., A.J.W., K.E.S., A.E.C., M.T.M., S.D., J.F.K., L.K., J.D.M., L.S., T.S.-N., M.Y.S., N.A.T., M.F.W., D.M.W., B.D., K.R., and M.W.S.; Validation, A.G.E.-S., M.T.M., and M.F.W.; Data Curation and Resources, S.J.S., A.J.W., A.G.E.-S., K.E.S., M.T.M., S.D., A.P.G., C.J., L.K., D.M.-D.-L., C.S.P., A.L.B., R.M.C., E.F., D.H.G., D.E.G., C.L., J.K.L., S.M.M., D.M.M., E.M.M., M.E.T., J.S.S., C.A.W., T.W.Y., A.S.C., D.H.L., C.L.M., E.H.C., J.D.B., M.J.D., B.D., K.R., and M.W.S.; Investigation, S.J.S., X.H., A.J.W., K.E.S., A.E.C., V.H.B., S.L.B., S.D., E.B.R., B.D., K.R., and M.W.S.; Writing, S.J.S., X.H., D.H.L., C.L.M., E.H.C., J.D.B., M.J.D., B.D., K.R., and M.W.S.
References
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaste P, Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, et al. Adjusting head circumference for covariates in autism: clinical correlates of a highly heritable continuous trait. Biol Psychiatry. 2013;74:576–584. doi: 10.1016/j.biopsych.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vultovan Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Desachy G, Croen LA, Torres AR, Kharrazi M, Delorenze GN, Windham GC, Yoshida CK, Weiss LA. Increased female autosomal burden of rare copy number variants in human populations and in autism families. Mol Psychiatry. 2015;20:170–175. doi: 10.1038/mp.2014.179. [DOI] [PubMed] [Google Scholar]
- Dobyns WB, Stratton RF, Parke JT, Greenberg F, Nussbaum RL, Ledbetter DH. Miller-Dieker syndrome: lissencephaly and monosomy 17p. J Pediatr. 1983;102:552–558. doi: 10.1016/s0022-3476(83)80183-8. [DOI] [PubMed] [Google Scholar]
- Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, Waqar Z, Gonzalez LE, Overton JD, Frahm S, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Dürr A, Wincker P, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge JC, Mitchell E, Pillalamarri V, Toler TL, Bartel F, Kearney HM, Zou YS, Tan WH, Hanscom C, Kirmani S, et al. Disruption of MBD5 contributes to a spectrum of psychopathology and neurodevelopmental abnormalities. Mol Psychiatry. 2014;19:368–379. doi: 10.1038/mp.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Coe BP, Hersch M, Duyzend MH, Krumm N, Bergmann S, Beckmann JS, Rosenfeld JA, Eichler EE. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Karakoc E, Mohajeri K, Nelson B, Vives L, Jacquemont S, Munson J, Bernier R, Eichler EE. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet. 2013;93:595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, Giuliano F, Stordeur C, Depienne C, Mouzat K, et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med. 1981;304:325–329. doi: 10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Li J, Shi M, Ma Z, Zhao S, Euskirchen G, Ziskin J, Urban A, Hallmayer J, Snyder M. Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol. 2014;10:774. doi: 10.15252/msb.20145487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sabo A, Neale BM, Nagaswamy U, Stevens C, Lim E, Bodea CA, Muzny D, Reid JG, Banks E, et al. Analysis of rare, exonic variation amongst subjects with autism spectrum disorders and population controls. PLoS Genet. 2013;9:e1003443. doi: 10.1371/journal.pgen.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lei J, Sanders SJ, Willsey AJ, Kou Y, Cicek AE, Klei L, Lu C, He X, Li M, et al. DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics. Mol Autism. 2014;5:22. doi: 10.1186/2040-2392-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013a;12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Sanders SJ, Willsey AJ, Mulle JG, Lowe JK, Geschwind DH, State MW, Martin CL, Ledbetter DH. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2013b;18:1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Moreno-De-Luca A, Cubells JF, Sanders SJ. Cross-Disorder Comparison of Four Neuropsychiatric CNV Loci. Curr Genet Med Rep. 2014;2:1–11. [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, Silhavy JL, Kara M, Khalil RO, Ben-Omran T, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, Samocha KE, Kosmicki JA, McGrath L, Neale BM, Perlis RH, Daly MJ. Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. Proc Natl Acad Sci USA. 2014;111:15161–15165. doi: 10.1073/pnas.1409204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ International Inflammatory Bowel Disease Genetics Constortium. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickel RD. Contiguous gene syndromes: a component of recognizable syndromes. J Pediatr. 1986;109:231–241. doi: 10.1016/s0022-3476(86)80377-8. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, Bjornsdottir G, Walters GB, Jonsdottir GA, Doyle OM, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, Miles J, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz JP, O’Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, Fromer M, Ruderfer D, Akterin S, Bergen SE, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, Zachariou A, Hanks S, O’Brien E, Aksglaede L, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.