Abstract
Regulator of chromatin condensation 1 (RCC1) is the only known guanine nucleotide-exchange factor for the Ran GTPase and has pivotal roles in nucleo-cytoplasmic transport, mitosis, and nuclear-envelope assembly1. RCC1 associates dynamically with chromatin through binding to histones H2A and/or H2B in a Ran-regulated manner2,3. Here, we report that, unexpectedly, the amino-terminal serine or proline residue of RCC1 is uniquely methylated on its α-amino group. Methylation requires removal of the initiating methionine, and the presence of proline and lysine at positions 3 and 4, respectively. Methylation-defective mutants of RCC1 bind less effectively than wild-type protein to chromatin during mitosis, which causes spindle-pole defects. We propose a bimodal attachment mechanism for RCC1 in which the tail promotes stable RCC1 association with chromatin through DNA binding in an α-N-methylation-dependent manner. These data provide the first known function for N-terminal protein methylation.
RCC1 has a propeller-like structure, one face of which binds to Ran4, whereas the other face binds chromatin. A flexible N-terminal tail contains a nuclear localization signal (NLS)5,6. Although the propeller seems rigid, Ran allosterically modulates RCC1 association with chromatin3, and the RCC1D182A mutant, which does not detectably bind Ran, shows reduced chromatin association7,8. Removal of the N-terminal tail also leads to exclusion of RCC1 from chromosomes7, whereas phosphorylation on Ser 2 and Ser 11 in human RCC1 reportedly permits stable association with mitotic chromosomes8,9. These phosphorylations have been proposed to reduce the affinity of the NLS for its nuclear transport receptor, importin-α3, which, when bound to RCC1, interferes with chromatin attachment9. However, Ser 2 is found only in simians and is a proline residue in other mammalian species.
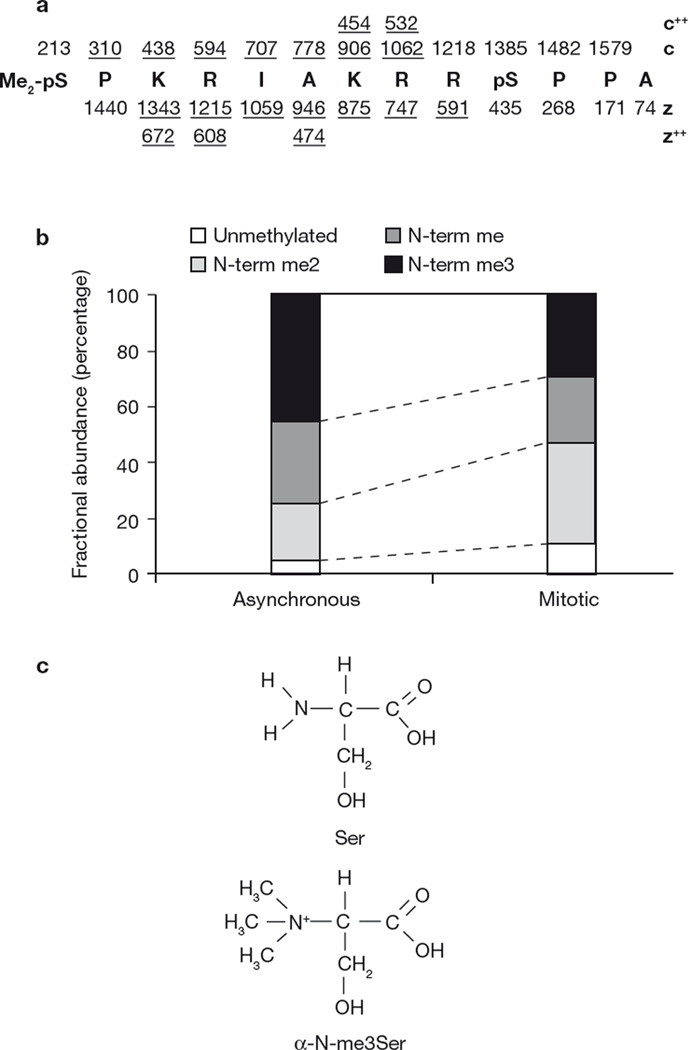
The N-terminus of RCC1 resembles histone tails, which also contain NLSs, and which are phosphorylated, acetylated and methylated in complex patterns that comprise a histone code10,11. We asked whether the RCC1 tail may also be post-translationally modified by mechanisms other than serine phosphorylation. To address this question, a stable tsBN2 cell line was generated that expressed a carboxy-terminal Flag-tagged human RCC1. TsBN2 cells contain a mutation in RCC1 that renders the protein unstable at elevated temperatures. Exogenous RCC1 can functionally replace the mutant12. To identify modifications of the tail, purified RCC1–Flag was subjected to tandem mass spectrometry. No lysine methylations or acetylations were observed. Remarkably, however, the initiating methionine residue was cleaved and the exposed α-amino group of Ser 2 was methylated (Fig. 1a, c, and see Supplementary Information, Fig. S1a). Mono-, di-, and trimethylated (α-N-me1-me2-me3Ser 2) forms of RCC1 were found in extracts from both interphase and mitotic cells (Fig. 1b). Ser 2 was also phosphorylated during mitosis (see Supplementary Iinformation, Figs S1, S2). In addition, phosphorylated Ser 11 was detected on RCC1 purified from cells at both interphase and mitosis (see Supplementary Information, Fig. S1b). The identity of the N-terminal peptide methylation was confirmed using a synthetic α-N-me2Ser 2 peptide corresponding to the sequence shown in Fig. 1a (data not shown).
Figure 1.
RCC1 N-terminal methylation. (a) Summary of ETD mass spectral data, recorded on the (M+4H)+4 ion corresponding to the 13-residue, N-terminal peptide of RCC1–Flag (residues 2–14 of the gene encoded sequence). RCC1 was isolated from mitotically arrested tsBN2 cells. Nominal m/z values for singularly charged ions of type c and z are shown above and below the peptide sequence, respectively. Those observed in the spectrum are underlined. Observed doubly charged c and z ions are also underlined. The spectra are shown in the Supplementary Information, Fig. S1a. m/z values for y2, y3 and y12 ions observed in the corresponding collision activated dissociation (CAD) mass spectrum also support the above sequence assignment (data not shown). We conclude that the peptide contains α-dimethyl- and phosphate-groups on the N-terminal-Ser 2 residue (Me2-pS2) and phosphate on Ser 11 (pS11). Experimental and calculated precursor m/z values for a peptide of this composition agree to within 2.5 p. p.m. (413.7190 and 413.7199, respectively). (b) Relative abundances of RCC1 N-terminal peptides (2–14) from asynchronous versus mitotically arrested tsBN2 cells as determined by semi-quantitative tandem mass spectrometry. Mono- and dimethylated RCC1 was also phosphorylated on Ser 2 in mitotic cells (data not shown). (c) Schematic representation of serine and me3Ser structures.
N-terminal methylation has been described for only a very limited number of proteins, and never on a serine residue13. Rabbit myosin light chains (MLC) LC-1 and LC-2 are the only proteins reported to be N-terminally methylated in chordates and no α-N-methyltransferase activity has been described in any metazoan13. The functional significance of N-terminal methylation is entirely unknown. Sequence comparisons of those few eukaryotic proteins containing N-terminal methylation suggest that the first three residues may function as recognition sites for an α-N-methyltransferase. In particular, for MLC LC-1, histone H2B (in Tetrahymena and Asterias), cytochrome c-557 (Crithidia)13 and RCC1, each protein has lost its initial methionine residue and is methylated on Ala 2, Pro 2 or Ser 2, followed by amino acids Pro 3 and Lys 4. However, there is no experimental evidence for an N-methylation motif. To address this issue, a series of RCC1 mutants (S2A, S2P, P3Q, K4Q, K4R and ASPK) were expressed as C-terminal Flag-tagged proteins in HeLa cells and purified for analysis by tandem mass spectrometry (Fig. 2a and see Supplementary Information, Fig. S2). Wild-type RCC1 and RCC1S2A proteins were each N-terminally methylated, indicating that the Ser 2 residue is not essential. Importantly, the Pro 2 variant, found in most mammals other than humans, was also stoichiometrically methylated (Fig. 2a and see Supplementary Information, Fig. S2). The K4Q mutation abolished N-terminal methylation (Fig. 2a), although Ser 11 was still phosphorylated (see Supplementary information, Fig. S1b). Importantly, the K4R mutant, which preserves the positive charge, was also only inefficiently methylated. The ASPK mutant has an alanine residue inserted at position 2 and N-methylation was absent from both this mutant and the P2Q mutants. We conclude that (Ser/Pro/Ala)2-Pro 3-Lys 4 serves as a substrate recognition motif for N-terminal methylation.
Figure 2.
Identification of N-terminal methylation motif, α-N-terminal methyltransferase activity and detection of endogenous RCC1 N-terminal methylation. (a) Relative abundances of RCC1 N-terminal peptide (2–14) from wild type (WT), S2A, S2P, ASPK, P3Q, K4Q and K4R mutants isolated from asynchronous HeLa cells as determined by tandem mass spectrometry. RCC1ASPK–Flag and RCC1P3Q–Flag are both N-terminally acetylated. (b) N-terminal methylation of RCC1–6xHis by soluble HeLa nuclear extract. Recombinant RCC1 or buffer was incubated with extract and 3H-SAM. Incorporation of 3H-methyl group into RCC1–6xHis was detected by filter binding and scintillation counting. (c, d) Immunoblots using an anti-N-me2Ser 2 antibody (c) and anti-N-me2/3Ser 2 antibody (d). Blots are shown for recombinant RCC1 +/− methylation by nuclear extract and SAM. Antibody specificities were determined by dot blots of the indicated peptides. Uncropped images of the blots are shown in the Supplementary Information, Fig. S5. (e) Detection of α-N-methylation on endogenous RCC1. Immunoblots for HeLa cell extracts transfected with RCC1 siRNAs or a control siRNA using the indicated antibodies. Ran was used as a loading control.
To identify an α-N-terminal methyltransferase activity for RCC1, in vitro methylation assays were conducted using 3H-S-adenosyl methionine (3H-SAM) as the methyl donor. The majority of bacterially expressed RCC1–6xHis lacks the initiating methionine, therefore this protein was used as a substrate. RCC1ASPK–6xHis, which is not N-terminally methylated in vivo, was used as a negative control. When incubated with a nuclear extract from HeLa cells, wild-type RCC1–6xHis was methylated, but the ASPK mutant was not, suggesting that the extract contains an α-N-specific methyltransferase activity (Fig. 2b). The absence of detectable methylation on the ASPK mutant shows that the soluble nuclear extract did not contain any lysine-specific methyltransferase activity that could recognize the RCC1 tail. Tandem mass spectrometry confirmed that wild-type RCC1 was uniquely mono- and dimethylated on the α-amino group of Ser 2 (data not shown). This is the first report of a metazoan α-N-terminal methyltransferase activity.
The studies described above all used exogenous RCC1. To determine whether the endogenous protein is methylated, antisera was raised against methylated peptides corresponding to the N-terminus of human RCC1. One antiserum targets α-N-me2Ser 2-RCC1, whereas the other targets α-N-me2 and α-N-me3Ser 2-RCC1 (Fig. 2c, d). Both antisera recognized recombinant RCC1–6xHis that had been methylated in vitro, but did not recognize non-methylated protein. They also both recognized a band of the expected size for RCC1 in HeLa whole-cell lysates (Fig. 2e), which was reduced after silencing of RCC1 expression by small interfering RNAs (siRNAs; Fig. 2e). These data support the conclusion that both antisera recognize bona fide endogenous methylated RCC1. At least one additional band was detected with each antiserum (Fig. 2e), suggesting that other N-terminally methylated proteins exist. Consistent with these results, the anti-N-me2/me3Ser 2 antibody stained both the cytoplasm and nucleus of HeLa cells, and only the nuclear staining was reduced by transfection with siRNAs against RCC1 (see Supplementary Information, Fig. S3a).
To investigate the functional significance of α-N-methylation, wild-type and RCC1K4Q–GFP fusions were expressed in MDCK cells and examined in mitotic cells. MDCK cells are advantageous for these studies because the cells orient their spindles in the plane of the monolayer, which facilitates scoring of mitotic defects. Tandem mass spectrometry confirmed that RCC1–GFP and RCC1–Flag, but not RCC1K4Q–Flag, were N-terminally methylated in MDCK cells (data not shown). Expression levels for wild-type and the K4Q mutant were similar (see Supplementary Information, Fig. S3b). In fixed mitotic cells, RCC1K4Q–GFP exhibited a more diffuse distribution than RCC1–GFP (Fig. 3a). To quantify this difference, GFP fluorescence-intensity ratios between the chromosomes and centrosomes were measured. Centrosomes were used as a fixed marker, spatially distinct from the chromosomes. The analysis confirmed that RCC1K4Q–GFP was more diffusely distributed than wild type, suggesting that loss of α-N-me1–me2–me3Ser 2 leads to decreased association of RCC1 with chromatin. This effect was not due to the loss of positive charge at position 4, as the K4R mutant was also mislocalized (Fig. 3a). RCC1ASPK–Flag was also expressed in MDCK cells, the cells were stained with anti-Flag and the fluorescence-intensity ratio between chromosomes and centrosomes was measured (Fig. 3a). The ASPK mutant, which has identical sequence to wild-type RCC1 except for the addition of a single alanine at the N-terminus, behaved similarly to K4Q and K4R mutants, which argues strongly that N-terminal methylation promotes RCC1 association with chromatin.
Figure 3.
Methylation of RCC1 regulates its interaction with chromosomes. (a) Mutations disrupt the localization of RCC1 in fixed mitotic MDCK cells. Cells expressing GFP or Flag fusions with either wild-type or mutant RCC1 were fixed and immunostained for tubulin and Flag (where indicated). Ratios of relative fluorescence intensity of RCC1 at chromosome versus centrosome were measured. Data were analysed by two-tailed independent t tests. Single asterisk indicates P ≤0.001, double asterisk indicates P ≤1E–08, three asterisks indicate P = 0.00011; n >30 cells per sample from three independent experiments. The error bars represent s.d. (b) Mutation disrupts the localization of RCC1 in live unfixed mitotic MDCK cells. Images of live cells expressing RCC1–GFP or RCC1K4R–GFP were collected. Sample pictures represent cells with two different protein expression levels for each construct. Ratios of relative fluorescence intensity of cytosolic versus chromatin bound RCC1 were measured. Data were analysed by two-tailed independent t tests (n ≥35 cells per sample from four independent experiments). The error bars represent s.d. (c) FRAP of RCC1–GFP in mitotic tsBN2 cells. Examples of representative cells expressing RCC1K4Q–GFP fusion proteins in metaphase are shown. The error bars represent s.d. from three independent experiments. Percentage recovery at 14 s was analysed by two-tailed independent t tests. For wild type (n = 11) versus RCC1K4Q (n = 10), P = 0.00022 and for RCC1S2A (n = 15) versus RCC1K4Q, P = 3.98E–05. (d) FLIP of RCC1–GFP in mitotic MDCK cells. Spots marked by a circle in mitotic cytosol were repeatedly photobleached. Representative FLIP images are shown. Data were analysed by 2-way ANOVA with posthoc comparisons by Tukey’s Honestly Significant Differences Test (HSD): wild type versus RCC1D182A (P <0.003, P <0.05 post hoc), wild type versus RCC1K4Q (P <0.00004, P <0.05 post hoc), wild type versus RCC1K4Q,D182A (P <0.0002, P <0.05 post hoc); n = 11 cells each with 100 time-points from three independent experiments. The scale bars in a, b and c represent 10 µm.
RCC1D182A, which does not detectably bind Ran, also associates only weakly with chromatin7,8 (and see Fig. 3a), presumably because Ran association is necessary to trigger a conformational change in RCC1 that stabilizes its interaction with chromatin. One possible explanation for the effect of methylation-defective mutations, therefore, is that they uncouple these two events. Strikingly, however, a double mutation of K4Q and D182A causes almost complete loss of chromosome localization (Fig. 3a). This additive effect of the K4Q and D182A suggests that α-N-methylation functions independently of Ran binding to promote RCC1 association with chromatin. To exclude the possibility that the diffuse localization in fixed cells is an artifact, GFP fluorescence ratios between mitotic chromosomes and cytosol were also measured in live unfixed cells expressing wild-type or RCC1K4R–GFP. RCC1K4R–GFP was significantly more cytosolic than the wild type (Fig. 3b), confirming that methylation is required for stable attachment to chromatin.
These data argue that the RCC1 mutants are more dynamic than the wild-type protein. To test this prediction, RCC1–GFP was photobleached on mitotic chromosomes in tsBN2 cells. Recovery kinetics were measured by sequential imaging (Fig. 3c). Because the bleached area often moved from its original location before fluorescence recovery was complete, only initial recovery rates between different proteins were compared. At 14 s post-bleach, recovery was ~65% for wild type or RCC1S2A–GFP, but ~80% for the RCC1K4Q–GFP, consistent with increased mobility of the methylation-defective mutant.
Next, fluorescence loss in photobleaching (FLIP) was used to determine the dissociation kinetics of RCC1–GFP from chromosomes. A spot (1 µm diameter) in the cytosol of mitotic MDCK cells was bleached repeatedly at maximum laser power, and the cell was imaged after each pulse to determine the loss of fluorescence from the chromosomes (Fig. 3d). Relative rates of fluorescence loss for RCC–GFP variants were K4Q + D182A > K4Q = D182A > wild type, consistent with the fluorescence recovery after photobleaching (FRAP) analyses and the changes in localization observed in fixed cells. Taken together, these data strongly argue that α-N-methylation is important for attachment of RCC1 to chromatin during mitosis.
A steep gradient of free Ran–GTP forms around chromosomes in Xenopus egg extracts and in mitotic HeLa cells14,15. Elevation of the RCC1 concentration in Xenopus egg extracts, which should dissipate this gradient, results in defective microtubule growth and spindle assembly15. Consistent with these results, ectopic expression of GFP–RCC1Δ27 or of GFP–RanQ69L, or microinjection of recombinant RanQ69L in HeLa cells, leads to an increased frequency of mitotic cells with multipolar spindles7,14. Because methylation-defective mutants of RCC1 have a reduced affinity for chromosomes, we asked whether these mutants would cause mitotic defects in MDCK cells. Wild-type RCC1–GFP itself caused a significant increase in abnormal mitoses — characterized by supernumerary spindle poles — probably because of the increased soluble pool of RCC1 caused by overexpression. The S2A mutant had no effect beyond the wild-type protein, but the frequency of abnormal mitoses was almost doubled by each of the methylation-defective mutants and was highly significant (P <0.01; Fig. 4a and Table 1). Quantification of cells with multipolar spindles is summarized in Table 1. Time-lapse fluorescence microscopy showed that in MDCK cells expressing GFP–tubulin and RCC1K4Q–mRFP the chromosomes congress normally then proceed through anaphase and telophase — even if they contain three or more spindle poles (see Supplementary Information, Movies 1 and 2), consistent with the fact that there is no checkpoint for centrosome defects16. Interestingly, the D182A mutant also produced mitotic defects, although at a lower frequency than the wild-type protein (see Supplementary Information, Table S1), even though it does not detectably catalyze nucleotide exchange on Ran7. Moreover, the effects of RCC1D182A and RCC1K4Q were additive, suggesting that they function through different mechanisms.
Figure 4.
Methylation of RCC1 is required for correct spindle assembly and chromosome segregation. (a) Examples of cells expressing RCC1–GFP with normal spindles and chromosomes or expressing RCC1K4Q –GFP with multiple spindle pole defects are shown. (b) MDCK cells expressing RCC1–Flag, RCC1K4Q–Flag, RCC1–Flag–H2A or RCC1K4Q–FLAG–H2A were fixed and stained. (c) Methylation of RCC1 facilitates its interaction with DNA. 35S-labelled recombinant RCC1–6xHis and 3H-labelled, N-terminally methylated RCC1–6xHis were combined, then incubated with DNA agarose or biotin-labelled recombinant histone H2A on streptavidin beads, or GST–importin-α3 on glutathione–Sepharose beads, or with agarose beads alone. Beads and supernatant were separated without washing by brief centrifugation and the ratio of 35S:3H was measured by scintillation counting. Data were analysed by two-tailed independent t test, n = 3. (d) Methylation promotes binding of the RCC1 tail to mitotic chromosomes in living unfixed MDCK cells. Cells expressing the indicated constructs were stained for DNA using Hoechst dye. Chromosomal:cytosolic GFP ratios were measured and analysed by two-tailed t-test. Asterisk indicates signficant differences from wild type (n = 14) for: RCC1K4R, P = 3.54E–05 (n = 14); RCC1K4Q, P = 1.9E–05 (n = 10); RCC1ASPK, P = 1.07E–05 (n = 10). (e) Methylated recombinant RCC1 tail (1–34) binds to nuclei with higher affinity than the unmethylated protein. Recombinant RCC11–34–CFP or RCC11–34–YFP were N-terminally methylated in vitro using HeLa nuclear extract. Indicated mixture of proteins was added to permeablized MDCK cells. After incubation, cells were incubated in PBS and imaged. Data were analysed by two-tailed independent t tests, n = 3. Single asterisk indicates P = 0.048; double asterisk indicates P = 0.027. The scale bars in a, b, c and e represent 10 µm. All error bars represent s.d. Y, YFP; C, CFP.
Table 1.
Quantification of mitotic defects in transfected MDCK cells
| Two spindle poles | Multispindle poles | Total | |
|---|---|---|---|
| RCC1 | 312 (90.2%) | 34 (9.8%) | 346 |
| RCC1ASPK | 318 (81.9%) | 70 (18.1%) | 388 |
| RCC1K4R | 369 (83.9%) | 71 (16.1%) | 440 |
| RCC1K4Q | 306 (82.7%) | 64 (17.3%) | 370 |
| RCC1S2A | 381 (91.3%) | 36 (8.6%) | 417 |
| RCC1–H2A | 363 (94.5%) | 21 (5.5%) | 384 |
| RCC1K4Q–H2A | 388 (96.5%) | 14 (3.5%) | 402 |
| GFP–H2A | 345 (95.8%) | 15 (4.2%) | 360 |
| GFP | 217 (96.9%) | 7 (3.1%) | 224 |
| Untransfected | 379 (99.2%) | 3 (0.8%) | 382 |
Data were collected from three independent experiments, one of which was double blind. Statistical analysis was performed using the χ2 test. Significant differences from the frequency of defects in wild-type RCC1 were found for RCC1ASPK (χ2 = 10.15, P ≤0.01, df = 1), for RCC1K4R (χ2 = 6.66, P ≤0.01, df = 1) and for RCC1K4Q (χ2 = 8.45, P ≤0.01, df = 1). Multipolar mitotic defects were significantly decreased in cells that expressed either RCC1–Flag–H2A or RCC1K4Q–FLAG–H2A. For RCC1 versus RCC1–H2A, χ2 = 4.96, P ≤0.01, df = 1; and for RCC1K4Q versus RCC1K4Q–H2A, χ2 = 40.5, P ≤0.01, df = 1.
To prove that the multispindle-pole phenotype induced by RCC1 overexpression, or by expression of methylation-defective mutants is caused by increased concentrations of cytosolic RCC1, we asked whether tethering RCC1 to chromatin could reverse the effect. As a tether, wild-type or RCC1K4Q was fused to Flag-tagged histone H2A. GFP–H2A is properly incorporated into nucleosomes in vivo and has very low mobility compared with RCC1 (refs 17, 18). In fixed mitotic cells, both fusion proteins colocalized with DNA (Fig. 4b), indicating proper incorporation into nucleosomes. As predicted, multipolar mitotic defects were dramatically decreased in cells that expressed either RCC1–Flag–H2A or RCC1K4Q–Flag–H2A (Table 1). Expression levels were similar to those of the untethered GFP-fusion proteins (see Supplementary Information, Fig. S3). We conclude that the mitotic defects are caused by increased cytosolic RCC1, which disrupts the Ran–GTP gradient. The data further demonstrate that, contrary to a previously proposed model3, RCC1 does not need to be released from chromatin during the Ran nucleotide-exchange cycle.
It was unclear how methylation could increase binding of RCC1 to chromatin. To answer this question, the ratio of methylated:unmethylated RCC1 binding to beads coupled to dsDNA, histones or importin-α3 was measured. Unmodified RCC1–6xHis was labelled with 35S-methionine, and a separate preparation of RCC1–6xHis was methylated in vitro using 3H-SAM. These two samples were mixed and incubated with the beads (Fig. 4c). The supernatants and bound fractions were then separated without washing, and counted simultaneously for 35S and 3H. In this equilibration assay, methylated RCC1 bound to the dsDNA beads significantly better than unmethylated RCC1 (P <0.01; Fig. 4c), but no significant difference in the 35S:3H ratio was observed using recombinant histone H2A or GST–importinα3 on beads. No binding was observed to beads alone. These results show that N-α-methylation confers an enhanced ability on wild-type RCC1 to bind dsDNA.
We next asked whether the isolated RCC1 tail could interact with chromatin. A previous study found no detectable association of the RCC1 tail with chromosomes, but the GFP was attached to the N-terminus, which would block methylation7. Therefore, to revisit this question, we attached the GFP to the C-terminus of the tail. Association of this fusion with mitotic chromosomes was clearly visible in live unfixed cells (Fig. 4d). Next, various RCC11–34 –GFP fusions were expressed and their distribution was examined in live mitotic cells. In each experiment, all methylated variants of the RCC1 tail showed significant concentration on the chromosomes, whereas all non-methylatable mutants showed little or no chromosome association (Fig. 4d).
One caveat of the cell-based experiments described above is that they depend on mutagenesis to block methylation. Therefore, the affinities of methylated versus unmethylated wild-type RCC1 N-terminal tails for intact nuclei were compared. Wild-type RCC1 tail was expressed as recombinant protein tagged at the C-terminus with either CFP or YFP and one of these fusions was methylated in vitro. The two versions of RCC1 were then mixed and incubated with permeabilized cells. Differential binding of the mixed proteins to chromatin was assessed by quantifying the ratio of CFP:YFP bound to the nuclei. Our data clearly show that the methylated RCC1 is retained preferentially on the chromatin (Fig. 4e). The result was independent of whether the CFP- or YFP-tagged RCC1 N-tail was methylated. We conclude, therefore, that chromatin association is regulated by N-α-methylation.
Taken together, our data unexpectedly show that RCC1 associates with nucleosomes through a bimodal attachment mechanism. The body of the protein interacts with histones H2A and H2B, and the methylated tail interacts through DNA. Di- and trimethylation abolish the nucleophilicity of the α-N and produce a permanent positive charge on the amino group, which may facilitate electrostatic binding to the phosphate groups on DNA13. We propose that the tail helps retain RCC1 on chromosomes during nucleotide exchange on Ran, as the affinity for histones drops when apo–Ran binds nucleotide3 (see Supplementary Information, Fig. S4). Failure of RCC1 to attach correctly to chromatin in mitosis causes a disruption of the Ran–GTP gradient, and a consequent increase in chromosome mis-segregation. RCC1 α-N-methylation is, therefore, a critical post-translational modification to prevent catastrophic genetic instability in mammalian cells. It will be of interest to determine if this unusual post-translational modification performs similar functions for other proteins.
METHODS
Constructs and antibodies
Human RCC1 DNA was amplified to introduce a 5′ XbaI restriction site, a C-terminal Flag tag and 3′ BamHI site, and subcloned into a mammalian expression vector (pRK7). RCC1–GFP was made by subcloning RCC1 into XbaI and BamH1 sites of a pKGFP vector. RCC1–mRFP was made by subcloning RCC1 into the XbaI site of pKmRFP. To generate pKRCC1–Flag–H2A, RCC1 was amplified to introduce a 5′ XbaI site and a C-terminal Flag tag followed by XhoI, H2A was amplified from pEGFP–H2A (a generous gift from S. Khochbin, INSERM, Grenoble, France) to introduce XhoI and BamH1 sites, and both inserts were subcloned into XbaI and BamH1 sites of pRK7 simultaneously. RCC11–34 –GFP was made by PCR amplification of 1–34 residues of RCC1 and subcloning into XbaI and BamH1 sites of a pKGFP vector. PRK7, pKGFP and pKmRFP are mammalian expression vectors previously described19, 20. ASPK, S2A, S2P, P3Q, K4R and K4Q mutants were generated by polymerase chain reaction using 5′ primers containing the designated mutations and subcloned as describe above. RCC1D182A was made using the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). To generate RCC11–34 –CFP/YFP, RCC11–34 was amplified to introduce a 5′ NdeI site and a 3′ BglII site, CFP/YFP was amplified to introduce 5′ BglII and 3′ XhoI sites, and both inserts were subcloned into NdeI and XhoI sites of pET30a simultaneously to express RCC11–34 –CFP/YFP–6xHis. Full-length human RCC1 was cloned into NdeI and XhoI sites of pET30a to express RCC1–6xHis, or NcoI–BglII sites of pQE60 to express RCC1ASPK–6xHis. Purification of RCC1–6xHis, RCC1ASPK–6xHis and RCC11–34 –CFP/YFP–6xHis were performed as described previously20. 35S-labelled RCC1–6xHis was prepared by growing Escherichia coli at 37 °C in 10 ml M9 medium plus kanamycin (10 µg ml−1). When cells reached an A600 value of 0.8, isopropyl β-d-thiogalactoside was added to 0.1 mM; 15 min later, rifampicin (Roche) was added to 0.25 mg ml−1; and another 15 min later, 1 mCi of the labelling 35S-methionine (MP Biomedicals, Solon, OH) was added to the culture. After 3 h, bacteria were harvested and RCC1–6xHis was purified as above.
Rabbit anti-N-me2Ser 2 and anti-N-me2/3Ser 2 antisera were produced against 2me-S-P-K-R-I-A-K-R-R-S-(8branch MAP)-C and 3me-S-P-K-R-I-A-K-R-R-S-C, respectively. The modified peptides were synthesized by Anaspec, Inc. (San Jose, CA). Peptides were coupled to activated keyhole limpet hemocyanin and injected into rabbits. The antisera were purified over beads to which the unmodified N-terminal RCC1 peptide had been attached, so as to remove antibodies against the non-methylated form of the protein. The supernatant from the beads was used without further purification, and diluted 1:1000 for immunoblots. Blots were developed using horseradish peroxidase-coupled secondary antibodies and enhanced chemiluminescence. Control and Smartpool siRNAs against human RCC1 were obtained from Dharmacon (Lafayette, CO). The sequences of the four RCC1 siRNAs in the pool were as follows: GGAGAACCGUGUGGUCUUA, CAGCAGCCCUCACCGAUGA, GCACAGAACCCGGCUUGGU and GGACAAUAACGGUGUGAUU.
Mass spectrometic analysis of RCC1 modification
RCC1 was immunoprecipitated, the buffer was exchanged with 100 mM ammonium bicarbonate at pH 8.0, and bound protein was digested with Asp-N (Roche, Palo Alto, CA) at an enzyme-to-protein ratio of 1:20 with shaking at room temperature for 6 h. The beads were removed by centrifugation, and the supernatant was transferred to a new tube and acidified using glacial acetic acid (Sigma, St Louis, MO). An aliquot of this peptide solution was loaded onto a capillary precolumn (360 µm o.d. × 75 µm i.d.) packed with irregular C18 packing material (5–20 µm). The precolumn was washed with 0.1% acetic acid and then connected to an analytical column packed with regular C18 packing material (5 µm) and a 5 µm emitter tip21. Samples were analysed using nanoflow HPLC-micro-electrospray ionization on a linear quadrupole ion trap-Fourier Transform mass spectrometer (LTQ-FT; Thermo Electron, Waltham, MA) for accurate mass and a Thermo LTQ instrument modified for electron transfer dissociation (ETD) for adequate MS/MS22. All spectra were manually interpreted.
Immunofluorescence microscopy
MDCK II, HeLa and tsBN2 cells were grown in DMEM supplemented with 5% calf serum plus 5% FCS, 100 U ml−1 penicillin and 100 U ml−1 streptomycin (GIBCO–BRL, Gaitherberg, MD). For transient transfections of MDCK II cells, 2–2.5 × 106 cells were electroporated using 2 µg DNA. Cells grown on Lab-Tek II chambers (Nunc, Naperville, IL) were fixed with 4% paraformaldehyde in PHEM buffer (60 mM PIPES at pH 7.2, 20 mM HEPES, 10 mM EGTA, 2 mM MgCl2), permeabilized with 0.5% Triton X-100, then blocked in 2% BSA–PHEM with 0.1% Triton X-100. The following primary antibodies were used: mouse anti-Flag (1:1000; Stratagene), mouse anti-α-tubulin (1:1000; Sigma), and rabbit anti-GFP (1:500; Molecular Probes, Eugene, OR). Alexa Fluor-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies were used at 1:1,000 (Molecular Probes). DAPI (1 ng ml−1), Draq5 (1:500, Alexis Corp., San Diego, CA) or Hoechst dye (1 µg ml−1) was used to stain DNA. Cells were imaged by confocal microscopy using an LSM510 (Carl Zeiss MicroImaging, Inc., Oberkochen, Germany) with a 100× oil immersion lens (na 1.3) and processed using Adobe Photoshop 7.0.
In vitro methylation assay
Soluble HeLa nuclear extract was purified as previously described23. In vitro methylation assays were performed in a volume of 200 µl of buffer (50 mM Tris at pH 8.0, 1 mM PMSF and 0.5 mM DTT), containing 16 µg RCC1–6xHis as substrate, 200 µg nuclear extract, and 1.1 µCi S-adenosyl-l-methyl-3H-methionine (0.55 mCi ml−1; MP Biomedicals, Inc.) as methyl donor. At the indicated times, 20 µl of reaction mixture was filtered through nitrocellulose. Incorporated methyl-3H-methionine was quantified by scintillation counting. To methylate RCC11–34 –CFP/YFP, the assay described above was scaled up with 500 µM unlabelled SAM (Sigma) and incubated at 30 °C for 2.5 h. The methylated protein was then repurified as described above.
Live-cell imaging, FRAP and FLIP
Cells were plated in Bioptek Delta-T dishes containing Ham’s F12 plus 10% FBS, 10 U ml−1 penicillin–streptomycin and 25 mM HEPES. An LSM510 confocal microscope was used for live-cell imaging, FRAP and FLIP experiments, with a 100× oil immersion lens (n.a. 1.3), except for the live-cell images in Fig. 3b. Cells were maintained at 37 °C with a stage heater. Gain and laser power were adjusted so that pixels were not saturated and photobleaching was negligible. For FRAP experiments, cells were scanned four times, followed by two bleach pulses of 245 ms using a spot 1 µm in diameter. Single section images (512 × 512 pixels) were then collected at 200 ms intervals for 25 time points, with the laser power attenuated to 9% of the bleach intensity. In FLIP experiments, cells were repeatedly imaged and bleached at intervals of 1 s with each bleaching for 250 ms and with imaging as for FRAP. For time-lapse imaging, MDCK cells stably expressing GFP–tubulin (a gift from J. Nelson, Stanford University, Stanford, CA) were transfected with RCC1SPQ–mRFP. Cells were imaged using the 488 nm and 543 nm laser lines at 1-min intervals, with the pinhole set to provide 1 µm z-sections. Data were converted to QuickTime format. For data in Fig. 3b, cells were plated as above but imaged using a wide-field microscope (Eclipse T200; Nikon, Tokyo, Japan) equipped with a 60× n.a. 1.2 Plan-achromatic water immersion lens and a charge-coupled device camera (Orca C4742-95-12NRB; Hamamatsu Photonics, Tsukuba, Japan). Images were collected at a 12-bit depth and 1,024 × 1,280 pixel resolution with 1 × 1 binning using Openlab 4.0 software (Improvision, Coventry, UK). Images were converted to grey scale and inverted using Openlab, then processed using Adobe Photoshop 7.0.
Nuclear-binding assay
MDCK cells growing on poly-L-lysine-coated cover-glass chamber slides were permeabilized with 0.2% Triton X-100 for 7 mins at room temperature. The indicated proteins were then added to the cells at 1 nM each and incubated for 10 mins. Then cells were incubated in PBS and imaged using an LSM510 (Carl Zeiss MicroImaging, Inc.) with a 20× lens, and processed as described above.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Macara laboratory for helpful suggestions, especially G. Riddick for conducting statistical tests. We also thank J. Nelson for MDCK cells stably expressing GFP–tubulin; S. Khochbin for GFP–H2A; L. Pemberton for core histones; R. Tsien for mRFP1; and T. Stukenberg, M. Smith and A. Spang for helpful suggestions and critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (NIH) to I.G.M (GM070902, CA040042) and D.F.H. (GM37537).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
T.C. made the RCC1 mutant proteins, identified the methyltransferase activity and its recognition motif, and performed the assays to identify its function. T.L.M. and J.S. performed the mass spectrometry. C.E.S.-T. made the antibodies against methylated RCC1 and performed the experiments with them. D.F.H. directed the mass spectrometry. I.G.M. directed the biochemical and cell biological studies. T.C. and I.G.M. wrote the paper. All authors discussed results and contributed to the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nature Cell Biol. 2002;4:E177–E184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- 2.Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- 3.Li HY, Wirtz D, Zheng Y. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 2003;160:635–644. doi: 10.1083/jcb.200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 5.Talcott B, Moore MS. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin α3/Qip. J. Biol. Chem. 2000;275:10099–10104. doi: 10.1074/jbc.275.14.10099. [DOI] [PubMed] [Google Scholar]
- 6.Nemergut ME, Macara IG. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 2000;149:835–850. doi: 10.1083/jcb.149.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore W, Zhang C, Clarke PR. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 2002;12:1442–1447. doi: 10.1016/s0960-9822(02)01076-x. [DOI] [PubMed] [Google Scholar]
- 8.Hutchins JR, et al. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr. Biol. 2004;14:1099–1104. doi: 10.1016/j.cub.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Li HY, Zheng Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 2004;18:512–527. doi: 10.1101/gad.1177304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosammaparast N, et al. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 12.Kai R, Ohtsubo M, Sekiguchi M, Nishimoto T. Molecular cloning of a human gene that regulates chromosome condensation and is essential for cell proliferation. Mol. Cell Biol. 1986;6:2027–2032. doi: 10.1128/mcb.6.6.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 14.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 15.Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 16.Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J. Cell Sci. 1997;110:421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- 17.Chen AE, Ginty DD, Fan CM. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–322. doi: 10.1038/nature03126. [DOI] [PubMed] [Google Scholar]
- 18.Perche PY, et al. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 2000;10:1531–1534. doi: 10.1016/s0960-9822(00)00832-0. [DOI] [PubMed] [Google Scholar]
- 19.Brownawell AM, Kops GJ, Macara IG, Burgering BM. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell Biol. 2001;21:3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol. Cell Biol. 2004;24:6608–6619. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 22.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.