Abstract
Inflammation is a common component of acute injuries of the central nervous system (CNS) and degenerative disorders such as Alzheimer’s disease. Glial cells play important roles in local CNS inflammation, and an understanding of the roles for microRNAs in glial reactivity in injury and disease settings may therefore enable novel therapeutic interventions. Here we show that the miR-181 family is developmentally regulated and present in high amounts in astrocytes compared to neurons. Over-expression of miR-181c in cultured astrocytes results in increased cell death when exposed to lipopolysaccharide (LPS). We show that miR-181 expression is altered in brain cells in vivo in response to LPS, a model of inflammation, in both wild-type mice and transgenic mice lacking both receptors for the inflammatory cytokine TNF-α. Knockdown of miR-181 enhanced LPS-induced production of pro-inflammatory cytokines (TNF-α, IL-6, IL-1β, IL-8) and HMGB1, while over-expression of miR-181 resulted in a significant increase in the expression of the anti-inflammatory cytokine IL-10. To assess the effects of miR-181 on the astrocyte transcriptome, we performed gene array and pathway analysis on astrocytes with reduced levels of miR-181b/c. To examine the pool of potential miR-181 targets, we employed a biotin pull-down of miR-181c and microarray analysis. We validated both MeCP2 and XIAP mRNAs as targets of miR-181. These findings suggest that miR-181 plays important roles in the response of astrocytes to inflammatory settings. Further understanding of the role of miR-181 in inflammatory events and CNS injury could lead to novel therapies for CNS disorders with an inflammatory component.
Keywords: miR-181, Inflammation, LPS, MeCP2, XIAP
Introduction
Both acute injury to the central nervous system (CNS) and chronic neurodegenerative disorders are characterized by an inflammatory response, a component of which is mediated by astrocytes (see Hamby et al., 2010; Sofroniew 2009 for review). Astrocytes are a heterogeneous population of glia with diverse functions in the uninjured CNS including maintenance and control of the blood-brain barrier, re-uptake of neurotransmitters and signaling through gap-junctions, among others (Sofroniew and Vinters 2010; Zhang et al., 2010). When acute or chronic injury occurs, astrocytes become “reactive” and migrate to the site of insult, change morphology, and up-regulate glial fibrillary acidic protein (GFAP) and cytokines (Sofroniew and Vinters, 2010).
Reactive gliosis is broadly characterized by an increase in expression of the cytoskeletal protein GFAP, activation of the transcription factor NF-κB, and the production of nitric oxide and inflammatory cytokines such as TNFα (Pekny et al., 2005; Kang et al., 2011). In stroke and traumatic injury to the CNS, reactive astrocytes form a protective barrier (glial scar) that compartmentalizes the damaged tissue, effectively separating it from the uninjured CNS (Sofroniew, 2009). Although a glial scar can be detrimental to the regeneration of damaged axons, it also prevents inflammatory cytokines, excess glutamate and other potentially harmful molecules from the damaged tissue from entering the healthy CNS. Despite these beneficial effects, the lasting nature of the glial scar makes it detrimental to long-term regeneration and recovery. Chronic inflammation observed in neurodegenerative disorders such as Alzheimer’s disease (AD) also involves reactive gliosis (Pike et al., 1994; Serrano-Pozo et al., 2011).
Pro-inflammatory stimuli such as lipopolysaccharide (LPS), an endotoxin component of gram-negative bacterial cell membranes, engage an innate immune response through TLR4 receptors, resulting in activation of astrocytes (Sofroniew et al., 2009b; Tang et al., 2007; Okun et al., 2012). TNFα is produced by astrocytes exposed to LPS (Chung et al., 1990); activation of NF-κB plays a pivotal role in the activation of astrocytes and their production of TNFα and other pro-inflammatory cytokines (O’Neill et al., 1997; Mattson et al., 2001). NF-κB signaling has important functions in all CNS cells including neurons, astrocytes and microglia (O’Neill et al., 1997; Bethea et al., 1998; John et al., 2003; Mattson et al., 2006)
Recent studies have demonstrated prominent roles for miRNAs in the activation of immune cells and the pathogenesis of a range of inflammatory disorders (Contreras and Rao, 2012). Depending upon the mRNAs they target, miRNAs may either promote or suppress inflammation (Boldin and Baltimore, 2012). The miR-181 family of miRNAs is expressed in astrocytes and is a candidate for post-transcriptional regulation of phenotypes found in neuroinflammation and reactive gliosis. miR-181 directly targets the mRNA encoding Bcl-2, a mitochondrial membrane-associated protein that inhibits apoptosis, and other mRNAs encoding proteins involved in responses to stress including heat-shock protein 70 (Chen et al., 2010; Ouyang et al., 2011a, b). miR-181 expression decreases with distance from the core of an ischemic insult in a rodent model of focal ischemic stroke (Ouyang et al., 2011b). This miRNA family also regulates differentiation in hematopoietic stem cells towards a CD44+ fate, and maintains ‘stemness’ through a feedback loop with let-7 (Chen et al., 2004; Li et al., 2012). miR-181 is also involved in glioblastoma, with expression inversely correlating with tumor grade; over-expression of miR-181a and b inhibits tumor cell proliferation and sensitizes tumor cells to radiation (Conti et al., 2009; Shi et al., 2008; Chen et al., 2010). Activation of Stat3, an important component of the NF-κB signaling pathway, has also been shown to increase miR-181 expression (Iliopoulos et al., 2010). Lastly, miR-181 targets Nemo-like kinase (NLK), a MAPK-like protein and repressor of Notch signaling which has been implicated in cellular stress responses in prostate tumors (Cichocki et al., 2011; Emami et al., 2009). In the present study we tested the hypothesis that the miR-181 family plays a role in the reactive phenotype of astrocytes in experimental models of neuroinflammation.
Materials and Methods
Cell Culture, Transfection and LPS Treatment
For mouse primary astrocyte cultures, the cerebral cortex was removed from the brains of newborn mice and placed in cold Hank’s balanced saline solution (HBSS). After removal of the meninges, cells of the cortical tissue were dissociated mechanically before being pelleted by centrifugation at 500 g for 5 minutes, washed with HBSS and re-pelleted. Cells were then re-suspended in Dulbecco’s minimum essential medium (DMEM) containing Anti-Anti (Invitrogen) and supplemented with 10% fetal calf serum (FCS) (Invitrogen). Cells were then passed through a 40-μm cell strainer mesh (BD Falcon) to obtain a single-cell suspension and plated at a density of 106 cells per ml on uncoated cell culture plates. Astrocyte cultures were passaged once at 106 cells per ml prior to experimentation to ensure greater culture purity.
For experiments involving transfection, Lipofectamine LTX (Invitrogen) was used to transfect cells to over-express (OE) either miR-181c or a control miRNA (the C. Elegans miR-67; CTRL), or to knock down (KD) miR-181 using miR-181c and miR-181b microRNA hairpin inhibitors (Dharmacon). All oligonucleotides were transfected at a final concentration of 20 nM. For KD microarray and validation experiments, samples were harvested 48 h after transfection to allow for degradation of miRNA targets. For pull-down (PD) microarray and validation experiments, samples were harvested 24 h after transfection.
Lactate dehydrogenase (LDH) assays (Promega) were used to quantify cell death. Medium from astrocyte cultures was collected 48 h after transfection and 6 hours after LPS treatment, and centrifuged at 600×g to remove any floating cell bodies and debris prior to analysis of LDH levels which was performed according to the manufacturer’s protocol. Reactions were quantified using a BioRad (Hercules, CA) microplate reader with a 492 nm excitation wavelength filter and normalized as a percent value to control wells in which cells were lysed with 0.01% Triton-X (maximum LDH release).
To assess cell proliferation, a colorimetric assay using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS; Promega Cell Titer 96AQueous One) was performed on cells 6 hours after treatment with LPS or vehicle control (54 hours after transfection). A stock solution of MTS reagent was added to each well and measured between 2–4 hours at 490 nm, according to the manufacturer’s protocol.
In-vivo LPS Treatment of TNF-α Receptor KO and Wild-Type Mice
All animal treatments were described previously (Kawamoto et al., 2012). Briefly, either adult 12 to 14 week-old male wild type (WT) C57BL/6J mice or TNFR1/TNFR2 double knockout (DKO) mice were maintained in a 12 h light/dark cycle with free access to food and water (DKO mice and methods for their breeding and genotyping are described in Bruce et al., 1996 and Kawamoto et al., 2012). LPS injections were administered between 08:00 and 11:00, with animals given a single intraperitoneal (IP) injection of LPS (0111:B4, Sigma-Aldrich) at a dose of 250 μg/kg. Mice were euthanized 4 h after LPS injection and dissection of brain regions was performed on ice. Guidelines established in the NIH Guide for Care and Use of Laboratory Animals were followed, and all procedures were approved by the Animal Care and Use Committee of the National Institute on Aging Intramural Research Program.
Preparation of Samples for qPCR, Gene Array and Gene Array Analysis
Total RNA was obtained from both astrocyte cultures and mouse cortex using Trizol reagent (Invitrogen), according to the manufacturer’s instructions. For validation of gene array results we used reverse transcription (RT) using random hexamers and SSIII reverse transcriptase (Invitrogen) to convert mRNA into cDNA, followed by RT-qPCR using SYBR Green master mix (Invitrogen). RT-qPCR reactions were performed using a MJ Research PTC-200 instrument and analyzed using the relative method with appropriate primers (see Supplemental Table 1 for primer sequences). The mRNAs expressing ‘housekeeping’ proteins HPRT, GAPDH and BACT were measured for normalization of samples before comparison to the control condition.
For assessment of miRNA expression, samples were prepared using Invitrogen’s NCODE miRNA qPCR assay and samples run using the manufacturer’s protocol. Samples were first polyadenylated prior to RT using Invitrogen’s Universal Primer followed by RT-qPCR using SYBR Green master mix (Invitrogen). The relative method of qPCR was used with samples first normalized to U6 before comparison to a control condition. For all miRNA experiments a C. elegans miR-67 mimic (Dharmacon), a miRNA with no known mammalian targets, was used.
Gene Array Methods and Data Analysis
RNA quality was assessed with an Agilent BioAnalyzer using RNA 6000 Nano Chips (Agilent Technologies, Santa Clara, CA). Transcriptional profiling was determined using Illumina Sentrix BeadChips. Total RNA was used to generate biotin-labeled cRNA with the Illumina TotalPrep RNA Amplification Kit. In short, 0.5 µg of RNA was first converted into single-stranded cDNA with reverse transcriptase using an oligo-dT primer containing the T7 RNA polymerase promoter site and then copied to produce double-stranded cDNA molecules. The double-stranded cDNA was cleaned, concentrated, and in vitro transcribed to generate single-stranded RNA (cRNA) containing biotin-16-UTP. A total of 0.75 µg of biotin-labeled cRNA was hybridized at 58 C for 16 hours to Illumina's Sentrix Human Ref-8 Expression BeadChips (Illumina, San Diego, CA). Each BeadChip has ~24,000 well-annotated RefSeq transcripts with approximately 30-fold redundancy. The arrays were washed and blocked, and the labeled cRNA was detected by staining with streptavidin-Cy3. Hybridized arrays were scanned using an Illumina BeadStation 500X Genetic Analysis Systems scanner and the image data extracted using Illumina’s GenomeStudio software, version 1.1.1.1. For statistical analysis, the expression data were filtered to include only probes with a consistent Illumina detection signal (p < 0.02).
Correlation analysis, sample clustering analysis and principal component analysis that included all of the probes was performed to identify/exclude any possible outliers. The resulting dataset was next analyzed with DIANE 6.0, a spreadsheet-based microarray analysis program using value statistics for Z-Score reliability below 0.05, and mean background-corrected signal intensity greater than zero. Ingenuity’s Pathway analysis (Ingenuity) was used to analyze both the KD and PD gene arrays using methods described previously (Lal et al., 2011). Input data included gene symbols, z-score, false discovery rate (FDR), and fold change.
Biotinylated Pull-Down of miR-181c Targets
Primary astrocytes were cultured in 6-well plates and transfected with either bi-miR-181c or bi-Cel-miR-67 (Dharmacon) as described above. Cells were detached using 0.025% Trypsin/1 mM EDTA solution 24 h after transfection, pelleted at 600×g and washed twice with PBS prior to re-suspension in 500 µl lysis buffer (100 mM KCl, 5 mM MgCl2, 20 mM Tris; pH 7.5). Lysis buffer was then added including 0.3% NP-40 (final concentration) and 50 U of RNAse OUT (Invitrogen) and cells were incubated on ice for 10 minutes. Cellular lysate was isolated by centrifugation at 10,000×g for 20 minutes at 4 C. Streptavidin-coated dynabeads (Invitrogen) were prepared according to protocol and then blocked for 2 h at 4 C in lysis buffer containing 1 mg/ml yeast tRNA. Following blocking, beads were washed 3 times in lysis buffer, and sample lysate was added and incubated overnight at 4 °C with agitation. Samples were then washed five times with 1 ml lysis buffer to remove unbound mRNAs. Both input RNA (10% of the lysate before hybridizing with the beads) and RNA bound to the beads (pull-down) were isolated with Trizol reagent (Invitrogen). Pull-down samples were then subjected to either gene array or RT-qPCR analysis.
Measurement of Protein from Cellular Lysate and Supernatant
To assess cytokine levels after altering miR-181 expression and treatment with LPS, astrocyte supernatant was collected 48 h after transfection and analyzed using Bio-Plex Cytokine assays according to the provided protocol (BioRad). Cytokines assessed were IL6, IL1β, FGF2, LIF, IL-10, IL-8, HMGB1 and TNFα. Multiplex assays were also performed for IL-5, IL-4, IL-13, IFN-γ, IL-12p70, IL-7, IL-2 and SAA, but all were below the detection threshold.
Heterologous Reporter Assay for Validation of miRNA Targets
To assess the direct interaction of the miR-181 family with XIAP, MeCP2 and SIRT1 mRNAs, heterologous luciferase reporters were prepared (Supplemental Table 2). A ~500-bp region spanning the predicted miRNA target site was cloned into the 3’ end of the Renilla luciferase (RL) reporter in the psiCHECK-2 plasmid (see Supplementary Table 2 for primer sequences and Figure 7 for miRNA binding sites). A second reporter protein, firefly luciferase (FL), is also expressed from the psiCHECK-2 plasmid under a separate promoter for use as an internal transfection control. Control reporters in which 2 or 3 nucleotides within the seed region of the miRNA binding sites were mutated using SDM were also constructed. Ratios of RL/FL luminescence in transfected 293 cells were measured to quantify miR-181 mediated repression of the target transcript.
Results
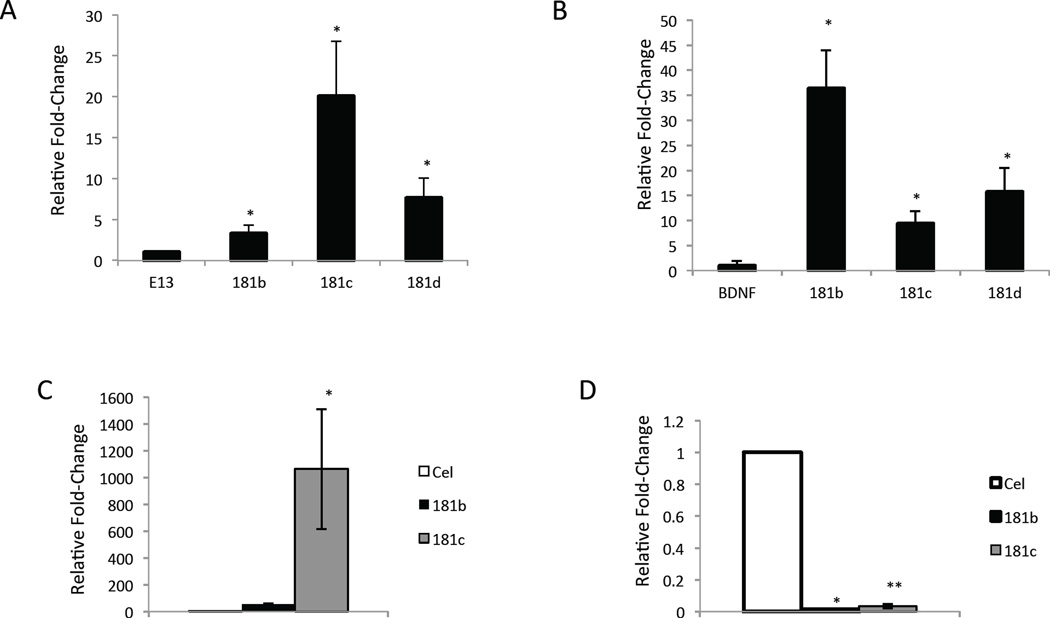
We first investigated the developmental specificity of miR-181 expression by comparing the levels of miR-181b, c and d in cerebral cortical tissue from mice at different developmental stages from embryonic day 13 (E13) to adult (Figure 1A). All three of these miR-181 family members were expressed at significantly higher levels in adult cortex relative to embryonic telencephalon. We next examined miR-181 expression in cultured astrocytes compared to E13 neural stem cells (NSCs) differentiated into neurons during a two-week culture period in the absence of fibroblast growth factor 2 (FGF2) and epidermal growth factor (EGF), and in the presence of brain-derived neurotrophic factor (BDNF) (Figure 1B). miR-181 family members were strongly enriched in cultured astrocytes relative to BDNF-differentiated neurons. In anticipation of subsequent experiments, we determined the efficacy of miR-181c overexpression (OE), or knockdown of miR-181b and miR-181c, on levels of these miRNAs in cultured cortical astrocytes. miR-181c was significantly elevated in astrocytes transfected with miR-181c (Figure 1C). Levels of miR-181b and miR-181c were significantly reduced in astrocytes transfected with miRNA hairpin inhibitors against these miRNAs (Figure 1D).
Figure 1. miR-181s are enriched in astrocytes, modify cell proliferation and vulnerability to degeneration in a model of neuroinflammation.
A. Expression levels of miR-181b-d in E13 telencephalon versus adult cortex (181b p<0.04, 181c p<0.02, 181d p<0.03, n=4). B. Expression of miR-181b-d in cultured astrocytes compared to BDNF-differentiated neurons (181b p<0.03, 181c p<0.01, 181d p<0.03, n=4). C. Levels of miR-181 b and c in cultured astrocytes 24 hours after transfection with a mirR-181c mimic (181c p<0.04, n=3). D. Levels of miR-181b and miR-181c in cultured astrocytes 48 hours after transfection with miR-181c and miR-181b microRNA hairpin inhibitors (181b p<0.04, 181c p<0.00002, n=4).
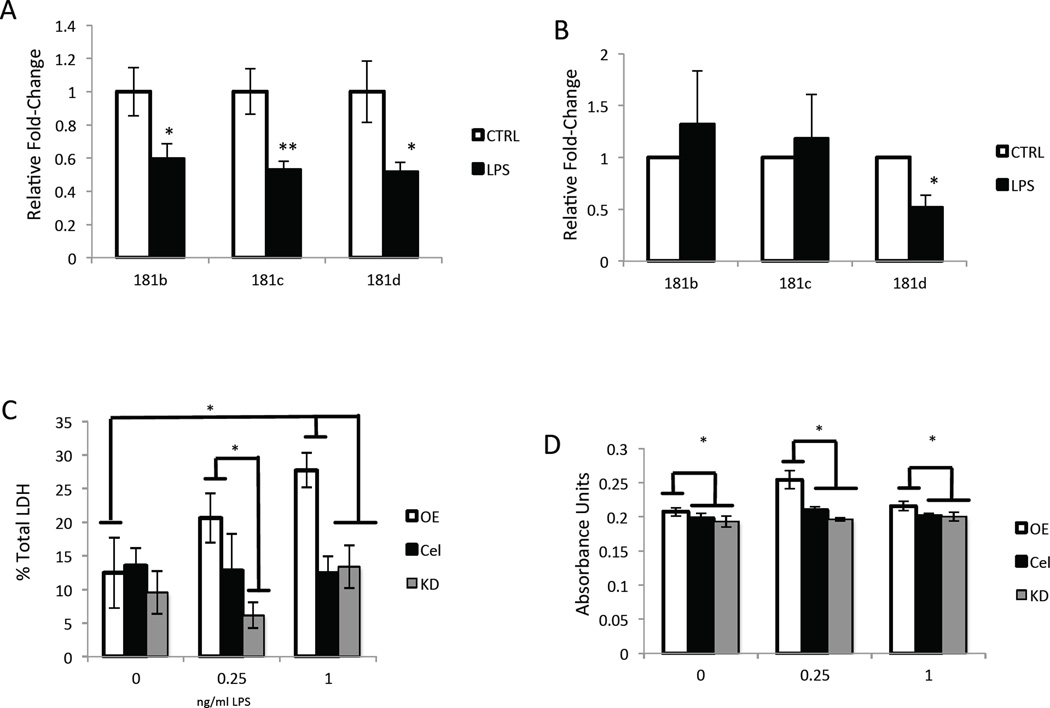
To explore the relationship between miR-181 expression and inflammatory responses, cortex samples from wild-type animals injected with either LPS or saline were examined for miR-181 expression. We found that expression of miR-181b, miR-181c and miR-181d were significantly decreased 4 hours after LPS injection (Figure 2A), a time point when TNFα production was increased in cortex (Kawamoto et al., 2012). To determine if TNFα signaling played a role in the observed decrease in miR-181 expression, we injected TNFR1/TNFR2 double knockout (DKO) mice with LPS and examined miR-181 expression in cortex. In contrast to WT mice, LPS did not cause a significant reduction of miR-181b and miR-181c in DKO mice (Figure 2B). However, there was a significant decrease of miR-181d levels in response to LPS in DKO mice (p<0.04; Figure 2B). OE of miR-181c resulted in increased cell death as measured by LDH release, regardless of LPS concentration or control treatment (Figure 2C). Interestingly, a small but significant increase in cellular proliferation as measured by MTS was observed in astrocytes over-expressing miR-181c relative to control, regardless of LPS concentration (Figure 2D).
Figure 2. Decline in R-181 levels following exposure to LPS in the cerebral cortex of mice in vivo and in cultured astrocytes.
A. miR-181b-d expression in cerebral cortex from LPS-treated wild-type mice versus vehicle-treated control mice, 4 h after LPS administration (181b p<0.02, 181c p<0.007, 181d p<0.02, n=4). B. miR-181b-d expression in cortex from LPS treated TNFα receptor knockout mice, 4 h after IP injection of LPS (181d p<0.04, n=3). C. Effects of miR-181 overexpression (OE) or knockdown (KD) on the vulnerability of cultured astrocytes to cell death caused by exposure to LPS, as measured by LDH levels in the culture medium 48 h after transfection and 6 h after LPS treatment (1 μg/ml LPS: OE p<0.03, OEvKD p<0.05; 1 μg vs. Basal: OE p<0.04, n=6; 250 ng/ml LPS: OEvKD p<0.02, n=4). D. In cultured astrocytes 48 h after miR-181 OE or miR-181 KD, followed by a 6 h LPS treatment, the relative levels of cellular metabolic activity were measured using an MTS reduction assay (1 μg/ml: n=8, OEvCTRL p<0.02; OEvKD p<0.01, OEVNoLPSOE p<0.04; 250 ng/ml: OEvCTRL p<0.03, KDvsCTRL p<0.01, OEvKD p<0.007, OEvNoLPSOE p<0.01, CTRLVSNoLPSCTRL p<0.04, n=4; 0 mg LPS: OEvCTRL p<0.003, OEvKD p<0.05, n=4).
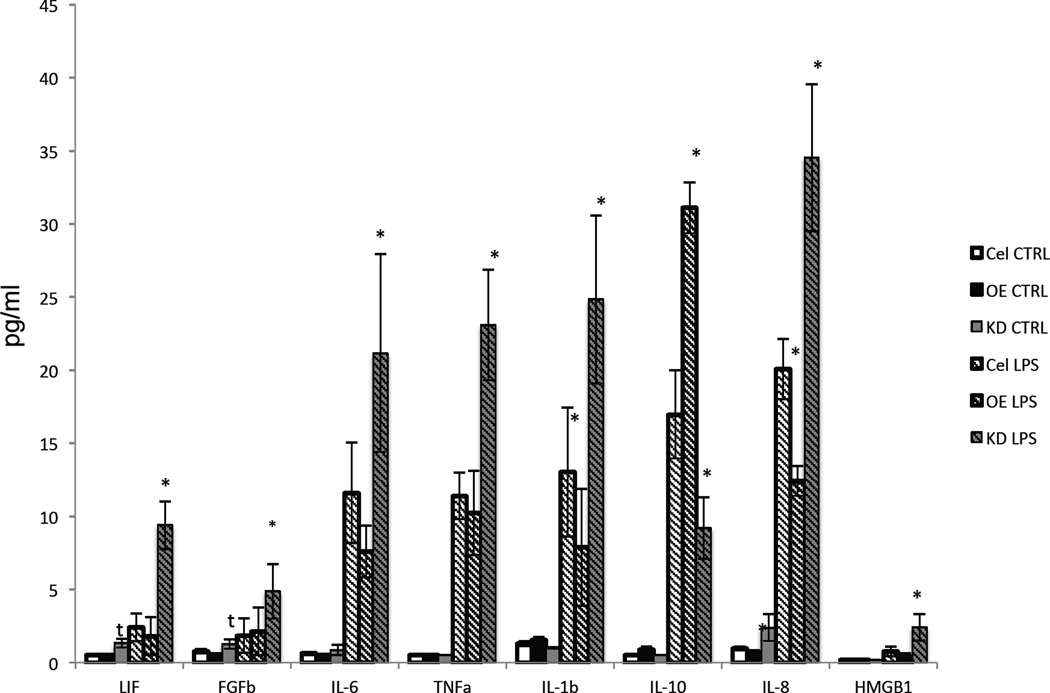
To ascertain the effects of reducing or over-expressing miR-181 on cytokine and growth factor production, we performed multiplex immunoassays for factors relevant to a reactive astrocyte phenotype, including LIF, FGF2, IL-6, TNF-α, IL-1β, IL-10, IL-8 and HMGB1 (Figure 3). Overexpression of miR-181c had no significant effect on LIF protein levels in control or LPS-treated astrocytes, whereas knockdown of miR-181b and miR-18c resulted in significant elevation of LIF levels in both control and LPS-treated astrocytes. Knockdown of miR-181s resulted in a trend towards elevated FGF2 levels in control astrocytes, and a significant elevation of FGF2 levels in LPS-treated astrocytes. A prominent result of this experiment was that miR-181 knockdown significantly enhanced the LPS-induced production of three different pro-inflammatory cytokines, TNF-α, IL-6 and IL-1β (Figure 3). In the case of the anti-inflammatory cytokine IL-10, overexpression of miR-181c enhanced the LPS-induced increase in IL-10 levels, whereas knockdown of miR-181s inhibited the LPS-induced production of IL-10. Finally, knockdown of miR-181s significantly enhanced the LPS-induced production of HMGB1 (Figure 3). These results suggest that miR-181s negatively regulate the production of multiple cytokines and FGF2 in astrocytes, effectively suppressing their production in an inflammatory environment.
Figure 3. Reduction of miR-181 levels alters both basal and LPS-induced cytokine and FGF2 production.
A. Multiplex immunoassay was used to detect levels of LIF (basal: KD p<0.055, OEvKD p<0.055; LPS: KD p<0.02, LPS KDvOE p<0.002; LPS vs. Basal: KD p<0.01), FGF2 (Basal: KD p<0.066, KDvOE p<0.07; LPS: KD p<0.02, KDvOE p<0.004), IL-6 (LPS: KD p<0.02, KDvOE p<0.004), TNF-α (LPS: KD p<0.02, KDvOE p<0.02; LPS vs Basal: OE p<0.04, CTRL p<0.01, KD p<0.01), IL-1β (LPS: OE p<0.03, KD p<0.006, KDvOE p<0.01; LPS vs Basal: CTRL p<0.057, KD p<0.02), IL-10 (LPS: OE p<0.02, KD p<0.007, OEvKD p<0.007; LPS vs. Basal: OE p<0.002, CTRL p<0.01, KD p<0.03), IL-8 (LPS: OE p<0.02, KD p<0.05, KDvOE p<0.02; Basal OE p<0.005; LPS vs Basal: OE p<0.003, CTRL p<0.004, KD p<0.01) and HMGB1 (LPS: KDvOE p<0.05, KDvCTRL p<0.06) in media from cultured astrocytes that had been treated with either 1 µg/ml LPS or vehicle (n = 3).
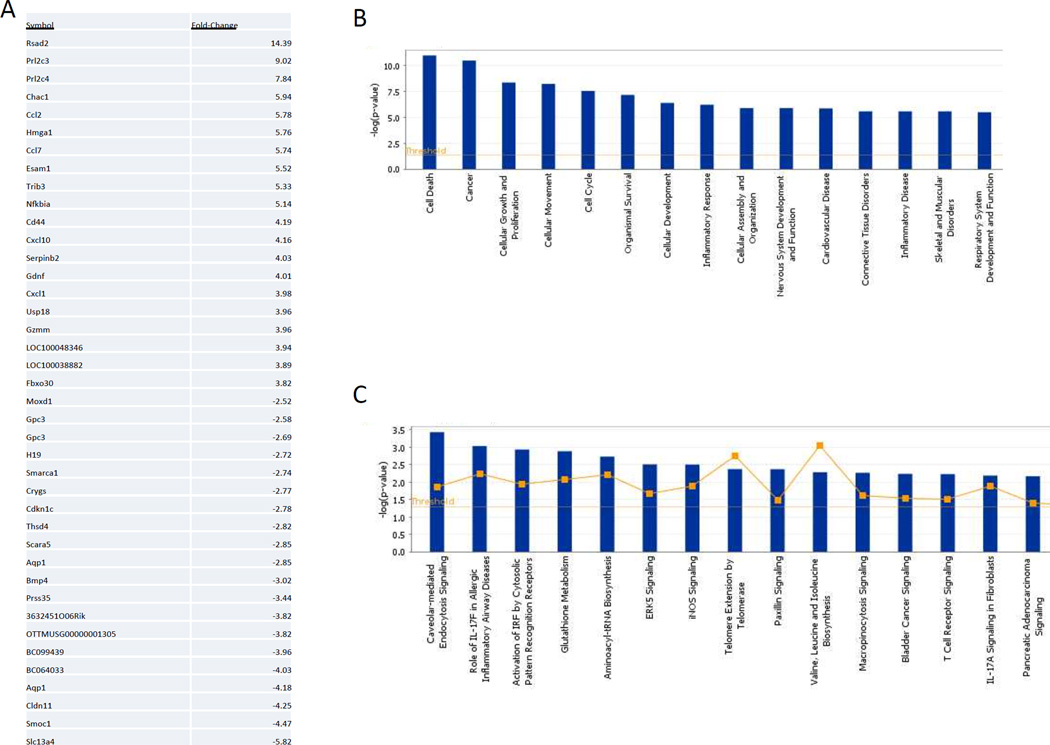
The effects of miR-181 on the transcriptome were assessed by microarray analysis of primary astrocytes in which miR-181b and c were knocked down (Figure 4, Supplemental Table 3). Pathway and signaling pathway analysis identified alterations of several major pathways related to cell death, cancer and the inflammatory responses (Figure 4B, 4C and Supplemental Table 4). These results were validated by qPCR analysis of a set of mRNAs of interest identified in the knockdown array (Figure 5).
Figure 4. Reduction of miR-181 expression alters levels of transcripts involved in cell death and inflammation responses.
A. A list of the twenty transcripts that increased by the greatest magnitude, and the twenty transcripts that decreased by the greatest magnitude in response to simultaneous knockdown of miR-181b and miR-181c. B. Pathway analysis of transcriptional changes demonstrated that alterations related to cell death, cell cycle regulation, cell differentiation, cancer and inflammatory responses, and related signaling pathways were affected. C. Signaling pathways of interest altered by miR-181b and c knock-down included iNOS signaling.
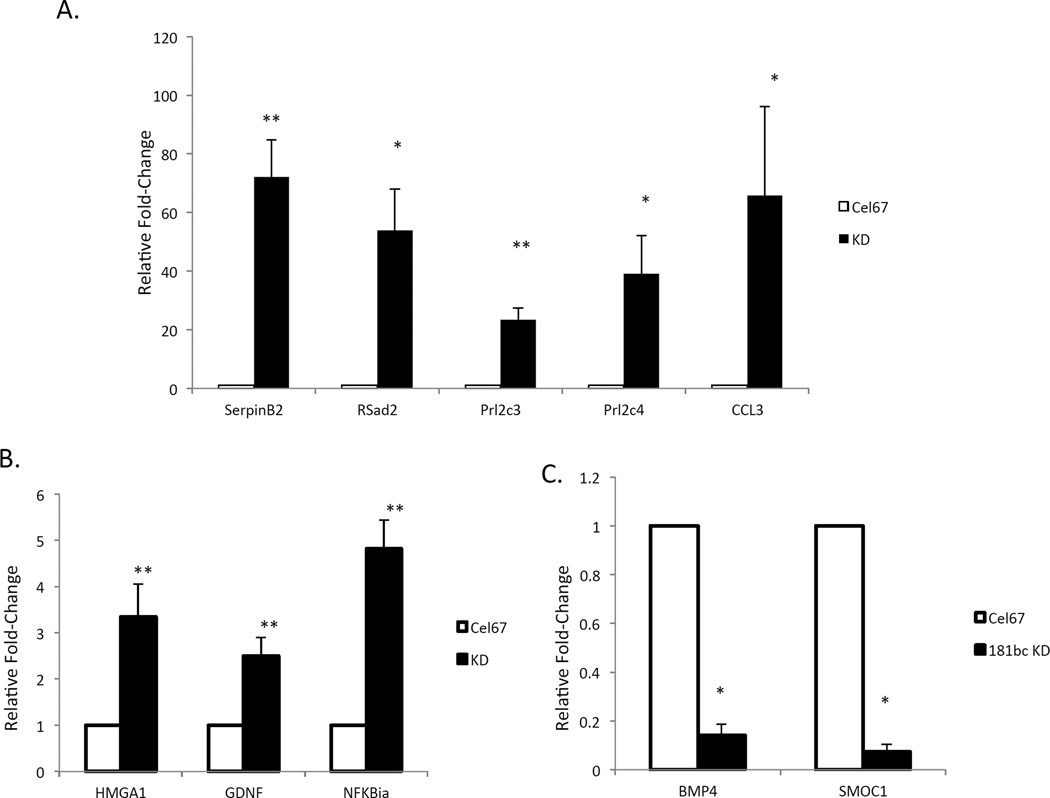
Figure 5. Evidence that miR-181 regulates diverse signaling pathways in astrocytes.
A. The mRNAs encoding SerpinB2 (p<0.007, n=4), RSad2 (p<0.02, n=4), Prl2c3 (p<0.007, n=4), Prl2c4 (p<0.03, n=4) and CCL3 (p<0.04, n=4) increased significantly after reducing miR-181b and c expression in cultured astrocytes. B. Increased levels of mRNAs encoding HMGA1 (p<0.007, n=4), GDNF (p<0.004, n=3) and NFKBia (p<0.008, n=4) after silencing miR-181b/c. C. Lower levels of BMP4 (p<0.05, n=4) and SMOC1 (p<0.04, n=4) mRNAs in astrocytes in which miR-181b/c levels were reduced.
To more directly assess potential targets of miR-181c in primary mouse astrocytes, we performed microarray analysis of transcripts preferentially enriched in a biotinylated miR-181c probe pulldown assay relative to a control C. Elegans-67 probe (Figure 6 and Supplemental Table 5). Function and signaling pathway analysis demonstrated an enrichment of transcripts involved in diverse functions including cell death, mTOR signaling and mitochondrial dysfunction (Figure 6B and 6C, and Supplemental Table 6). We examined several transcripts that overlapped between the pull-down and knock-down arrays by qPCR and found that HMGA1 mRNA positively associates with the miR-181c probe (Figure 7A). While not predicted by Targetscan, RNAhybrid analysis (Rehmsmeier et al., 2004) demonstrates extensive pairing between the mature miR-181b and a segment on the 5’ end of the HMGA1 mRNA. Bcl-2, a previously validated miR-181 target (Ouyang et al., 2012a) and the predicted target NAMPT mRNA were also enriched in the pull-down (Figure 7A). Reporter constructs for MeCP2, SIRT1 and XIAP were generated to assess if these were direct targets of miR-181 (Figure 7B). The SIRT1 reporter demonstrated no repression by miR-181c (not shown). The predicted MeCP2 binding site was validated as a miR-181 target, as the Psi-CHECK2 MeCP2 reporter showed a 72% reduction in light units (Figure 7C). The XIAP predicted binding site was also validated as a miR-181 target, with a roughly 63% decrease in relative light units for the Psi-CHECK2 XIAP reporter (Figure 7D).
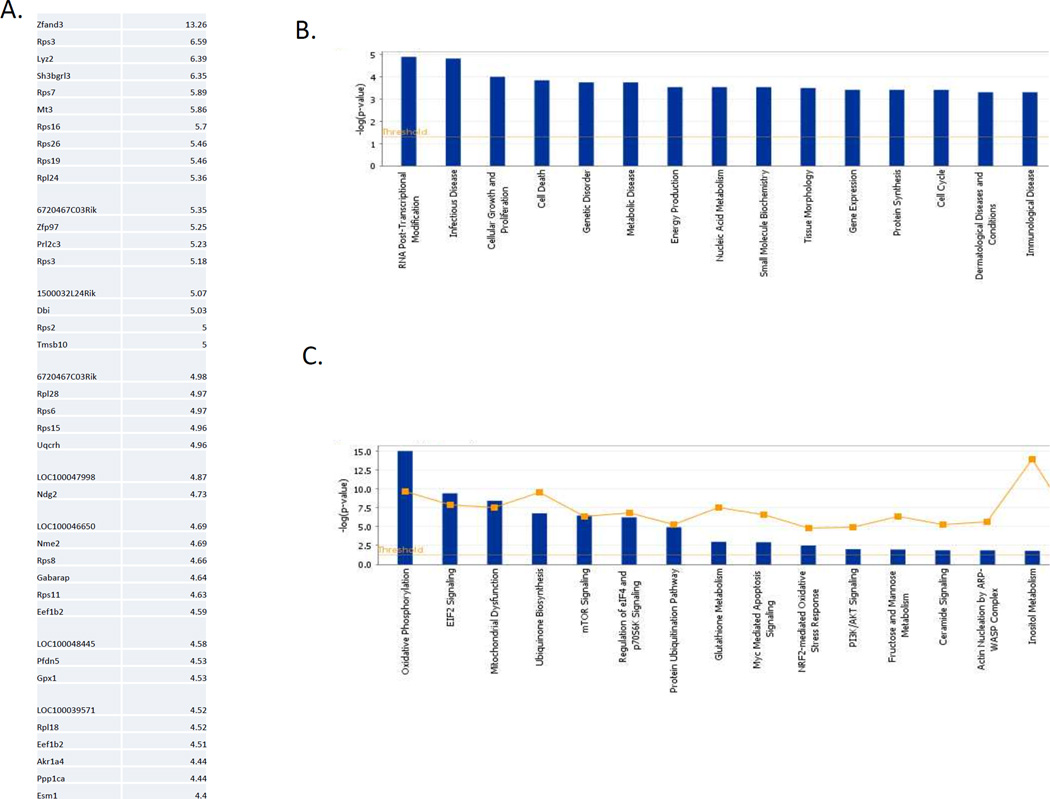
Figure 6. Direct interaction of miR-181 with transcriptome.
A. Top 40 most highly enriched transcripts after pull-down of biotinylated miR-181c. B. Pathway analysis indicating that transcripts that interact with miR-181c encode proteins implicated controlling cell death. C. Putative miR-181c targets include proteins implicated in EIF2 signaling, mTOR signaling and mitochondrial dysfunction.
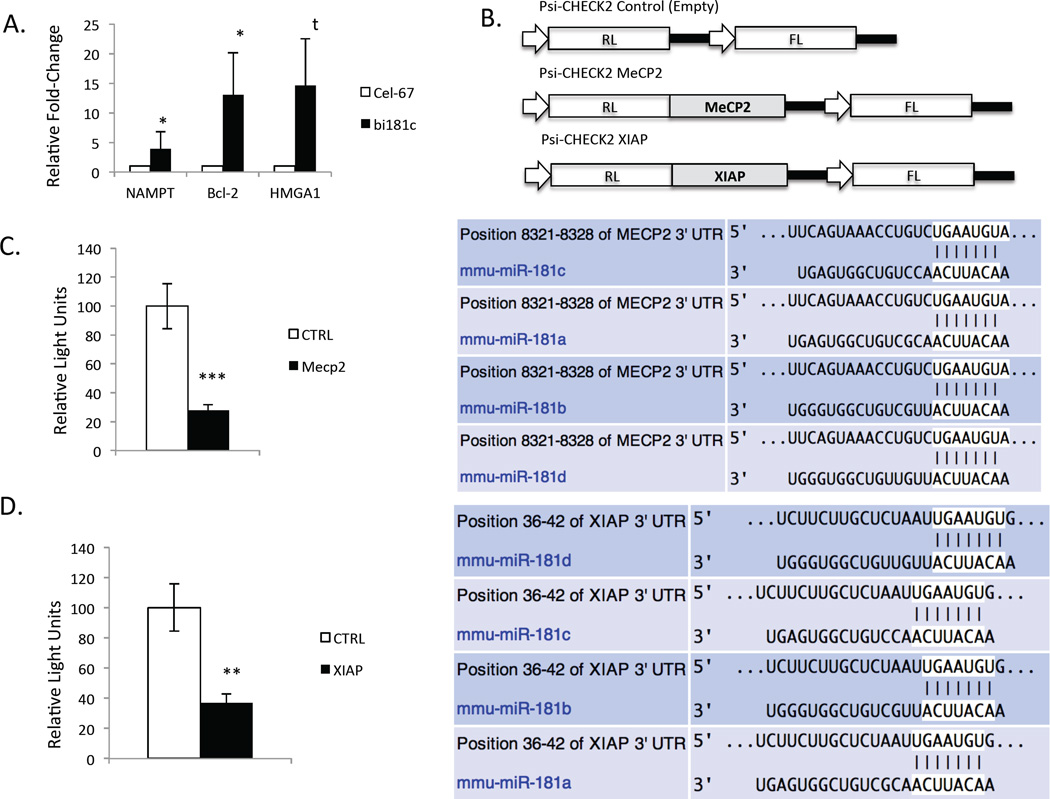
Figure 7. MeCP2 and XIAP mRNAs are direct targets of miR-181.
A. NAMPT, Bcl-2 and HMGA1 mRNAs are all enriched in biotin pull-down of miR-181c (HMGA1 p<0.08, n=3, Bcl-2 p<0.02, n=4, NAMPT p<0.07, n=4). B. Schematic representation of Psi-CHECK2 reporter constructs generated to test putative miR-181 targets MeCP2 and XIAP mRNAs. C. Analysis of Psi-CHECK2 MeCP2 reporter activity 48 h after miR-181 OE (p<0.0004, n=6). D. Analysis of Psi-CHECK2 XIAP reporter activity 48 h after miR-181 OE (p<0.001, n=6).
Discussion
We found that miR-181 expression is developmentally regulated and that miR-181s are enriched in astrocytes compared to neurons. It was previously reported that miR-181s play roles in cell fate specification in the hematopoietic system and developmental patterning of muscle cell fate (Chen et al., 2004; Naguibneva et al., 2006). Expression of miR-181 is also implicated in the differentiation of stem cells by repressing Lin28 (Li et al., 2011). Taken together with our observation that miR-181b, c and d are present at high levels in the mature CNS, but not early in development prior to astrogenesis, these data support a possible role for miR-181 in developmental specification of cell fate in the CNS and warrant further investigation.
Our data suggest that in the adult brain miR-181 expression is dynamically regulated by inflammatory stimuli and modifies the proliferation of astrocytes and their sensitivity to death in an experimental model of neuroinflammation. Previous studies have suggested roles for miR-181s in modifying the sensitivity of cells to death in ischemic brain injury and glioblastoma, with particular emphasis on its regulation of the Bcl-2 transcript (Chen et al., 2010; Ouyang et al., 2011a). It should be noted that miR-181 regulates other transcripts involved in cell survival and responses to stress including hsp-70, MCL-1 and Bim (Ouyang et al., 2011a, 2011b). We found that an inflammatory stimulus, the TLR4 receptor ligand LPS, decreased the levels of the miR-181s in vivo, suggesting that the previously observed alteration in miR-181 expression in ischemic stroke models could be due to a TNF-α–independent component of the injury. Our results therefore suggest that miR-181s are involved in a general cell stress response and cell survival-related changes in transcriptional profiles.
The most striking phenotype that we observed as a consequence of miR-181 reduction was a dramatic increase in the production of pro-inflammatory cytokines including LIF, IL-6, TNF-α, IL-1β, IL-8 and HMGB1. On the other hand, we found that levels of the anti-inflammatory cytokine IL-10 (Murray, 2005; de Vries, 1995) were increased in astrocytes when miR-181 was over-expressed. This result suggests that miR-181 might play an anti-inflammatory role when expressed while reduction of miR-181, as observed in-vivo in animals exposed to LPS, might contribute to the production of inflammatory cytokines. We also found that levels of FGF2, a potent mitogen for astrocytes (Joy et al. 1997), were also increased as a result of knocking down miR-181 and exposing astrocytes to LPS. FGF2 is also known to protect neurons against conditions associated with inflammation including oxidative stress and excitotoxicity (Mattson et al., 1989; Mark et al., 1997), suggesting that suppression of FGF2 production by miR-181s could render neurons vulnerable to such adverse conditions.
We have identified two novel targets of miR-181, MeCP2 and XIAP mRNAs, as well as a non-canonical potential target, HMGA1. HMGA1 is typically restricted to embryonic tissues and is frequently observed in both benign and malignant tumors (see Cleynen et al., 2008 for a review). HMGA1 is a transcription factor that binds to AT-rich regions of the genome, and increased HMGA1 expression is typically observed in glioblastoma, with expression levels increasing with tumor grade (Donato et al., 2004; Fan et al., 2011; Fang et al., 2012). It was previously demonstrated that HMGA1 and HMGA2 are subject to repressive regulation through elements in the 3’UTR by an as-yet unknown mechanism (Borrmann et al., 2001). HMGA1 has been observed to enhance NF-κB binding, as in the case of the IFN1-γ promoter (Thanos et al., 1992). Particularly relevant to the role of miR-181 repression of HMGA1 in the inflammatory response, HMGA1 has been shown to promote an inflammatory transcriptional profile, at least in part by increasing Cox-2 and STAT-3 levels (Resar, 2010 for a review; Schuldenfrei et al., 2011). HMGA1 activation of STAT3, which results in an increase of miR-181 expression, could represent an important feedback loop to regulate expression of HMGA1 in adult tissues (Hillion et al., 2008).
MeCP2, the methyl CpG-binding protein 2, is frequently observed in neurons and plays an important role in the neurodegenerative disorder Rett Syndrome (Jung et al., 2003). Although MeCP2 is normally only observed in adult neurons, recent evidence has suggested that astrocytes from MeCP2-deficient mice have abnormalities that alter neuronal morphology, and that rescuing MeCP2 expression in astrocytes leads to a partial recovery in Rett Syndrome mouse models (Ballas et al., 2009; Lioy et al., 2011). In the present study we validated MeCP2 mRNA as a direct target of miR-181 by reporter assay. miR-181 repression of MeCP2 in astrocytes could therefore play a physiological role in reactive gliosis, the developmental differentiation of astrocytes, and possibly Rett Syndrome.
The mRNA encoding X-linked inhibitor of apoptosis (XIAP) is another miR-181 target that we validated by reporter assay. XIAP is a member of the Inhibitors of apoptosis (IAP) family of proteins and acts by binding to and inactivating caspases 3, 7 and 9 (Eckelman et al., 2006). XIAP inhibits TNF-α-induced cell death and its expression is regulated by NF-κB (Chu et al., 1997; Rothe et al., 1995; Stehlik et al., 1998). Other studies have demonstrated that miR-181 represses important antiapoptotic targets such as Bcl-2 and stress-induced proteins such as hsp70, the addition of XIAP as a miR-181 target could help explain the pro-apoptotic effects of this miRNA family. Additionally, we have demonstrated that miR-181 is regulated by inflammatory signaling through the innate immune system receptor TLR4, and the reduction of miR-181 levels as a response to inflammatory signaling could be a pro-survival mechanism. Taken together with the other confirmed targets of the miR-181 family, our findings argue that miR-181s are fundamentally involved in major programs of posttranscriptional regulation of cell differentiation and stress responses in astrocytes, with increasing miR-181 expression resulting in increased differentiation and vulnerability to environmental stress.
Several other potential miR-181 targets were identified in the knockdown/microarray and/or biotinylated miR-181 pulldown experiments that are known to play prominent roles in the responses of brain cells to injury and stress. These include glial cell line-derived neurotrophic factor (GDNF) and vascular endothelial cell growth factor (VEGF), two proteins secreted from astrocytes in response to metabolic and oxidative stress. GDNF can protect dopaminergic neurons against oxidative and metabolic stress in experimental models of Parkinson’s disease (Ramaswamy et al., 2009). VEGF stimulates cerebral angiogenesis and neurogenesis (MacKenzie and Ruhrberg, 2012). By enabling the production of GDNF and VEGF, a reduction in miR-181s might therefore be expected to reduce neuronal vulnerability and enhance neurogenesis. Four other mRNAs identified as potential miR-181 targets in the pulldown array analysis encode antioxidant enzymes: glutathione peroxidases 1 and 4 (Gpx1, Gpx4); peroxiredoxin 2 (Prdx2); and a presumptive glutaredoxin (SH3BGRL3; Mazzocco et al., 2001). When taken together with the regulation of proteins involved in inflammation, the latter findings suggest that miR-181s regulate cellular redox status in astrocytes. This possibility is intriguing because of the well-established concomitant production of pro-inflammatory cytokines and reactive oxygen species in reactive microglia and macrophages (Teismann and Schulz, 2004). Our findings therefore suggest that miR-181s may normally suppress the production of a similar collection of oxidative/pro-inflammatory proteins in astrocytes.
Supplementary Material
Acknowledgement
This research was supported by the Intramural Research Program of the National Institute on Aging.
Abbreviations
- FGF2
fibroblast growth factor 2
- HMGB1
high-mobility group protein 1
- IL
interleukin
- LIF
leukemia inhibitory factor
- TNF
tumor necrosis factor
- GDNF
glial cell-line derived neurotrophic factor
- MeCP2
methyl CpG binding protein 2
- NAMPT
Nicotinamide phosphoribosyltransferase
- Bcl-2
B-cell lymphoma 2
References
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baune BT, Wiede F, Braun A, Golledge J, Arolt V, Koerner H. Cognitive dysfunction in mice deficient for TNF-α and its receptors. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1056–1064. doi: 10.1002/ajmg.b.30712. [DOI] [PubMed] [Google Scholar]
- Bethea JR, Castro M, Keane RW, Lee TT, Dietrich WD, Yezierski RP. Traumatic spinal cord injury induces nuclear factor-kappaB activation. J. Neurosci. 1998;18:3251–3260. doi: 10.1523/JNEUROSCI.18-09-03251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-κB. Immunol Rev. 2012;246:205–220. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- Bruce AJ, Boling W, Kindy MS, Peschon J, Kraemer PJ, Carpenter MK, Holtsberg FW, Mattson MP. Altered neuronal and microglial responses to excitotoxic and ischemic brain injury in mice lacking TNF receptors. Nat Med. 1996;2:788–794. doi: 10.1038/nm0796-788. [DOI] [PubMed] [Google Scholar]
- Borrmann L, Wilkening S, Bullerdiek J. The expression of HMGA genes is regulated by their 3'UTR. Oncogene. 2001;20:4537–4541. doi: 10.1038/sj.onc.1204577. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chung IY, Benveniste EN. Tumor necrosis factor-alpha production by astrocytes. Induction by lipopolysaccharide, IFN-gamma, and IL-1 beta. J Immunol. 1990;144:2999–3007. [PubMed] [Google Scholar]
- Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kB control. Proc. Natl. Acad. Sci. USA. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions. Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- Conti A, Aguennouz M, La Torre D, Tomasello C, Cardali S, Angileri FF, Maio F, Cama A, Germanò A, Vita G, Tomasello F. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol. 2009;93:325–332. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia. 2012;26:404–413. doi: 10.1038/leu.2011.356. [DOI] [PubMed] [Google Scholar]
- de Vries JE. Immunosuppressive and anti-inflammatory properties of interleukin 10. Ann Med. 1995;27:537–541. doi: 10.3109/07853899509002465. [DOI] [PubMed] [Google Scholar]
- Della-Morte D, Dave KR, DeFazio RA, Bao YC, Raval AP, Perez-Pinzon MA. Resveratrol pretreatment protects rat brain from cerebral ischemic damage via a sirtuin 1-uncoupling protein 2 pathway. Neuroscience. 2009;159:993–1002. doi: 10.1016/j.neuroscience.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato G, Martinez Hoyos J, Amorosi A, Maltese L, Lavano A, Volpentesta G, Signorelli F, Pentimalli F, Pallante P, Ferraro G, Tucci L, Signorelli CD, Viglietto G, Fusco A. High mobility group A1 expression correlates with the histological grade of human glial tumors. Oncol Rep. 2004;11:1209–1213. [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Brown LG, Pitts TE, Sun X, Vessella RL, Corey E. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate. 2009;69:1481–1492. doi: 10.1002/pros.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Guo H, Zhang IY, Liu B, Luan L, Xu S, Hou X, Liu W, Zhang R, Wang X, Pang Q. The different HMGA1 expression of total population of glioblastoma cell line U251 and glioma stem cells isolated from U251. Brain Res. 2011;1384:9–14. doi: 10.1016/j.brainres.2011.01.105. [DOI] [PubMed] [Google Scholar]
- Hamby ME, Sofroniew MV. Reactive astrocytes as therapeutic targets for CNS disorders. Neurotherapeutics. 2010;7:494–506. doi: 10.1016/j.nurt.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillion J, Dhara S, Sumter TF, Mukherjee M, Di Cello F, Belton A, Turkson J, Jaganathan S, Cheng L, Ye Z, Jove R, Aplan P, Lin YW, Wertzler K, Reeves R, Elbahlouh O, Kowalski J, Bhattacharya R, Resar LM. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res. 2008;68:10121–10127. doi: 10.1158/0008-5472.CAN-08-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GR, Lee SC, Brosnan CF. Cytokines: powerful regulators of glial cell activation. Neuroscientist. 2003;9:10–22. doi: 10.1177/1073858402239587. [DOI] [PubMed] [Google Scholar]
- Joy A, Moffett J, Neary K, Mordechai E, Stachowiak EK, Coons S, Rankin-Shapiro J, Florkiewicz RZ, Stachowiak MK. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene. 1997;14:171–183. doi: 10.1038/sj.onc.1200823. [DOI] [PubMed] [Google Scholar]
- Jung BP, Jugloff DG, Zhang G, Logan R, Brown S, Eubanks JH. The expression of methyl CpG binding factor MeCP2 correlates with cellular differentiation in the developing rat brain and in cultured cells. J Neurobiol. 2003;55:86–96. doi: 10.1002/neu.10201. [DOI] [PubMed] [Google Scholar]
- Kang W, Hébert JM. Signaling pathways in reactive astrocytes, a genetic perspective. Mol Neurobiol. 2011;43:147–154. doi: 10.1007/s12035-011-8163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto EM, Scavone C, Mattson MP, Camandola S. Curcumin Requires Tumor Necrosis Factor α Signaling to Alleviate Cognitive Impairment Elicited by Lipopolysaccharide. Neurosignals. 2012 May 9; doi: 10.1159/000336074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, Deutsch A, Hofmann O, Ventura A, Hide W, Lieberman J. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011 Nov;7(11):e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, Owen LB, Piazza GA, Xi Y. miR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012;19:378–386. doi: 10.1038/cdd.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, Mandel G. A role for glia in the progression of Rett's syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie F, Ruhrberg C. Diverse roles for VEGF-A in the nervous system. Development. 2012;139:1371–1380. doi: 10.1242/dev.072348. [DOI] [PubMed] [Google Scholar]
- Mark RJ, Keller JN, Kruman I, Mattson MP. Basic FGF attenuates amyloid beta-peptide-induced oxidative stress, mitochondrial dysfunction, and impairment of Na+/K+-ATPase activity in hippocampal neurons. Brain Res. 1997;756:205–214. doi: 10.1016/s0006-8993(97)00196-0. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Murrain M, Guthrie PB, Kater SB. Fibroblast growth factor and glutamate: opposing roles in the generation and degeneration of hippocampal neuroarchitecture. J Neurosci. 1989;9:3728–3740. doi: 10.1523/JNEUROSCI.09-11-03728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Camandola S. NF-kappaB in neuronal plasticity and neurodegenerative disorders. J Clin Invest. 2001;107:247–254. doi: 10.1172/JCI11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- Mazzocco M, Arrigo P, Egeo A, Maffei M, Vergano A, Di Lisi R, Ghiotto F, Ciccone E, Cinti R, Ravazzolo R, Scartezzini P. A novel human homologue of the SH3BGR gene encodes a small protein similar to Glutaredoxin 1 of Escherichia coli. Biochem Biophys Res Commun. 2001;285:540–545. doi: 10.1006/bbrc.2001.5169. [DOI] [PubMed] [Google Scholar]
- Murray PJ. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc Natl Acad Sci U S A. 2005;102:8686–8691. doi: 10.1073/pnas.0500419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Okun E, Griffioen KJ, Mattson MP. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci. 2011;34:269–281. doi: 10.1016/j.tins.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Kaltschmidt C. NF-kappa B: a crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20:252–258. doi: 10.1016/s0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12:213–219. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Xu LJ, Xiong XX, White RE, Sun X, Giffard RG. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012;45:555–563. doi: 10.1016/j.nbd.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B, Fan H, Zhang IY, Liu B, Feng B, Meng L, Zhang R, Sadeghi S, Guo H, Pang Q. HMGA1 expression in human gliomas and its correlation with tumor proliferation, invasion and angiogenesis. J Neurooncol. 2012;106:543–549. doi: 10.1007/s11060-011-0710-6. [DOI] [PubMed] [Google Scholar]
- Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia. 2005;50:427–434. doi: 10.1002/glia.20207. [DOI] [PubMed] [Google Scholar]
- Pike CJ, Cummings BJ, Monzavi R, Cotman CW. Beta-amyloid-induced changes in cultured astrocytes parallel reactive astrocytosis associated with senile plaques in Alzheimer's disease. Neuroscience. 1994;63:517–531. doi: 10.1016/0306-4522(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Soderstrom KE, Kordower JH. Trophic factors therapy in Parkinson's disease. Prog Brain Res. 2009;175:201–216. doi: 10.1016/S0079-6123(09)17514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resar LM. The high mobility group A1 gene: transforming inflammatory signals into cancer? Cancer Res. 2010;70:436–439. doi: 10.1158/0008-5472.CAN-09-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Schuldenfrei A, Belton A, Kowalski J, Talbot CC, Jr, Di Cello F, Poh W, Tsai HL, Shah SN, Huso TH, Huso DL, Resar LM. HMGA1 drives stem cell, inflammatory pathway, and cell cycle progression genes during lymphoid tumorigenesis. BMC Genomics. 2011;12:549. doi: 10.1186/1471-2164-12-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Pozo A, Mielke ML, Gómez-Isla T, Betensky RA, Growdon JH, Frosch MP, Hyman BT. Reactive glia not only associates with plaques but also parallels tangles in Alzheimer's disease. Am J Pathol. 2011;179:1373–1384. doi: 10.1016/j.ajpath.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32:638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik C, de Martin R, Kumabashiri I, Schmid JA, Binder BR, Lipp J. Nuclear factor (NF)-kB-regulated X-chromosome-linked IAP gene expression protects endothelial cells from tumor necrosis factor -induced apoptosis. J. Exp.Med. 1998;188:211–216. doi: 10.1084/jem.188.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SC, Arumugam TV, Xu X, Cheng A, Mughal MR, Jo DG, Siler DA, Chigurupati S, Ouyang X, Magnus T, Camandola S, Mattson MP. Pivotal role for neuronal Toll-like receptors in ischemic brain injury and functional deficits. Proc Natl Acad Sci USA. 2007;104:13798–13803. doi: 10.1073/pnas.0702553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teismann P, Schulz JB. Cellular pathology of Parkinson's disease: astrocytes, microglia and inflammation. Cell Tissue Res. 2004;318:149–161. doi: 10.1007/s00441-004-0944-0. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol. 2010;20:588–594. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.