Abstract
The cerebral cortex, the brain structure responsible for our higher cognitive functions, is built during embryonic development in a process called corticogenesis. During corticogenesis, neural stem cells generate distinct populations of progenitors and excitatory neurons. These new neurons migrate radially in the cortex, eventually forming neuronal layers and establishing synaptic connections with other neurons both within and outside the cortex. Perturbations to corticogenesis can result in severe neurodevelopmental disorders, thus emphasizing the need to better understand molecular regulation of brain development. Recent studies in both model organisms and humans have collectively highlighted roles for post-transcriptional regulation in virtually all steps of corticogenesis. Genomic approaches have revealed global RNA changes associated with spatial and temporal regulation of cortical development. Additionally, genetic studies have uncovered RNA-binding proteins (RBPs) critical for cell proliferation, differentiation, and migration within the developing neocortex. Many of these same RBPs play causal roles in neurodevelopmental pathologies. In the developing neocortex, RBPs influence diverse steps of mRNA metabolism, including splicing, stability, translation, and localization. With the advent of new technologies, researchers have begun to uncover key transcripts regulated by these RBPs. Given the complexity of the developing mammalian cortex, a major challenge for the future will be to understand how dynamic RNA regulation occurs within heterogeneous cell populations, across space and time. In sum, post-transcriptional regulation has emerged as a critical mechanism for driving corticogenesis and exciting direction of future research.
INTRODUCTION
The cerebral cortex is the most complex biological “machine” known to man. Part of this complexity resides in the web of coordinated functional units, the cortical areas. Cortical areas are radially organized within layers, each of which contain neurons with similar molecular, electrophysiological, and connectivity characteristics.1 The cytoarchitecture of an area and thus the number of neurons in each layer is paramount to specify its post-natal function. Additionally, glial cells (astrocytes, oligodendrocytes, and microglia) play a key role in the homeostasis of the cortex. Defects in cortical development can cause acute neurological disorders affecting brain size and function such as microcephaly or lissencephaly. Therefore, the developmental mechanisms that regulate neuronal number and positioning together with glial cells are crucial to build a healthy brain. This review will focus on the molecular regulation of neuronal generation and positioning during embryonic neocortical development.
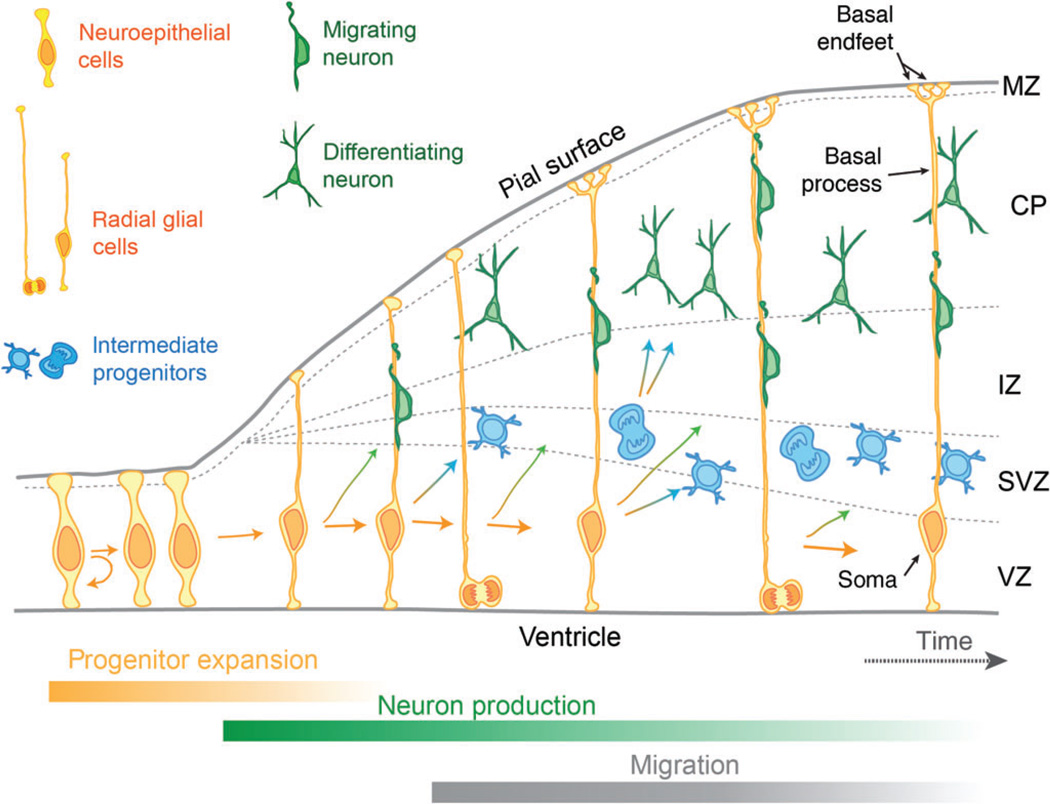
During embryonic development, excitatory neurons are generated from neural progenitor populations in a process termed neurogenesis.2 The germinal zones include the ventricular zone (VZ), located at the border of the cerebral ventricles, and the subventricular zone (SVZ) located beside the VZ (Figure 1). During early cortical development the predominant neural progenitors are neuroepithelial cells (NE cells), which mainly undergo symmetric proliferative divisions to self-renew. NE cells are later replaced by radial glial cells (RGCs), which primarily undergo asymmetric divisions to generate a new RGC and a more differentiated cell, either a neuron or an intermediate progenitor (IP)3,4. Both NEs and RGCs extend processes from the ventricular border to the pial surface while their cell body (nucleus) resides in the VZ. IPs are lineage-restricted multipolar progenitors which divide in the SVZ to amplify the neuronal population.5–7 Hence neurons are directly generated by both RGCs and IPs. In mice, the most widely utilized model for studying corticogenesis, the proliferative period begins around embryonic day (E) 10.0, and the neurogenic period begins about E11.5 and continues to E18.5. Neurons of different layers are born in a sequential fashion, with deep layer neurons born between E11.5 and E14.5 and superficial layer neurons born between E14.5 and E18.5.8,9
FIGURE 1.
Schematic representation of cortical development. Shown are three different progenitor populations (neuroepithelial cells, radial glial cells, and intermediate progenitors) and neurons (both migrating and differentiating). Progenitors residing within the VZ undergo self-renewal divisions to generate new progenitors (curved arrow) as well as divisions to generate either neurons or progenitors (straight arrows). As corticogenesis proceeds, progenitors initially expand their population, shift to neuron, and intermediate progenitor production. Intermediate progenitors within the SVZ also generate neurons. Neurons migrate through the IZ to the CP to form layers of the cerebral cortex. MZ, marginal zone; CP, cortical plate; IZ, intermediate zone; SVZ, sub-ventricular zone; VZ, ventricular zone
After their generation, newborn neurons migrate toward the pial surface of the cortex, using the basal process of RGCs as their scaffold. Their route passes across the intermediate zone (IZ) in the middle of the cortex and ends in the cortical plate (CP), the final location of neuronal layers (Figure 1). During normal development young neurons migrate up to the pial surface, bypassing neurons born earlier.10,11 Thus, deep layer neurons born earlier in development eventually reside closer to the ventricle, whereas superficial layer neurons are ultimately found near the pial surface. Upon reaching their final position within the cortex, the excitatory neurons then establish connections with other neurons both within and outside of the cortex. Hence the fate and final function of projection neurons is ultimately defined by their birth and subsequent migration to distinct layers of the brain.
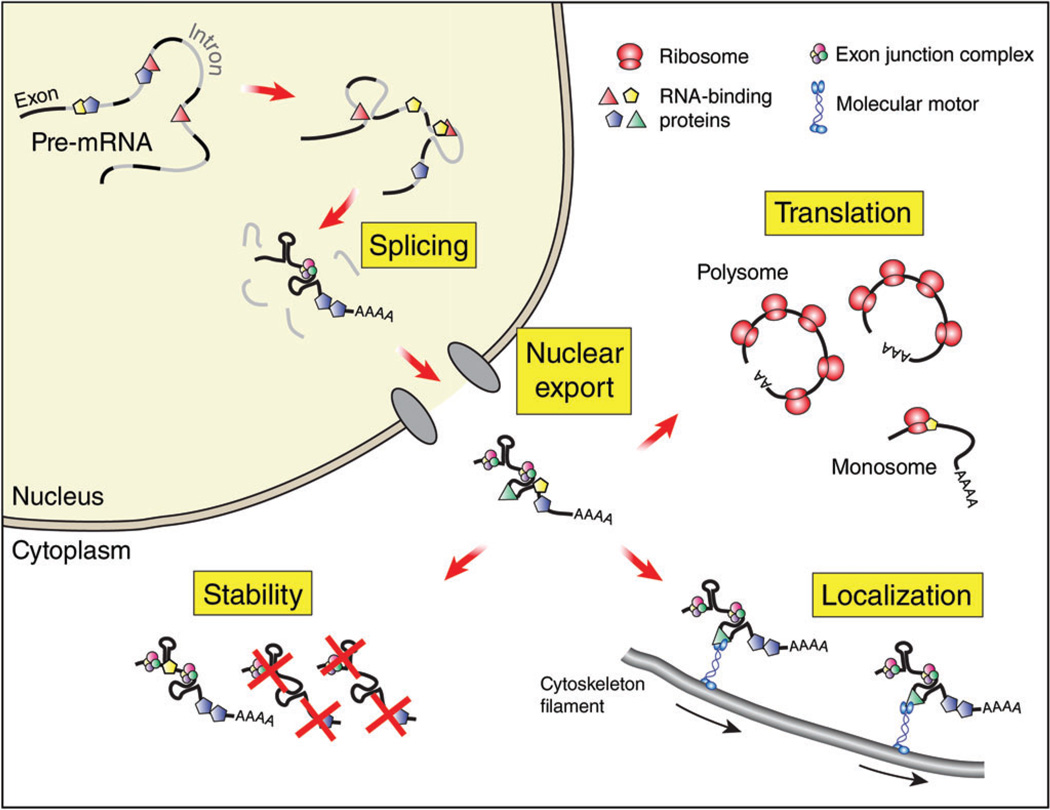
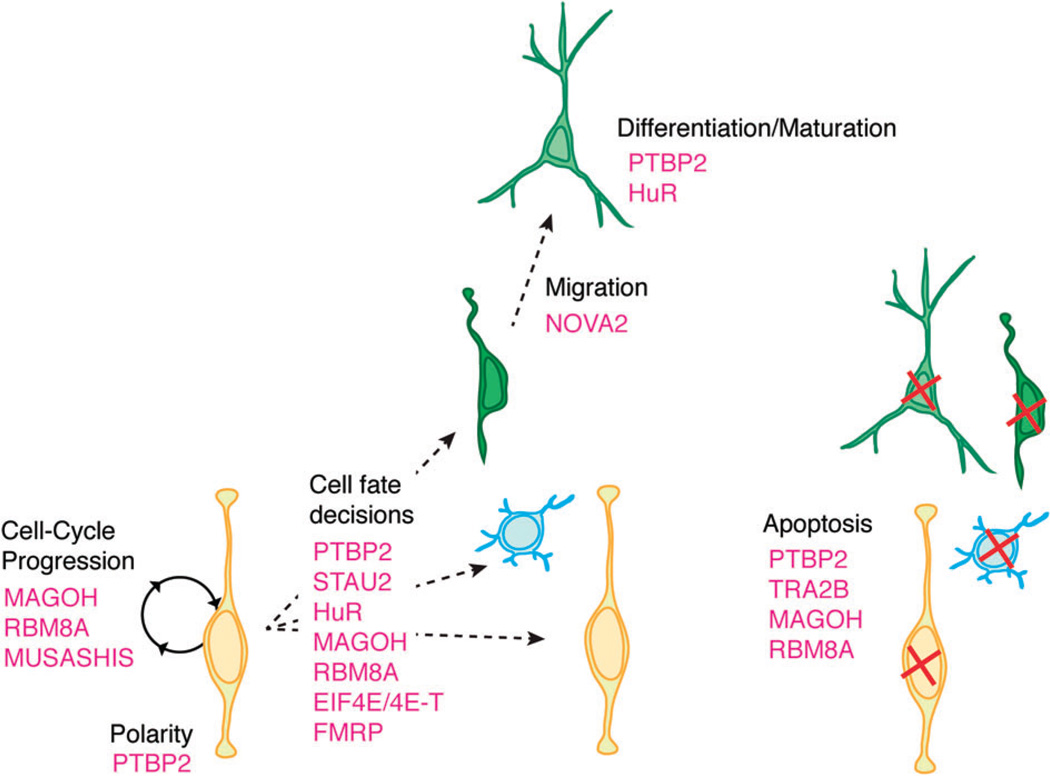
Although much is known about transcription factors and signaling molecules in corticogenesis, only recently have we begun to appreciate the widespread roles of RNA-binding proteins (RBPs) in neocortical development. Specific RBPs are expressed across different developmental stages of the developing neocortex as evidenced by quantitative RT-PCR analyses, in situ hybridization and transcriptome – analyses.34–36 However only a small number of RBPs have actually been tested for a functional impact upon corticogenesis (Table 1). Those RBPs important for neocortical development impact diverse steps of RNA metabolism, and thus collectively reveal that modulation of all stages of the RNA life cycle is necessary for cortical development (Figure 2). In this review we highlight critical RBPs implicated in embryonic corticogenesis, including the production, differentiation and migration of excitatory neurons. We describe their known functions in RNA regulation, cortical development, and in relevant cases disease pathogenesis. These include both RBPs strictly expressed in the developing cortex and ubiquitous RBPs with enriched neocortical expression. We organize our review by discussing neocortical RBPs relevant for each major step of posttranscriptional regulation: alternative splicing, nucleo-cytoplasmic transport, RNA stability and translation, and localization (Figures 2 and 3).
TABLE 1.
RNA-binding Proteins Required for Corticogenesis
| RNA-binding Protein |
Cortical Function | Cortical Expression | RNA Function | Key RNA Targets | Associated Neuro-diseases |
References |
|---|---|---|---|---|---|---|
| NOVA2 | Neuronal migration | Neuron specific | Alternative splicing | Dab1 | Paraneoplastic neurologic disorder |
12,13 |
| TRA2B | Neuronal and progenitor survival |
Progenitors and neurons |
SR pre-mRNA splicing |
? | 14,15 | |
| PTBP2 | Progenitor polarity, proliferation, neuronal maturation |
Neuron specific | Alternative splicing, exon inclusion |
Psd-95 | 16–18 | |
| MAGOH | Progenitor proliferation | Ubiquitous progenitor enriched |
splicing, RNA localization NMD, translation |
Lis1 | 19 | |
| RBM8A | Progenitor proliferation | Ubiquitous progenitor enriched |
splicing, RNA localization, NMD, translation |
? | TAR syndrome | 20 |
| HuR | Progenitor proliferation, neuronal identify and maturation |
Neuron specific | Translation | Dll1 | 21,22 | |
| MSI1,2 | Progenitor proliferation | Ubiquitous, enriched in progenitors |
Translation |
Numb, Jag1, Prfpf3, Kirrel3, Rbm22, Dhx37 |
23–25 | |
| FMRP | Progenitor maintenance | Progenitors and neurons |
Translation |
Pfn1, Cdh2, NOS1 |
Fragile X | 26–30 |
| Eif4E/4E-T | Progenitor maintenance | Progenitors and neurons |
Translation |
Neurog1, Neurog2, Neurod1 |
31 | |
| STAU2 | Radial glia maintenance | Progenitors and neurons |
RNA localization |
Prox1, Bsb2, Trim32 |
32,33 |
FIGURE 2.
Cartoon depicting various stages of mRNA life cycle when RBPs function. Different stages of posttranscriptional regulation are shown along with their nuclear-cytoplasmic location. This review discusses roles for RNA-binding proteins (shown as geometric shapes) in these various aspects of mRNA metabolism.
FIGURE 3.
Summary of known RNA-binding proteins and the aspects of corticogenesis they regulate. Different aspects of neural progenitor function (cell cycle progression, cell fate decision, apoptosis) and neuronal function (migration, differentiation, maturation, apoptosis) are indicated along with the RBPs discussed in this review.
ALTERNATIVE SPLICING
Alternative splicing (AS) is a powerful mechanism to amplify the output diversity of the genome through the editing of primary transcripts (Figure 2). The excision or inclusion of intronic and exonic sequences of pre-mRNA produces distinct transcripts that may be translated into biochemically diverse proteins. AS of the 5’ and 3’ UTR, or coding regions of a pre-mRNA can also impact downstream steps of mRNA metabolism including stability, nuclear export, nonsense mediated decay (NMD), and RNA localization, largely by exposing binding sequences for RBPs or miRNAs. Hence, AS is a powerful mechanism to differentially manipulate gene expression between cells, and recent studies reviewed below underscore its relevance for cortical development.
Genomic Analyses of AS in the Developing Neocortex
With the advent of transcriptome techniques, we are now beginning to appreciate the contribution of genome-wide RNA splicing for cortical development. Studies of both mouse and human cortical models have collectively revealed both spatial and temporal AS differences during cortical development (Table 2). One of the first studies to dissect AS came from Nenad Sestan’s group, who utilized whole-genome exon microarrays to reveal region specific differences in AS in the human brain at mid-gestation.37 This comprehensive study discovered that at mid-fetal development, 28% of expressed genes are alternatively spliced between different human brain structures. Among those genes showing robust AS is ROBO1, which is involved in axon guidance and neural progenitor proliferation, and is implicated in various neurodevelopmental disorders.38,39 Distinct AS ROBO1 transcripts might promote establishment of connectivity and/or the generation of the appropriate number of neurons in distinct layers of mature cortical areas.
TABLE 2.
Genomic Studies Highlighting Alternative Splicing in Cortical Development
| Stage | Organism | Analysis | References |
|---|---|---|---|
| Mid-gestation | Human | Spatial differences between brain structures | 37 |
| E14.5 | Mouse | Spatial differences in cortical layers(VZ/SVZ vs IZ vs CP) | 36 |
| E15.5-P1 | Mouse | Temporal and spatial differences in sorted excitatory neurons: callosal, subcerebral, corticothalamic/subplate neurons | 40 |
| E16-P30 | Mouse | Temporal differences in in vivo cerebral cortices | 41 |
| In vitro | Human | Temporal differences in in vitro differentiated ES cells | 42 |
A similar spatial analysis of AS was applied within the developing embryonic mouse cortex at mid-gestation (E14.5) by Ayoub et al., who coupled RNA sequencing with laser-capture microdissection to demonstrate the existence of differential AS between different embryonic cortical zones (VZ vs SVZ + IZ vs CP).36 This study revealed that some genes, such as Wdr61, show no significant difference in overall expression levels between zones but do exhibit differential expression of splice variants. For other genes, such as Mfge8, differential expression of just one splice variant across cortical zones can explain overall shifts in expression. In addition for other classes of genes, such as Cugbp2 and Hes6, the relative ratio of spliceoform expression is similar between zones indicating AS may be less relevant. The three embryonic cortical zones assessed by Ayoub et al. contain largely different populations of progenitors and post-mitotic neurons. These spliceoform expression differences imply there are cell-specific AS within the developing mouse neocortex.
As embryonic development proceeds, the repertoire of progenitors and neurons also changes. Hence it is not surprising that in addition to spatial differences in AS, temporal differences in AS are evident across different stages of cortical development. Dillman et al. compared cortical samples from embryonic day (e) 16 to those from postnatal day 30 in the mouse, and discovered AS differences. Amongst these they noted that spliceforms more highly expressed postnatally encode actin-related proteins.41 This finding is of interest as actin metabolism is paramount in maturing neurons during early postnatal stages when neuronal connections are being established.43 Temporal patterns of AS have also been discovered using in vitro models of corticogenesis, in which human embryonic stem cells are differentiated into neurons.42 This longitudinal analysis (termed Cortecon by the authors) revealed widespread AS of 5017 genes during in vitro corticogenesis. Interestingly a significant fraction of these AS genes were associated with cancer or nervous system diseases.
A significant limitation of the aforementioned analyses is that most samples analyzed to date contain heterogeneous cell populations, which collectively may contribute to AS differences, thus complicating interpretation. A recent comprehensive study by Molyneaux et al. significantly overcame this hurdle, using deep sequencing to probe transcriptome and AS changes in sorted excitatory neuronal populations from various stages of corticogenesis.40 This group discovered 1181 genes with shifts in isoform expression during corticogenesis. From transcriptome analyses the authors identify genes showing uniform expression at the gene-level but significant differences at the isoform level, an observation also made by Ayoub et al. as described above for Wdr61.36 Altogether, these analyses provide evidence that AS is at play in the developing cerebral cortex across multiple dimensions (tangential, radial, and temporal). The current studies collectively highlight new candidate genes that may regulate corticogenesis. Future studies that similarly apply cell sorting and/or single cell transcriptome analysis will be valuable for further discovery of AS differences in cortical development.
SPLICING FACTORS REGULATING CORTICOGENESIS
The spatio-temporal regulation of AS relies on the differential expression and function of trans-splicing factors including RBPs. Both McKee et al. and DeBoer et al. uncovered splicing factors expressed within the developing cortex.34,35 Several of these RBPs have been experimentally shown to be critical for cortical development.
NOVA2
NOVA1 and 2 are members of KH-domain RBPs and are among the best-characterized RBPs in the brain. Nova proteins bind RNA via YCAY clusters and UCAU sequences, and regulate AS in vitro. Of these, Nova2 is highly expressed in the neocortex.44 Consistent with biochemistry data, Nova2 knock out mice contain a significant number of splicing anomalies in the postnatal brain.12 Interestingly, the vast majority of mis-spliced genes (34 of 40) encode proteins localized to the synapse. During prenatal cortical development, NOVA2 binds to a large number of transcripts (27,576).13 Yano et al. showed Nova2 is necessary for the proper migration of upper-layer neurons toward the CP. The authors used HITS-CLIP of Nova2-deficient brains to identify key downstream splicing targets of Nova2. They focused on 20 genes of the Reelin pathway, because Reelin is a migration cue secreted by early born Cajal Retzius (CR) neurons. Remarkably, the authors identified significant splicing changes in only one transcript of this pathway, finding that NOVA2 regulates excision of exons 7b and 7c of Dab1. Using in utero electroporation of minigene constructs, the authors then elegantly showed that Nova2 is essential for the proper expression of Dab1 spliceforms and that this splicing mediates neuronal migration in the neocortex. In the future it will be of interest to identify additional splicing targets genome-wide that may also be regulated by Nova2 in the developing cerebral cortex.
PTBP2
Poly-pyrimidine tract-binding proteins (PTBP proteins) are involved in multiple steps of RNA metabolism including splicing. Biochemical studies of PTBP1 demonstrated this family of proteins binds introns (recognizing CU-repeats and UCUY-rich elements).44 Depending on its relative positioning with respect to certain exons (either upstream or downstream), PTBP1 can either promote or inhibit exon inclusion.16 Although PTBP1 is minimally expressed in the brain, PTBP2 (also called nPTB) is highly expressed in the brain.16 In the developing mouse brain, PTBP2 binds thousands of RNAs.16 Among these mRNAs, splicing of Psd-95, which encodes a synaptic protein, is repressed by PTBP1 and PTBP2 during development.17 To further understand the role of PTBP2 in cortical development, two groups recently generated PTBP2-deficient mice. Licatolosi et al. discovered that E14.5 PTBP2 mutant brains had defective neural progenitor polarity, accompanied by defects in proliferation and neuronal differentiation.16 Li et al. observed only postnatal cortical defects in their mutant mice, related to a role in neuronal differentiation, maturation, and survival.18 The phenotypic differences may be because of the nature of the mouse mutation, as the former study used germline knockouts and the latter study used conditional expression with Nestin-Cre and Emx1-Cre lines. Regardless, in both studies, PTBP2-deficiency was associated with significant alterations in AS, including mRNAs involved in regulation of the actin cytoskeleton, proliferation and cell fate. Although RNA-targets of PTBP2 involved in the regulation of neurogenesis have not yet been identified, these studies establish PTBP2 as a key regulator of AS in both neural progenitors and immature neurons.
TRA2B
TRA2B splicing factor is a member of the Serine-/Arginine-Rich (SR) protein family, which have well-established roles in constitutive and AS.45 SR proteins recognize exonic splice enhancers, as well as interact with other splicing factors to promote splice site recognition. Once splicing is complete, SR proteins may or may not remain on the mRNA. This retention of SR proteins on the mature mRNA can impact nuclear export and downstream RNA regulation. TRA2B regulates splicing of Tau and Smn2 mRNAs, involved in Alzheimer’s disease and spinal muscular atrophy, respectively.46,47 Hence PTBP2 is relevant for human diseases of the nervous system. Constitutional loss of Tra2b in the mouse leads to embryonic lethality by E7.0, precluding the use of this model for the study of its role in cortical development.48 However conditional knock-out embryos using either Emx1-Cre14 or Nestin-Cre15 point toward an essential role of Tra2b in the survival of neural progenitors and neurons. Strikingly, conditional Tra2b knockout in the cerebral cortex leads to almost complete absence of the cortex at adulthood, following a massive wave of apoptosis during embryonic cortical development. These studies establish the fundamental requirement of Tra2b in survival of neural cells, and highlight the importance of future studies to determine key mRNA targets of TRA2B in the embryonic cortex. Altogether, these data suggest that AS regulation plays a critical role in cortical development. Given the abundance of splicing factors in the developing brain, clearly these studies are just the tip of the iceberg.
From the Nucleus to the Cytoplasm: The Exon Junction Complexes
As splicing proceeds, spliced transcripts become decorated by exon junction complexes (EJC), which bind primarily at the junctions where introns are excised.49,50 The EJC remains bound to the spliced mRNA as the RNA is exported into the cytoplasm. The core EJC is composed of the heterodimer, Magoh and Rbm8a, the helicase, Eif4a3, and the cytoplasmic component Casc3. This core complex interacts transiently with other proteins to mediate various aspects of mRNA metabolism, including mRNA splicing, localization, nonsense-mediated decay, and – translation initiation.51–53
Magoh was recently shown to be essential for corticogenesis in mice.19,54 Haploinsufficiency for Magoh causes a severe microcephaly, associated with depletion of IPs and massive apoptosis of new neurons. Silver et al. showed that Magoh regulates proper cell division of radial glia and hypothesized that defective mitosis induces aberrant production of progenitors and neurons.19 Microarray analysis of Magoh haploinsufficient cortices revealed only 147 transcripts with significant differential expression. Given that the EJC decorates >80% of exon–exon junctions49 this indicates Magoh haploinsufficiency may not globally impact RNA stability although this remains to be formally tested. The authors identified protein changes downstream of Magoh, including one physiologically relevant target, Lis1, a microtubule-associated protein also involved in brain development. Future studies will be useful to assess how Magoh impacts radial glia divisions either via translation and/or some other step in mRNA metabolism.
The same group recently probed whether haploinsufficiency for the Magoh heterodimer, Rbm8a, impacts corticogenesis.20 Similar to Magoh, Rbm8a is also highly expressed in the developing cortex. Conditional haploinsufficiency for Rbm8a induced microcephaly, even more severe than Magoh loss. This phenotype was associated with depletion of progenitors and dramatic apoptosis especially of neurons. Rbm8a mutant embryos showed precocious neuron production and faster cell cycle exit of progenitors. Thee phenotypic similarities induced by Magoh and Rbm8a haploinsufficiency support a model where Magoh and Rbm8a act together as part of the EJC to regulate corticogenesis. The observed differences in severity of phenotypes could reflect distinct functions outside of the EJC or redundancies with other proteins, such as MagohB.55 In addition to these roles of core EJC components in the brain, the peripheral EJC component involved in NMD, Upf1, is also expressed in the developing neocortex and promotes a stem cell state in primary cells.56 Future genetic and molecular studies of these mutants will help establish which aspect(s) of EJC regulation are critical to development of the brain.
Recent mouse and human genetic studies have collectively implicated EJC dosage in neurodevelopmental pathologies associated with aberrant cortical development, including autism, schizophrenia, and – intellectual disability.57–59 UPF3B, an EJC component required for nonsense-mediated decay, is mutated in X-linked intellectual disability, schizophrenia, and autism. Copy number variations in several EJC components, including UPF3B, EIF4A3, RBM8A, and MAGOH, are found in patients with intellectual disability frequently accompanied by brain malformations.57 RBM8A is within the proximal 1q21.1 microdeletion/duplication associated with microcephaly.58 EIF4A3 is mutated in Richeiri-Costa Syndrome, a developmental disorder which can also be associated with brain malformations.60 Continuing genetic studies in model organisms will help establish if roles for these EJC components in corticogenesis are the root causes for these neurodevelopmental disorders.
RNA Stability and Translational Control
Outside of the nucleus, RNA stability and translational regulation offer yet another layer of control for gene expression. The role for RNA stability in corticogenesis is poorly defined. A number of RNAs have been shown to have short-half lives but so far this has been attributed to oscillations in transcription. Translational control can be exerted at different steps: initiation, elongation, or termination. These steps are regulated by the coordination between ribosomal complexes and a vast set of RBPs. RBPs which impact translation control have recently been shown to influence development of the cerebral cortex.
HuR
HuR is a well-characterized RBP, part of a family of Hu-related proteins that preferentially bind the 3’UTR of its RNA targets to influence multiple aspects of RNA metabolism including RNA stability and translation.61,62 While several Hu proteins have been implicated in neuron differentiation and post-mitotic function,63,64 so far only HuR has been formally shown to regulate normal embryonic corticogenesis. HuR is expressed early in neuroepithelial cells, when these progenitors are undergoing primarily proliferative divisions.21 Garcia-Dominguez et al. postulated HuR influences the Notch pathway by regulating mRNA levels of the ligand Delta.21 The expression of Delta ligand promotes proliferation and prevents differentiation in neighboring cells via a mechanism called lateral-inhibition.65 The authors discovered that HuR interacts with Dll1 mRNA. HuR depletion in neural precursors leads to reduced Dll mRNA levels and less differentiation. This differentiation phenotype can be rescued by overexpression of Dll1. Hence the authors propose that HuR regulates Dll1 stability to promote lateral inhibition and thus influence early cell fates in the cortex.
Later in development when neuroepithelial cells have been replaced by radial glial progenitors, HuR is expressed in radial glia, IPs, and newborn neurons.22 In an elegant study evaluating translation, Kraushar et al. used genome-wide polysome-profile analysis in conditional HuR knockout murine cortices at E13 and P0 to uncover a large pool of HuR-regulated mRNAs redistributed to different polysomal fractions during development. HuR-dependent RNAs were enriched for regulators of transcription, translation and layer specific pathways. The transcripts regulated by HuR were dramatically different in E13 and P0 brains, perhaps reflecting different biological processes occurring at these distinct ages. The authors also make the novel discovery that HuR interacts with the Eif2 kinase, Eif2ak4, which regulates the presence of distinct ribosomal proteins in active sites of translation at polysomes. The authors argue that HuR coordinates the translation of a network of mRNAs encoding proteins that share common functions, akin to the RNA regulon model first proposed by Jack Keene.62 Consistent with a functional requirement of HuR in cortical development, phenotypic anatomical analyses of P0 HuR conditional knockout mice revealed that HuR regulates the position, identity and maturation of post-mitotic glutamatergic neurons. Future work will be valuable to further identify the molecular mechanisms by which HuR regulates these developmental processes. Moreover this study sets the stage for future identification of signals that influence temporal control of mRNA translation.
Musashi
The translational regulators, Musashi 1 and 2 (Msi-1 and Msi-2), are highly expressed in NSCs throughout the central nervous system, including the mammalian cortex.66 Cortical Msi-1−/− dissociated NSCs transfected with antisense peptide-nucleic acid against Msi-2 showed decreased neurosphere formation and proliferative capacity, perhaps linked to impaired cell-cycle progression.23 Hence Msi-1 and Msi-2 have redundant functions in neural stem cells. Musashis (Msis) are reported to act as both positive and negative regulators of translation, effects that are mediated through binding to the 3’UTRs of target mRNAs, including mammalian Numb.24 Although this translational relationship between Msi and Numb has so far been shown in fibroblasts it is tempting to speculate that it may also hold true in NSCs, where numb is important for influencing neurogenesis.67,68 Msi targets have not yet been identified in NSCs; however, a recent study discovered translational targets from primary NSCs overexpressing Msi, using Ribosome profiling and RNA-seq.25 Among several transcripts with reduced translation efficiency was Jag1, a ligand for Notch receptors, as well as a number of RBPs including Prpf3, Kirrel3, Rbm22, and Dhx37. Interestingly these targets have abundant Msi-binding sites in their 3’ UTRs, thus Msi is thought to directly bind these targets. Katz et al. also demonstrated that Msi overexpression impacted AS while not perturbing overall RNA levels extensively.25 Because Msi is primarily cytoplasmic, these changes are thought to be a secondary consequence of translational regulation of splicing factors, and not because of a direct role in AS or RNA stability per se. With identification of Msi translational targets, it will be of interest in upcoming studies to assess the role of these Msi targets upon NSC behavior in the cortex.
FMRP
FMRP (Fragile-X mental retardation protein) is an RBP encoded by the Fmr1 gene. FMRP has been largely characterized as a translational inhibitor. Fmr1 null mutations result in the Fragile-X syndrome (FXS) in humans, which is the most prevalent intellectual disorder caused by mutations in one single gene.69,70 Postnatally, FMRP localizes at the synapses between neurons, where it inhibits the translation of a subset of localized mRNAs encoding proteins involved in synaptic plasticity.69,71 In response to neuronal activity, FMRP translational inhibition can be alleviated to allow for local, fast and massive, production of proteins necessary for structural modifications of the postsynaptic compartment.69,72,73
Recent studies of humans and mice support a role for FMRP in regulation of prenatal cortical development. An analysis of brain region volumes of 1-to 3-year-old boys with FXS showed that several cortical areas display enlarged gray matter volume, suggesting a possible regional increase in neurons.74 Indeed, analysis of P5 Fmr1−/− mouse pups revealed increased neuronal density in the somatosensory cortical area.26 Moreover cultured NSCs derived from either pre- or postnatal Fmr1−/− mice generate more neurons than those derived from comparably aged WT mice.27 Interestingly, the density of Tbr2+ IPs is higher in Fmr1-KO cortices, compared to controls, indicating increased neurons could be produced by more IPs.26 Hence FMRP may control neuron production either by regulating IP differentiation or IP generation from radial glial cells. In support of the latter possibility, Saffary et al. used in utero electroporation to knockdown Fmr1 in the developing cortex and demonstrate FMRP is required for IP generation.28 The authors identified the candidate mRNA-target Profilin1 (Pfn1) as a mediator of this process, finding that Pfn1 over-expression rescues the overproduction of IPs in Fmr1 mutant brains.
Another recent study revealed a role for FRMP in neuronal migration in the cortex. In a study by La Fata et al., newborn neurons labeled by in utero electro-poration in Fmr1 knockout brains showed defective neuronal migration.29 These defects eventually lead to abnormal neuronal networks in the postnatal brain, which could be rescued by the overexpression of N-Cadherin, an mRNA target of FMRP. In addition to its well-established role in translational repression, FMRP has also been implicated as a pro-translation regulator in young neurons of the human neocortex.30 Kwan et al. showed that FMRP expression enhances the translation of NOS1, an important regulator of synapse formation and spine maintenance.30,75 Interestingly FMRP-mediated regulation of NOS1 translation was not evident in mouse projection neurons, highlighting potentially interesting evolutionary differences in FMRP function. Altogether these two studies suggest that defective neuronal circuits induced by defects in immature neurons could be at the origin of Fragile-X pathology in the adult.
Pfn1, NOS1, and N-Cadherin are likely part of a vast FMRP-regulated mRNA network involved in the regulation of the NSC-to-IP transition, early neuronal differentiation, and migration. Although FMRP targets have been elucidated in adult brains,76 it still remains an outstanding question which RNAs are FMRP targets in neural stem cells of the developing neocortex and whether their translation is repressed or activated by FMRP. Future work is needed to identify those potential targets, and to assess their contribution to behavior of NSCs and neurons.
Eif4E/4E-T Complex
The EIF4E protein family, composed of Eif4E1, 2, and 3 (Eukaryotic Initiation Factor 4E), is part of a supercomplex docked to the 5’ cap of mRNAs.77 Once bound to mRNAs this complex can either promote or inhibit translation, depending on its composition. These functions are mediated via interactions with additional translation factors. For example, EIF4E1 association with EIF4G initiates translation whereas EIF4E1 binding to 4E-T blocks translation or promotes mRNA decay by targeting mRNAs to P bodies.78
A role for translational regulators in corticogenesis was recently revealed using in utero knockdown in embryonic brains.31 Yang et al. discovered that decreased levels of either Eif4e1 or 4E-T in neural progenitors lead to more neurons and fewer neural progenitors. This is accompanied by an increase in the number of cells with high protein levels for Ngn1, Ngn2, and Neurod1, basic-Helix-Loop-Helix (bHLH) pro-neuronal transcription factors. The authors discovered that in neural progenitors, EIF4E1 binds to Neurog1 and Neurog2 and NeuroD1 mRNAs. These biochemical results, along with rescue experiments using constructs deficient in RNA binding or protein–protein interactions, collectively revealed the Eif4e1/4E-T complex may repress translation of key neurogenic transcripts. Conversely, knockdown of Eif4G in neural progenitors (a positive regulator of translation when associated with EIF4E1) promotes fewer neurons and more progenitors. Altogether, these results suggest that certain neural progenitors are predisposed to the generation of neurons through the transcription of proneural bHLH transcription factors, but are stalled in a proliferating state by EIF4E1/4E-T-mediated translational repression of these target mRNAs. The authors speculate that NSCs are preloaded with mRNAs encoding prodifferentiation factors; however, translation of these mRNAs is repressed by Eif4E1 binding. This is a compelling hypothesis and it will be exciting in the future to test this model. It will also be valuable to demonstrate the direct role of Eif4E1/4E-T on translation of key mRNAs in NSCs, to rule out potential roles in nuclear export or sequestration of mRNAs in RNA-processing bodies.
RNA Localization
RNA localization plays a critical role in neurons both pre- and postnatally. When paired with translational regulation, RNA localization allows for local protein synthesis in the cytoplasm in response to intra- or extracellular signals (see Buxbaum et al., for a recent comprehensive review on this topic).79 In immature neurons, RNA localization and translation, in response to extracellular guidance molecules, is paramount for axon guidance and synaptic function.80 In mature neurons, RNA localization at the synapse may be involved in the precise, fast response of cells to integrate signals from other neurons, in order to consolidate or suppress memories.72,81 Recent studies have now highlighted roles for mRNA localization in mammalian neural progenitors, within both the cell body and basal process.
RNA Localization to the Basal Endfeet
Recently, it was shown that mRNAs encoding CyclinD2 accumulate in structures called basal endfeet, located at the end of basal process.82 CyclinD2 is an outstanding candidate for the maintenance of neural progenitor proliferation as it is a well-characterized G1-phase regulator, and G1 phase is strongly linked to neural progenitor proliferation (Figure 1).83,84 Osumi’s group identified a region in the 3’UTR of the Cyclin D2 mRNA that is sufficient for its translocation to the basal endfeet. They also used a GFP reporter construct to argue that these mRNAs are locally translated, although diffusion of GFP proteins from the cell body could not be ruled out from this experiment. During asymmetric division of NSCs, the daughter cell which adopts proliferative behavior inherits the basal process, whereas the daughter cell that does not inherit the basal process proceeds toward differentiation.85 This led to the hypothesis that following cell division, Cyclin D2 mRNA is locally translated and newly generated proteins subsequently migrate back to the soma through the basal process to promote proliferation. Identification of the transmachinery, including RBPs that bind CyclinD2 will be useful for understanding why it is asymmetrically localized in NSCs, and for identifying additional localized RNAs.
Stau2
Stau2 is a double-stranded RBP, which in neurons has been well characterized as a translational repressor and a regulator of subcellular localization.86 In neurons, Stau2-positive RNA granules aggregate to form heterogeneous RNA granules that subsequently associate with motor proteins to translocate along microtubules to distal regions. Inspired by the Drosophila literature which established a role for Stau in neuroblasts, two independent groups recently showed that Stau2 plays a key role in mammalian cortical development.32,33 In mitotic progenitors, Stau2 is enriched at one pole of the cell, and becomes asymmetrically localized to only one of the post-mitotic progeny. Kusek et al. showed Stau2 was specifically inherited by the daughter cell that differentiates into an intermediate precursor cell following mitosis.33 Both groups showed that downregulation of Stau2 by shRNA knockdown leads to increased differentiation and a depletion of radial glia both in vitro and in vivo. Additionally, Vessey et al. showed that Stau2 acts in coordination with at least two other RBPs: the helicase Ddx1 and the translational repressor Pum2.32 RIPs were employed to identify Stau2 targets, with Sally Temple’s group identifying genome-wide targets in the entire cortex and Freda Miller’s group focussing on specific candidates. Gene ontology (GO) analysis showed Stau2 RNA targets were enriched in transcripts encoding regulators of cell-cycle exit and cilia.33 Of note, several of these targets, Prox1, Bbs2 and Trim32, are asymmetrically localized in a Stau2-dependent fashion. Altogether, these results suggest that Stau2 plays a preponderant role in the selective transmission of pro-differentiation mRNAs in progeny. In parallel, given other RNA regulatory roles of Stau2, the RBP may additionally repress translation of pro-differentiation mRNAs in NSCs. The authors speculated that this translational repression would be relieved in differentiated daughter cells.
These studies indicate that RNA localization may serve as a cell fate determinant to help two daughter cells adopt different fates. Utilizing the “RNA medium” to segregate cell fate determinants represents a certain advantage. As translation is largely paused during mitosis87 the inheritance of select mRNA molecules in daughter progeny might allow for the rapid and massive synthesis of this determinant immediately after completion of mitosis. This mechanism involves a multistep process which includes: (1) the production of an mRNA and its associated RBPs prior to mitosis, (2) the translational repression of this mRNA until the completion of mitosis, (3) the precise localization of this mRNA to a cellular region which will be specifically inherited by one daughter cell, and (4) the derepression of translation following mitosis. Future studies and identification of asymmetrically localized mRNAs and RBPs in mitotic neural progenitors will help define whether this mechanism is broadly used for cell fate determination in the mammalian cortex.
CONCLUSION
This review highlighted key players in posttranscriptional RNA regulation with fundamental roles in corticogenesis. One theme that emerges from this review is that we have just scratched the surface in terms of a comprehensive understanding of how RBPs influence cortical development and which RBPs are important. A second theme that emerges is that virtually all aspects of posttranscriptional regulation are implicated in corticogenesis. Many fundamental questions now remain to be answered. How is RNA regulation coordinated within rapidly dividing populations across stages of embryonic development? What additional RBPs influence corticogenesis, how do they do so, and what are their key targets? What role does RNA stability play in modulating cell fate choices in the developing brain? Answering these questions in a complex tissue such as the embryonic mammalian cortex is challenging and will require multidisciplinary approaches encompassing bioinformatics, biochemistry, and genetics.
Identifying RNA targets for RBPs is critical to gain a mechanistic understanding of how these RBPs help shape the developing brain. Techniques in RNA immunoprecipitation have been critical for uncovering RNA targets for many RBPs within immortalized cells. With variations on RIP approaches, such as HITs-CLIP, it has now become possible to identify RNA targets within whole mouse brains. However, as noted throughout this review, approaches using entire tissue only give a superficial understanding of RBP targets, given the heterogeneous nature of the developing brain over time and space. Future studies which couple optimized RNA immunoprecipitation approaches with single cell resolution will be ideal. These will inform our understanding of how RBPs function in progenitors versus postmitotic neurons, and in early versus late development. Moreover, the use of Ribotag-sequencing approaches or ribosomal profiling within neural stem cells will be extremely valuable to assess genome-wide translation profiles for RBPs. On an individual transcript basis, the direct visualization of RNA targets in situ is now also possible, using single-molecule FISH probes. Using the MS2-tagging approach, one can also now follow single RNA movements in the cell.79 Similarly, live-imaging can be used to evaluate translational targets, by use of reporter constructs in which regulatory mRNA sequences are tethered to photo-convertible protein, such as Kaede or Dendra.88
Over 800 RBPs have now been bioinformatically and empirically identified, and among these many have annotated expression in the developing cerebral cortex.34,35,89 Yet it remains an outstanding question as to which of these RBPs are critical. These questions can be addressed using traditional genetics, CRISPR/Cas9 approaches, or in utero electroporation, the latter of which allows one to rapidly manipulate gene expression within the developing brain. Moreover, the ability to utilize primary cell culture of progenitors and ex vivo embryonic brain slice culture provide tools that make this developing organ amenable to testing candidates involved in posttranscriptional regulation. The future is exciting for RNA regulation in corticogenesis as the advent of new technologies will lead to great advances in this field of research.
ACKNOWLEDGMENTS
We thank members of the Silver lab for helpful discussions. Work in this lab is supported by an R01NS0830897 (to DLS). We apologize to all those whose work we could not include in this review.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 4.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5363. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 5.Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasistha NA, García-Moreno F, Arora S, Cheung AFP, Arnold SJ, Robertson EJ, Molnár Z. Cortical and clonal contribution of Tbr2 expressing progenitors in the developing mouse brain. Cerebral Cortex. 2014 Jun 13; doi: 10.1093/cercor/bhu125. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polleux F, Dehay C, Moraillon B, Kennedy H. Regulation of neuroblast cell-cycle kinetics plays a crucial role in the generation of unique features of neocortical areas. J Neurosci. 1997;17:7763–7783. doi: 10.1523/JNEUROSCI.17-20-07763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 10.Lodato S, Shetty AS, Arlotta P. Cerebral cortex assembly: generating and reprogramming projection neuron diversity. Trends Neurosci. 2015;38:117–125. doi: 10.1016/j.tins.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angevine JB, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 12.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts JM, Ennajdaoui H, Edmondson C, Wirth B, Sanford JR, Chen B. Splicing factor TRA2B is required for neural progenitor survival. J Comp Neurol. 2013;522:372–392. doi: 10.1002/cne.23405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storbeck M, Hupperich K, Gaspar JA, Meganathan K, Martínez Carrera L, Wirth R, Sachinidis A, Wirth B. Neuronal-specific deficiency of the splicing factor Tra2b causes apoptosis in neurogenic areas of the developing mouse brain. PLoS One. 2014;9:e89020. doi: 10.1371/journal.pone.0089020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Zheng S, Han A, Lin CH, Stoilov P, Fu XD, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao H, Pilaz L-J, McMahon JJ, Golzio C, Wu D, Shi L, Katsanis N, Silver DL. Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J Neurosci. 2015;35:7003–7018. doi: 10.1523/JNEUROSCI.0018-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Dominguez DJ, Morello D, Cisneros E, Kontoyiannis DL, Frade JM. Stabilization of Dll1 mRNA by Elavl1/HuR in neuroepithelial cells undergoing mitosis. Mol Biol Cell. 2011;22:1227–1239. doi: 10.1091/mbc.E10-10-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraushar ML, Thompson K, Wijeratne HRS, Viljetic B, Sakers K, Marson JW, Kontoyiannis DL, Buyske S, Hart RP, Rasin MR. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc Natl Acad Sci. 2014;111:E3815–E3824. doi: 10.1073/pnas.1408305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakakibara S-I, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci USA. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz Y, Li F, Lambert NJ, Sokol ES, Tam W-L, Cheng AW, Airoldi EM, Lengner CJ, Gupta PB, Yu Z, et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3:e03915. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tervonen TA, Louhivuori V, Sun X, Hokkanen M-E, Kratochwil CF, Zebryk P, Castrén E, Castrén ML. Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol Dis. 2009;33:250–259. doi: 10.1016/j.nbd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Castrén ML, Tervonen T, Kärkkäinen V, Heinonen S, Castrén E, Larsson K, Bakker CE, Oostra BA, Akerman K. Altered differentiation of neural stem cells in fragile X syndrome. Proc Natl Acad Sci USA. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.La Fata G, Gärtner A, Domínguez-Iturza N, Dresselaers T, Dawitz J, Poorthuis RB, Averna M, Himmelreich U, Meredith RM, Achsel T, et al. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat Neurosci. 2014;17:1693–1700. doi: 10.1038/nn.3870. [DOI] [PubMed] [Google Scholar]
- 30.Kwan KY, Lam MMS, Johnson MB, Dube U, Shim S, Rasin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang G, Smibert CA, Kaplan DR, Miller FD. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron. 2014;84:723–739. doi: 10.1016/j.neuron.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Vessey JP, Amadei G, Burns SE, Kiebler MA, Kaplan DR, Miller FD. An asymmetrically localized Staufen2-dependent rna complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell. 2012;11:517–528. doi: 10.1016/j.stem.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Kusek G, Campbell M, Doyle F, Tenenbaum SA, Kiebler MA, Temple S. Asymmetric segregation of the double-stranded rna binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell. 2012;11:505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeBoer EM, Kraushar ML, Hart RP, Rasin MR. Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons. Neuroscience. 2013;248:1–30. doi: 10.1016/j.neuroscience.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borrell V, Cárdenas A, Ciceri G, Galcerán J, Flames N, Pla R, Nóbrega-Pereira S, García-Frigola C, Peregrín S, Zhao Z, et al. Slit/robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 2012;76:338–352. doi: 10.1016/j.neuron.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Bendito G, Flames N, Ma L, Fouquet C, Di Meglio T, Chedotal A, Tessier-Lavigne M, Marín O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J Neurosci. 2007;27:3395–3407. doi: 10.1523/JNEUROSCI.4605-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molyneaux BJ, Goff LA, Brettler AC, Chen H-H, Brown JR, Hrvatin S, Rinn JL, Arlotta P. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, Cookson MR. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci. 2013;16:499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Leemput J, Boles NC, Kiehl TR, Corneo B, Lederman P, Menon V, et al. NeuroResource. Neuron. 2014;83:51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Svitkina T, Lin W-H, Webb DJ, Yasuda R, Wayman GA, Van Aelst L, Soderling SH. Regulation of the post-synaptic cytoskeleton: roles in development, plasticity, and disorders. J Neurosci. 2010;30:14937–14942. doi: 10.1523/JNEUROSCI.4276-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Wang J, Gao L, Lafyatis R, Stamm S, Andreadis A. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c.SRp55 complex that either recruits or antagonizes htra2beta1. J Biol Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 49.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 51.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shibuya T, Tange TØ, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, Murigneux V, Le Hir HE. Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol. 2014;15:551. doi: 10.1186/s13059-014-0551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMahon JJ, Shi L, Silver DL. Generation of a Magoh conditional allele in mice. Genesis. 2014;52:752–758. doi: 10.1002/dvg.22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh KK, Wachsmuth L, Kulozik AE, Gehring NH. Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay. RNA Biol. 2013;10:1291–1298. doi: 10.4161/rna.25827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 2014;6:748–764. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gécz J. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–1825. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 58.Rosenfeld JA, Traylor RN, Schaefer GB, McPherson EW, Ballif BC, Klopocki E, Mundlos S, Shaffer LG, Aylsworth AS 1q21.1 Study Group. Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur J Hum Genet. 2012;20:754–761. doi: 10.1038/ejhg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alachkar A, Jiang D, Harrison M, Zhou Y, Chen G, Mao Y. An EJC factor RBM8A regulates Anxiety behaviors. Curr Mol Med. 2013;13:887–899. doi: 10.2174/15665240113139990019. [DOI] [PubMed] [Google Scholar]
- 60.Favaro FP, Alvizi L, Zechi-Ceide RM, Bertola D, Felix TM, de Souza J, Raskin S, Twigg SR, Weiner AM, Armas P, et al. A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am J Hum Genet. 2014;94:120–128. doi: 10.1016/j.ajhg.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 2012;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeBoer EM, Azevedo R, Vega TA, Brodkin J, Akamatsu W, Okano H, Wagner GC, Rasin MR. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J NeuroscI. 2014;34:3674–3686. doi: 10.1523/JNEUROSCI.3703-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murciano A, Zamora J, López-Sánchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- 66.Sakakibara S-I, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 67.Rasin MR, Gazula V-R, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- 68.Zhong W, Feder J, Jiang M, Jan L, Jan Y. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 69.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 71.Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 73.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoeft F, Carter JC, Lightbody AA, Hazlett HC, Piven J, Reiss AL. Region-specific alterations in brain development in one-to three-year-old boys with fragile X syndrome. Proc Natl Acad Sci USA. 2010;107:9335–9339. doi: 10.1073/pnas.1002762107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nikonenko I, Boda B, Steen S, Knott G, Welker E, Muller D. PSD-95 promotes synaptogenesis and multi-innervated spine formation through nitric oxide signaling. J Cell Biol. 2008;183:1115–1127. doi: 10.1083/jcb.200805132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 77.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E - from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 78.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoen-berg DR, Sonenberg N. A role for the eIF4E–binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2014;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leung K-M, van Horck FPG, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan S-S, Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012;31:1879–1892. doi: 10.1038/emboj.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pilaz L-J, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doer-finger N, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 85.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 87.Sivan G, Elroy-Stein O. Regulation of mRNA Translation during cellular division. Cell Cycle. 2008;7:741–744. doi: 10.4161/cc.7.6.5596. [DOI] [PubMed] [Google Scholar]
- 88.Hörnberg H, Holt CE. RNA-binding proteins and translational regulation in axons and growth cones. Front Neurosci. 2013;7:81. doi: 10.3389/fnins.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]