Abstract
Antigens recognized by T cells are expressed as peptides bound to major histocompatibility complex (MHC) molecules. Microcapillary high-performance liquid chromatography–electrospray ionization–tandem mass spectrometry was used to fractionate and sequence subpicomolar amounts of peptides isolated from the MHC molecule HLA-A2.1. Of 200 different species quantitated, eight were sequenced and four were found in cellular proteins. All were nine residues long and shared a distinct structural motif. The sensitivity and speed of this approach should enhance the analysis of peptides from small quantities of virally infected and transformed cells as well as those associated with autoimmune disease states.
Cytotoxic T lymphocytes (CTLs) are a part of the immune system concerned with recognition of host cells that express new antigens as a result of viral infection or transformation. CTLs do not recognize new antigens directly, but only as short peptides bound to a deep cleft in class I molecules of the MHC (1–3). Newly synthesized viral and cellular proteins are degraded into peptides in the cytoplasm, transported to the endoplasmic reticulum where they bind to class I molecules, and then expressed on the cell surface (4–7). Each of the allelic forms of the class I MHC molecule binds to a complex mixture of structurally distinct peptides (8, 9). Information on the nature of these peptides has been obtained from studies with synthetic peptides (10–12) and from Edman degradation applied to unfractionated mixtures of peptides extracted from five different class I MHC molecules (8). Sequences of 11 peptides extracted from HLA-B27 were identified after high-performance liquid chromatography (HPLC) fractionation and Edman degradation (9). Because HPLC was unable to completely resolve the complex mixture, this analysis could only be applied to the few fractions that contained one or two dominant peptides. Declining PTH (phenylthiohydantoin)–amino acid yields made it difficult to determine the exact number of residues in several peptides.
We have applied microcapillary HPLC–electrospray ionization–tandem mass spectrometry to circumvent the above problems. In a matter of hours, this technique determines the molecular mass and therefore maximum length of each peptide component, and the approximate number and quantity of individual peptides. Sequence information can be also obtained on subpicomolar amounts of peptides. We analyzed the naturally processed peptides bound to HLA-A2.1, one of the most widely distributed class I molecules within the human population. The three-dimensional structure of this molecule allows modeling of the complex (2).
HLA-A2.1 molecules were purified by immunoprecipitation from the human B lymphoblastoid cell line C1R-A2.1. The associated peptides were released by acid extraction and separated from HLA-A2.1 and antibody by filtration. A “mock” extract was prepared from an equal number of Hmy2.C1R (C1R) cells, which do not express any HLA-A or -B products (13). Since C1R-A2.1 is a transfectant of C1R, these cell lines should be identical except for the expression of HLA-A2.1. Extracts were fractionated by microcapillary reversed-phase HPLC and eluted directly into the electrospray ion source of a triple quadrupole mass spectrometer (14). A broad band of material was observed in the extract from C1R-A2.1 (Fig. 1A), but not in the extract from C1R (Fig. 1B). If all HLA-A2.1 molecules contain bound peptide, the total peptide yield from 108 C1R-A2.1 cells should be roughly 20 pmol; we estimate that our total yield of peptide was about 70 to 80% of the amount expected. This provides evidence that the material isolated for C1R-A2.1 cells by this method is specifically associated with the HLA-A2.1 molecule.
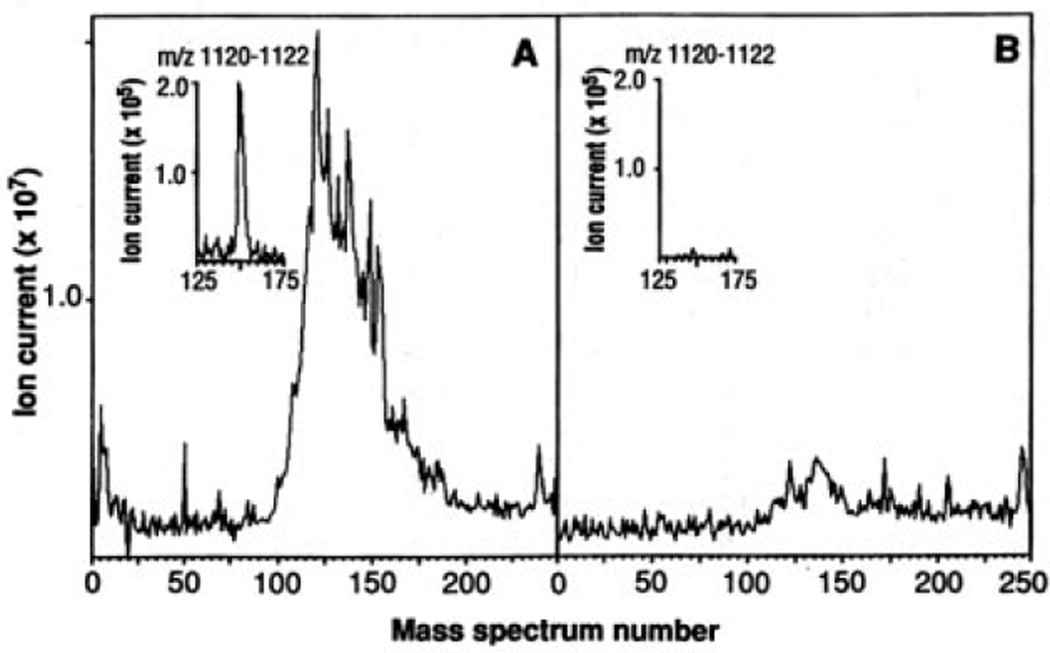
Fig. 1.
Total ion chromatograms of HLA-A2.1–associated peptides. C1R-A2.1 (A) or C1R (B) cells (2 × 109) were solubilized in 20 mM tris, pH 8.0, 150 mM NaCl, 1% NP-40, 2 mM PMSF, 100 µM iodoacetamide, Aprotinin (5 µg/ml), Leupeptin (10 µg/ml), Pepstatin A (10 µg/ml), EDTA (3 ng/ml), and 0.2% sodium azide. After sample centrifugation at 100,000g for 1 hour, supernatants were immunoprecipitated with the HLA-A2.1–specific monoclonal antibody BB7.2 (30) and protein A–Sepharose (Sigma). Precipitates were extracted with 0.2 N acetic acid. The supernatants were brought to pH 2.1 with glacial acetic acid and boiled for 5 min, centrifuged through an Ultrafree-Cl unit (millipore) with a norminal limit of 5 KD, and concentrated by vacuum centrifugation. Mass spectra were recorded on a Finnigan-MAT TSQ-70 (san Jose, California) triple quadrupole mass spectrometer equipped with an electrospray ion source and a C-18 microcapillary-HPLC column (75 µm by 10 cm) (14). The column was eluted with a gradient of 0 to 80% acetonitrile in 0.5% acetic acid over 10 min at 1 to 2 µl/min. The electrospray needle was operated with a voltage differential of 4 to 5keV and a sheath flow of 2 to 4 µl/min of 3:1 mixture of methanol:0.5% acetic acid. Total ion signal from samples equivalent to 108 cells was obtained by scanning the range of masses corresponding to m/z values between 300 and 1500 every 2 s and then summing each of the spectra. Insets are plots of ion current for the peptide (M+H)+ ions at m/z 1121 (nominal mass).
Material with a nominal mass-to-charge ratio (m/z) of 1121 was present in the extract from C1R-A2.1 cells but not that from C1R cells (Fig. 1 insets). By a similar analysis, approximately 200 distinct ions having a signal-to-noise ratio (S/N) of at least 2 were observed in the material eluted from C1R-A2.1 cells. Of these, 47% were singly charged, 45% were doubly charged, and 8% were triply charged. Calculated molecular masses ranged between 737 and 1370. Known quantities of synthetic peptides added to cell extracts allowed quantitation of peptides, and defined the lower limit of detection (S/N = 2) as 30 fmol. Of the 200 peptides detected, 10% were present at 150 to 600 fmol per 108 cells, whereas 90% were present at 30 to 150 fmol. By this measure, individual peptides in the mixture represent between 0.08 and 1.2% of the total peptide bound. Depending on the cell, this would correspond to as few as 100 complexes per cell or as many as 3000. These numbers are in accord with estimates of the number of peptide-MHC complexes required to stimulate T cells (12, 15, 16). However, 67 of the 200 peptides detected carried two-thirds of the ion current observed for this group. Thus, HLA-A2.1 seems to be preferentially occupied by a relatively small set of peptides. On the other hand, the number of peptides detected accounts for only 50% of the total ion current. If the remaining undetected peptides are all present at amounts near that of the background, the total number of different peptides presented by HLA-A2.1 could easily exceed 1000.
Table 1 shows a partial list of data obtained on peptides chosen at random from the more than 200 species observed. Fourteen of the peptides were in extracts of immunoprecipitates from both C1R-A2.1 and another HLA-A2.1-expressing cell line, JY, but not in extracts from C1R cells. Five others afforded relatively weak signals in extracts from the JY cells but were undetected in extracts from either C1R-A2.1 or CIA cells. To obtain sequence information, (M+H)+ and (M+2H)2+ ions were subjected to collision-activated dissociation (CAD) on the triple quadrupole mass spectrometer (17). Representative results for the (M+H)+ ion at m/z 1121 (peptide 19) are shown in Fig. 2. Complete sequences for 8 peptides in Table 1 have been obtained; partial sequences have been obtained on 11 others. Each of the completely sequenced peptides is a nonarmer and contains either Leu or Ile at position 2. Since Ile and Leu are of identical mass, they cannot be differentiated on the triple quadrupole instrument. Leu was considered a “dominant residue” at position 2 in unfractionated peptide mixtures isolated from HLA-A2.1 (8). For peptide 19, a definitive assignment of Leu at position 2 was established by adding synthetic peptides containing Leu or Ile at this position to the cell extract, and demonstrating that only the Leu-containing peptide coeluted from a microcapillary HPLC column with the naturally processed species. For the 19 peptides for which COOH-terminal information is available, all end in residues with aliphatic hydrocarbon side chains: nine terminate in Val, eight in Leu/Ile, and two in Ala [Table 1 and (18)]. Only Val was categorized as “dominant” based on the pool sequencing technique (8). No other residues were present at any position in more than 50% of the peptides. None of the sequences in Table 1 contains Met at position 2, Lys at position 4, Val at position 6, or Lys at position 8, assignments that were considered “strong” in analysis of pooled peptides (8).
Table 1.
Partial listing of data for peptides extracted from HLA-A2.1. HLA-A2.1 was purified and acid extracts prepared from the cell lines JY, C1R-A2.1, and C1R, as outlined in Fig. 1. Aliquots corresponding to 108 cells were analyzed by microcapillary HPLC–electrospray ionization–mass spectrometry. Signals represent measured ion current. Synthetic peptides corresponding to sequences S and 14, doped at 1 pmol into samples, gave signals of 60 × 105 to 70 × 105, whereas background noise gave a signal of 1 × 105. Sequences were deduced as in Fig. 2.
| Peptide | (M+H)+ (m/z) |
JY | C1R-A2.1 | C1R | Sequence* | Database match |
||
|---|---|---|---|---|---|---|---|---|
| Signal (×105) |
Yield (fmol) |
Signal (×105) |
Yield (fmol) |
Signal (×105) |
||||
| 1 | 786 | 9.1 | 137 | 2.0 | 30 | -† | SXPSGGXGV | |
| 2 | 868 | 2.7 | 41 | 7.9 | 119 | - | ||
| 3 | 871 | 2.9 | 44 | 6.1 | 98 | - | ||
| 4 | 898 | 11.9 | 179 | 15.8 | 237 | - | XXDVPTAAV | LLDVPTAAV |
| 5 | 903 | 4.7 | 71 | 11.8 | 177 | - | ||
| 6 | 930 | 4.9 | 74 | - | - | - | GXVPFXVSV | |
| 7 | 945 | 11.8 | 177 | 3.1 | 47 | - | ||
| 8 | 954 | 11.9 | 179 | 2.5 | 38 | - | SXXPAXVEX | SLLPAIVEL |
| 9 | 971 | 8.5 | 128 | 4.1 | 66 | - | ||
| 10 | 994 | 6.2 | 93 | - | - | - | ||
| 11 | 999‡ | 3.5 | 53 | - | - | - | SXXVRAXEV | |
| 12 | 1018‡ | 4.3 | 65 | 2.5 | 38 | - | ||
| 13 | 1037‡ | 10.0 | 150 | 7.3 | 110 | - | KXNEPVXXX | |
| 14 | 1038‡ | 39.4 | 591 | 8.1 | 122 | - | YXXPAXVHX | YLLPAIVHI |
| 15 | 1064 | 8.1 | 122 | 3.5 | 53 | - | ||
| 16 | 1095 | 5.2 | 78 | - | - | - | ||
| 17 | 1098 | 2.3 | 35 | - | - | - | ||
| 18 | 1115 | 11.4 | 171 | 2.9 | 44 | - | ||
| 19 | 1121 | 6.4 | 320§ | 2.0 | 100§ | - | TLWVDPYEV | TLWVDPYEV |
Single letter abbreviations for the amino acid residues are: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Leu and Ile are not distinguishable by tandem mass spectrometry, and are designated X.
Signal was at or below background.
Observed as a double charged, (M+H)2+ ion at half of this m/z value.
Signal obtained from 1 pmol of this synthetic standard added into sample was 2 × 106.
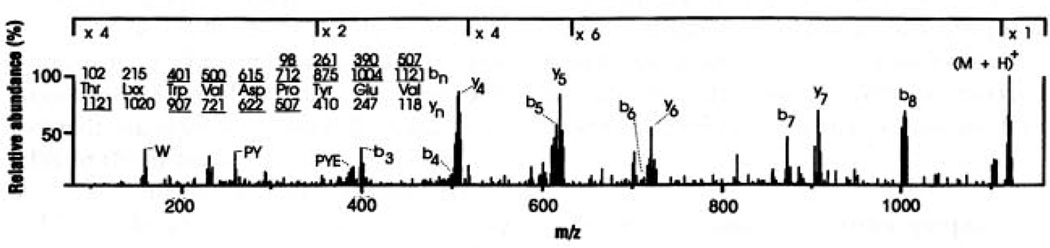
Fig. 2.
CAD mass spectrum of peptide (M+H)+ ions at m/z 1121 using 100 to 300 fmol of material derived from 2 × 108 cells. Predicted masses for fragment ions of types b and y (17) are shown, respectively, above and below the deduced amino acid sequence. Ions observed in the spectrum are underlined. Subtraction of m/z values for any two fragments that differ by a single amino acid generates a value that specifies the mass and thus the identity of the extra residue in the larger fragment. Residues 5 to 9 were deduced from fragments of type b, whereas residues 3 to 5 were defined by fragments of type y. CAD spectra recorded on additional aliquots of the sample mixture that had either been subjected to a single cycle of Edman degradation (17) or treated with methanolic-HCl to convert peptides to their corresponding methyl esters (17) confirmed the order of the first two amino acids as Thr and Leu/Ile, and the presence and location of the two acidic residues in the peptide. Total sample consumed in these analyses corresponded to the extract from 2 × 109 cells. Since Ile and Leu are of identical mass, they cannot be differentiated on the triple quadrupole instrument and are specified here as Lxx.
The length of the peptides bound to HLA-A2.1 is identical to that of peptides bound to FILA-B27 (9) and inferred for other class I molecules (12). This could reflect the mechanism by which these peptides were generated, or their ability to bind with particularly high affinity (3, 19). However, the motif for peptides bound to HLA-A2.1 is different from that of peptides bound to HLA-B27. Position 2 was also the most conserved in the peptides isolated from HLA-B27, but was an Arg (9). This residue is suggested to interact with negatively charged Glu45 in the B pocket of the HLA-B27 antigen binding site (3). In HLA-A2.1, this pocket is hydrophobic and is capable of accommodating a Leu or an Ile side chain (2). In addition, positively charged residues predominate at position 9 in peptides associated with HLA-B27, and this residue was suggested to interact with Asp116 in the F pocket. In HLA-A2.1, position 116 is a Tyr, rendering the F pocket smaller and more hydrophobic. Based on these considerations and the similarity of the electron density corresponding to peptide in these two molecules (2, 3), we suggest that interactions of Leu or Ile at position 2 with the B pocket, an aliphatic side chain at position 9 with the F pocket, and perhaps a nine-residue length are the primary determinants of peptide binding to HLA-A2.1.
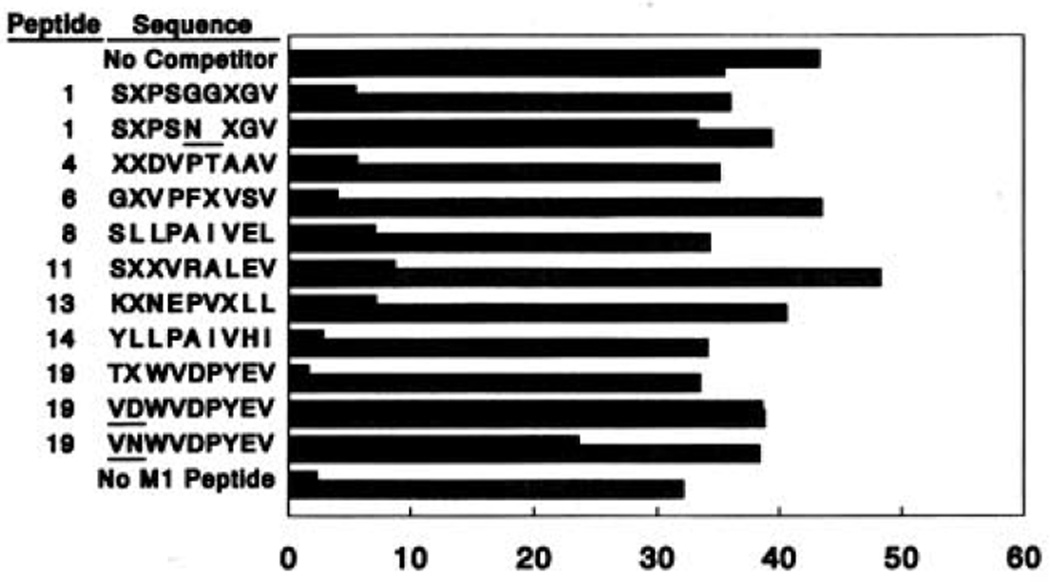
The eight completely sequenced peptides in Table 1 were synthesized in order to demonstrate that the peptides identified actually bound to HLA-A2.1. When no complete match to a sequence in the protein or gene databases was found (below), a mixture of Leu and Ile was incorporated at positions specified by X. Cells of the HLA-A2.1–expressing line JY were incubated with the M157–68 peptide of influenza A virus in the absence or presence of a 16- to 20-fold molar excess of one of the competitor peptides and assayed for their ability to be lysed by an M1 peptide–specific, HLA-A2.1–restricted CTL done. All eight peptides inhibited recognition under these circumstances but had no effect on recognition by a human alloreactive CTL line that is specific for HLA-A2.1 (Fig. 3). Thus, these synthetic peptides bind to HLA-A2.1 in vitro, which confirms their association with HLA-A2.1 in intact cells. Additional peptides were synthesized in which two Gly residues at positions 5 and 6 in peptide 1 were replaced by a single Asn to produce an octamer, or the Thr and Leu in positions 1 and 2 of peptide 19 were substituted with Val and Asn or Val and Asp. None of these peptides blocked recognition, indicating that a nine-residue length, the presence of Leu or Ile at position 2, and perhaps a polar residue at position 1 are important features for peptide binding.
Fig. 3.
Synthetic peptides corresponding to sequenced peptides bind to HLA-A2.1. Synthetic peptides are referenced by the numbers in Table 1. JY cells were incubated with the M157–68 peptide of influenza A (8 µg/ml), in the presence or absence of one of the synthetic peptides (200 µg/ml) as a competitor, in a total volume of 250 µl of RPMI 1640 containing 5% new-born calf serum and 50 µCi of51 Cr for 3 hours. Washed target cells (2 × 103) were incubated in 200 µl with either an M1 peptide–specific, HLA-A2.1–restricted CTL, done 5.2–11 (31) (filled bars) or an HEA-A2.1–specific human alloreactive CTL line (stippled bars) at an effector to target ratio of 1.25:1. Data are the means of duplicate samples with an SD of less than 5% and are representative of at least three independent experiments.
To identify possible precursor proteins for these peptides, gene and protein sequence databases were searched in a manner that allowed X residues to be assigned as either Leu or Ile. Only four peptides gave 100% matches with proteins in the library (Table 1). All of the sequences contain Leu at position 2, although Ile is often found at other positions. A match for peptide 8 occurs in the 65-kD α and β regulatory subunits of human protein phosphatase 2A and in a highly homologous human transformation-associated protein, p61. Both are cytoplasmic proteins and p61 represents only 0.01 to 0.02% of the total cellular protein (20). A match for peptide 14 appears in the human nuclear protein p68, which exhibits RNA-dependent adenosine triphosphatase activity and RNA unwinding activity (21). A different sequence from this protein is found in a peptide bound to HLA-B27 (9). A match for peptide 19 is found in TIS21, a protein of unknown function that is a member of the primary response group of genes induced by growth factors (22). Finally, a match for peptide 4 is found in the human protein IP-30, the expression of which is induced by gamma interferon (23). This precursor sequence is unique among all naturally processed peptides described to date in that it is located in the signal peptide domain of the protein. This peptide is also bound to HLA-A2.1 molecules expressed on CEMx721.174(T2) (24), a mutant cell line defective in the normal antigen processing pathway (6, 7). This result suggests a second pathway through which peptides can enter the endoplasmic reticulum and associate with MHC class I molecules.
Peptides derived from the processing of normal cellular proteins located in the cytoplasm have been implicated in the correct folding and intracellular transport of class I molecules (6, 7) and in the epitopes recognized by alloreactive T cells (25–28). The use of tandem mass spectrometry for the direct analysis of these peptides has enhanced our knowledge of peptide-MHC interactions and the cellular processes that regulate formation of these complexes and should allow the direct identification of peptides that are alloreactive T cell epitopes. Peptides associated with class II MHC molecules can also be characterized by the above approach (29). This same methodology should also facilitate identification of peptide antigens associated with viral infection, cellular transformation, and autoimmunity.
Acknowledgments
Supported by USPHS grants GM37537 (to D.F.H) and AI20963 (to V.H.E) and by National Science Foundation predoctoral fellowship RCD 9154717 (to N.S.).
Contributor Information
Donald F. Hunt, Department of chemistry, University of Virginia, Charlottesville, VA 22903
Robert A. Henderson, Department of Microbiology and Beirne Carter Center for Immunological Research, University of Virginia School of Medicine, Charlottesville, VA 22908
Jeffrey Shabanowttz, Department of Chemistry, University of Virginia, Charlottesville, VA 22903.
Kazuyasu Sakaguchi, Laboratory of Cell Biology, National Cancer Institute, NIH, Bethesda, MD 20892.
Hanspeter Michel, Department of Chemistry, University of Virginia, Charlottesville, VA 22903.
Noelle Sevilir, Department of Microbiology and Beirne Carter Center for Immunological Research, University of Virginia School of Medicine, Charlottesville, VA 22908.
Andrea L. Cox, Department of Chemistry, University of Virginia, Charlottesville, VA 22903
Ettore Appella, Laboratory of Cell Biology, National Cancer Institute, NIH, Bethesda, MD 20892.
Victor H. Engelhard, Department of Microbiology and Beirne Carter Center for Immunological Research, University of Virginia School of Medicine, Charlottesville, VA 22908
REFERENCES AND NOTES
- 1.Townsend A, Bodner H. Anau. Rev. Immunol. 1989;7:601. doi: 10.1146/annurev.iy.07.040189.003125. [DOI] [PubMed] [Google Scholar]
- 2.Saper MA, Bjorkman PJ, Wiley DC. J. Mol. Biol. 1991;219:277. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 3.Madden DR, Gorga JC, Strominger JL, Wiley DC. Nature. 1991;353:321. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- 4.Morrison LA, et al. J. Exp. Med. 1986;163:903. doi: 10.1084/jem.163.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore MW, et al. Cell. 1988;54:777. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 6.Cerundolo V, et al. Nature. 1990;345:449. doi: 10.1038/345449a0. [DOI] [PubMed] [Google Scholar]
- 7.Spies T, et al. ibid. 1990;348:744. doi: 10.1038/348744a0. [DOI] [PubMed] [Google Scholar]
- 8.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee H-G. ibid. 1991;351:290. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 9.Jardetsky TS, Lane WS, Robinson RA, Madden DR, Wiley DC. ibid. 1991;353:326. doi: 10.1038/353326a0. [DOI] [PubMed] [Google Scholar]
- 10.van Bleek GM, Nathenson SG. ibid. 1990;348:213. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- 11.Rotzschke O, et al. ibid. :252. [Google Scholar]
- 12.Falk K, et al. J. Exp. Med. 1991;174:425. doi: 10.1084/jem.174.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storkus WJ, et al. J. Immunol. 1987;138:1657. [PubMed] [Google Scholar]
- 14.Hunt DF. In: Techniques in Protein Chemistry II. Villafranca JJ, editor. New York: Academic Press; 1991. pp. 441–454. [Google Scholar]
- 15.Demotz S, Grey HM, Sette A. Science. 1990;249:1028. doi: 10.1126/science.2118680. [DOI] [PubMed] [Google Scholar]
- 16.Christnick ER, Luscher MA, Barber BH, Williams DB. Nature. 1991;352:67. doi: 10.1038/352067a0. [DOI] [PubMed] [Google Scholar]
- 17.Hunt DF, et al. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6233. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt DF, Michel H, Shabonowitz J, Cox A. unpublished data. [Google Scholar]
- 19.Cerundolo V, et al. Eur. J. Immunol. 1991;21:2069. doi: 10.1002/eji.1830210915. [DOI] [PubMed] [Google Scholar]
- 20.Walter G, Ferre F, Espiritu O, Carbone-Wiley A. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8669. doi: 10.1073/pnas.86.22.8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirling H, Scheffner M, Restle T, Stahl H. Nature. 1989;339:562. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher BS, et al. J. Biol. Cheat. 1991;266:14511. [PubMed] [Google Scholar]
- 23.Luster AD, Weinshank RL, Feinman R, Ravetch JV. ibid. 1988;263:12036. [PubMed] [Google Scholar]
- 24.Henderson RA, et al. Science. 1992;255:1264. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 25.Heath WR, Sherman LA. Eur. J. Immunol. 1991;21:153. doi: 10.1002/eji.1830210123. [DOI] [PubMed] [Google Scholar]
- 26.Heath WR, et al. Proc. Natl. Acad. Sci. U.S.A. 1991;88:5101. [Google Scholar]
- 27.Man S, Salter RD, Engelhard VH. Int. Immunol. doi: 10.1093/intimm/4.3.367. in press. [DOI] [PubMed] [Google Scholar]
- 28.Crumpacker DB, Alexander J, Cresswell P, Engelhard VH. preparation [Google Scholar]
- 29.Hunt DF, et al. preparation [Google Scholar]
- 30.Parham P, Brodsky FM. Hum. Immunol. 1981;3:277. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 31.Engelhard VH, Lacy E, Ridge JP. J. Immunol. 1991;146:1226. [PubMed] [Google Scholar]