Abstract
Rapid growth in electronic health records (EHRs) use has led to an unprecedented expansion of available clinical data in electronic formats. However, much of the important healthcare information is locked in the narrative documents. Therefore Natural Language Processing (NLP) technologies, e.g., Named Entity Recognition that identifies boundaries and types of entities, have been extensively studied to unlock important clinical information in free text. In this study, we investigated a novel deep learning method to recognize clinical entities in Chinese clinical documents using the minimal feature engineering approach. We developed a deep neural network (DNN) to generate word embeddings from a large unlabeled corpus through unsupervised learning and another DNN for the NER task. The experiment results showed that the DNN with word embeddings trained from the large unlabeled corpus outperformed the state-of-the-art CRF’s model in the minimal feature engineering setting, achieving the highest F1-score of 0.9280. Further analysis showed that word embeddings derived through unsupervised learning from large unlabeled corpus remarkably improved the DNN with randomized embedding, denoting the usefulness of unsupervised feature learning.
Keywords: Clinical natural language processing, Named entity recognition, Neural network, Deep learning, Chinese clinical text
Introduction
The wide use of health information technologies has led to an unprecedented expansion of electronic health record (EHR) data. EHR data have been used not only to support operational tasks in clinical practice (e.g., clinical decision support systems), but also to enable clinical and translational research. However, much of the important patient information is dispersed in narrative clinical documents, which are not directly accessible for computerized applications that rely on structured data. Therefore, clinical natural language processing (NLP) technologies, which can extract important patient information from narrative clinical text, have been introduced to the medical domain and have demonstrated great utility in many applications [1].
Originating from the Sixth Message Understanding Conference (MUC-6) [2], Named Entity Recognition (NER), which aims to identify boundaries and types of entities in text, has been one of the well established and extensively investigated tasks in NLP. In the medical domain, NER for important clinical concepts (e.g., problems, treatments, or lab tests) is also a fundamental step for many clinical NLP systems. Many existing clinical NLP systems use dictionaries and rule-based methods to identify clinical concepts, such as MedLEE [3] - one of the earliest and most comprehensive clinical NLP system developed by Carol Friedman et al. at Columbia University, the MetaMap system [4, 5] - a general biomedical NLP system developed by Aronson et al. at National Library of Medicine, as well as cTAKES [6] - an open source comprehensive clinical NLP system developed based on the Unstructured Information Management Architecture (UIMA) framework and OpenNLP natural language processing toolkit. More recently, a number of challenges on NER involving shared tasks in clinical text have been organized, including the 2009 i2b2 (the Center of Informatics for Integrating Biology and the Bedside) challenge [7] on medication recognition, the 2010 i2b2 challenge [8] on recognizing medical problems, treatments, and tests entities, 2013 Share/CLEF challenge [9, 10] on disorder mention recognition and normalization, and the 2014 Semantic Evaluation (SemEval) challenge on disorder mention recognition and normalization. For the 2009 i2b2 challenge on medication recognition, although different approaches including rule-based methods, machine learning (ML)-based methods, as well as hybrid methods have been reported, seven out of the top ten ranked systems were rule-based systems that rely on existing biomedical vocabularies to identify medication concepts [7], such as the MedEx system developed at Vanderbilt University [11]. At the 2010 i2b2 challenge for recognizing problems, treatments and lab tests, the organizers provided a large annotated corpus. Therefore, many participating teams including all the top five systems used ML-based methods [12-14]. Many state-of-the-art clinical NER systems implement supervised ML algorithms, such as Conditional Random Fields (CRFs) [15] and Structural Support Vector Machines (SSVMs) [16], with extensive investigation on comprehensive features [17].
In addition to previous NER work on clinical text written in English, there is a growing interest in studying NER of clinical text written in Chinese. With the rapid growth of EHRs implemented in China, there is an urgent need to extract important patient information from Chinese clinical text, to accelerate clinical research in China. Conventional ML-based methods have been applied to Chinese clinical NER tasks. Wang et al. [18] conducted a study using CRF, support vector machines (SVM), and maximum entropy (ME) to recognize symptoms and pathogenesis in Chinese medical records. Another study by Wang et al. [19] investigated CRF and different feature sets for recognizing symptom names from clinical notes of traditional Chinese medicine. In 2004, Xu et al. [20] proposed a joint model that integrates segmentation and NER simultaneously to improve the performance of both tasks in Chinese discharge summaries. A more recent work by Lei et al. [21] compared different machine learning algorithms and various types of features for NER in Chinese admission notes and discharge summaries. In summary, current efforts on NER in Chinese clinical text primarily focus on investigating different machine learning algorithms or optimizing combinations of different types of features via human engineering.
Recently, there is increasing interest in designing deep learning based NLP systems that could automatically learn useful feature representations from large-scale unlabeled corpora through unsupervised learning [22-24]. Deep learning [25, 26] is a research area of machine learning that can learn high-level feature representations by designing deep neural networks. It has achieved state-of-the-art performances in a number of different applications across multiple domains, such as image processing [27], automatic speech recognition [28] and machine translation [29]. Instead of spending a great amount of time on selecting task-specific features, NLP researchers have developed deep neural networks (DNNs) to automatically learn useful features from vast amounts of unlabeled data. Researchers have shown that deep learning approaches can learn useful linguistic features as well as capture semantic meanings through word embedding [30], hence improving the performances of a number of NLP tasks in general English domain. A DNN-based system developed by Dr. Ronan Collobert [23] successfully achieved the state-of-the-art performances on a number of NLP tasks, including POS tagging, Chunking, NER and Semantic Role Labeling, using only one single deep neural network.
In this study, we propose to investigate the use of deep neural network in NER from Chinese clinical text. We developed a deep neural network (DNN) approach for NER in Chinese clinical text and compared it with the traditional CRF-based NER system at the minimal feature setting. To the best of our knowledge, this is the first study to investigate deep neural network in Chinese clinical NER.
Methods
Datasets
Two Chinese clinical corpora were used in this study. The first dataset is an annotated corpus from a previous study by Lei et al.[21], which contains 400 randomly selected admission notes from the EHR database of Peking Union Medical College Hospital in China. For each admission note, four types of clinical entities - problems, lab tests, procedures, and medications were annotated by following the annotation guidelines developed in the study. Details about creation of this dataset can be found in Lei et al. [21] In summary, a total of 24,433 problems, 2,171 procedures, 11,168 tests and 1,201 medications were annotated in 400 Chinese admission notes. We further divided the 400 admission notes into a training set of two-thirds (266) of the notes and a test set of one-third (134) of the notes. Table 1 shows the distribution of the clinical concepts among the training and test set.
Table 1.
Description of annotated Chinese admission notes
| Training | Test | |
|---|---|---|
| # Notes | 266 | 134 |
| # Sentences | 20,506 | 10,287 |
| # Characters | 277,701 | 139,885 |
| # Problems | 16,253 | 8,180 |
| # Procedures | 1,500 | 671 |
| # Tests | 7,414 | 3,754 |
| # Medication | 840 | 361 |
Another dataset, which includes 36,828 unlabelled admission notes (383 Megabytes in total) from the same institute in China, was used for learning word embeddings. Only minimal pre-processing steps (e.g., removing the empty lines) were applied to these notes. As we trained the embedding matrix using individual Chinese characters, word segmentation was not included in the preprocessing.
Experiments and evaluation
In this study, we compared three different NER approaches: 1) the traditional CRF-based NER method, as described in Lei et al. [21]; 2) a DNN-based NER method that uses a randomly initialized word embedding matrix; and 3) another DNN-based NER method that uses the word embedding matrix derived from the unlabelled corpus. All three models were trained using the training set and their performance on the test set are reported. Similar to other deep learning studies, we evaluated the three NER systems at the minimal feature setting, which uses only the word feature. The details of the CRFs model and the DNN models are introduced in the following subsections.
We used the standard micro-average precision, recall and F1-score to evaluate all the three NER systems. All scores were calculated using the Conll 2000 challenge official evaluation script1. Wilcoxon signed-ranks test was used to test the statistical significance between two classifiers.
CRF vs. DNN Appraoches
CRF-based NER
The CRFs model decodes the sequence labeling problem by undirected Markov chain and Viterbi algorithm with a training criteria of maximizing the likelihood estimation of conditional probability of the output variable y given the observation x. Here, the observations are the word and its context words in the sentence and the output is its label (e.g, B, I, or O: B - beginning of an entity; I - inside an entity; O - outside of an entity). CRFs was intrinsically designed for sequence labeling problem as it models the relationships between neighboring tokens in sequence. Thus, it has been widely used in various NER tasks and has achieved state-of-the-art performances in both open domains and the biomedical domain. Therefore, CRFs serves as a strong baseline for comparing with the DNN-based NER approaches. More specifically, we used CRF++ package, one of the most popular implementations of CRF (http://crfpp.googlecode.com/svn/trunk/doc/index.html). The parameters for CRFs were optimized using the training set. The details of the CRF-based approach implemented in this study can be found in Lei et al. [21].
DNN-based NER
Researchers have proposed different approaches of designing deep neural networks for NLP systems. In this research, we adopted one of the popular architectures from Dr. Ronan Collobert – the sentence level log-likelihood approach[23], which consists of a convolutional layer, a non-linear layer using the hard version of the hyperbolic tangent (HardTanh), and several linear layers. This structure has been widely used in various NLP tasks and achieved state-of-the-art performances. Figure 1 shows the DNN architecture as well as the propagate function for each layer.
Figure 1.
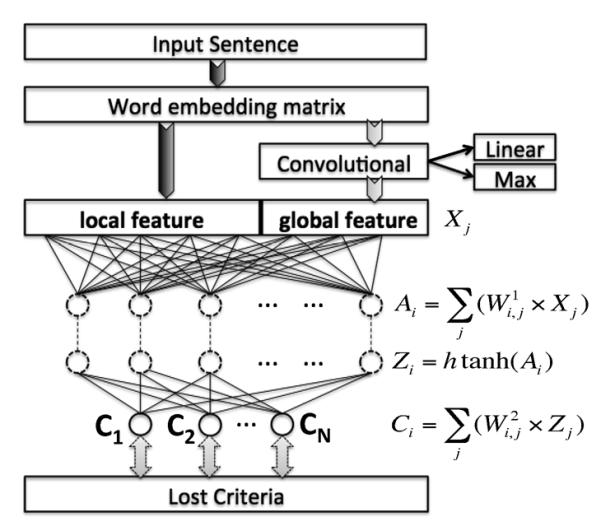
The sentence approach DNN
When calculating the classification score for a word, the context words within a specific window size W of the target word are taken as inputs. For the words near the beginning or the end of a sentence, a pseudo padding word will be used to form a fixed length input vector. Each word in the input window can be mapped to an N-dimension vector (N is the embedding dimension) using an embedding matrix. Then, a convolutional layer generates the global features represented as a number of global hidden nodes. Both the local features and the global features are then fed into a standard affine network trained using back propagation. The lost function is defined using the following sentence level log likelihood:
| (1) |
where, S(X,T) is the sentence level log-likelihood score that the sequence of tag T was asigned to the input sequemce X, H(Tt-1,Tt) is a global transition score from tag Tt-1 to tag Tt, and the DNN(Xt, Tt) is the score of asigning the tag Tt to an input word Xt assigned by the DNN.
Word embedding is a popular method to enrich the traditional bag-of-word representation through mapping the words into real value vectors. Previous research shows that the embedding space is more powerful than the one-hot representation (e.g., bag-of-words) as it conveys more semantic meanings. We adopted the ranking-based embedding method developed by Collobert. The ranking-based embedding treats a sequence of words naturally occurring in the free text as a positive sample. For example, we can form a fixed length positive sample X={wL2, wL1, w0, wR1, wR2} for each word in a sentence given the window size of 5, where, w0 is the target word, wR1 and wR2 are the right-side context words, wL1 and wL2 are the left-side context words. The embedding procedure will generate a negative sample X*={wL2, wL1, w*, wR1, wR2} by replacing the central word (w0) with another word (w*) that is randomly picked from the vocabulary and try to minimize the ranking criteria with respect to:
| (2) |
The DNN parameters were updated following the standard stochastic gradient descent, as shown in equation 3.
| (3) |
Where, λ is the learning rate and Δθ is the gradient.
We implemented two DNN-based NER approaches for Chinese clinical text. The first model starts with a random initialized word embedding matrix, which will then be updated during the back propagation training procedure. The other DNN model starts with the word embedding matrix derived from the unlabeled notes. The DNN parameters are tuned by splitting one-fifth of the training samples as a validation set using early stopping strategy. Following the work by Collobert [23], we fixed the learning rate at 0.01 and the word embedding dimension at 50. The hidden node number was set to 100, as we tested the numbers from 50 to 500 and noticed that 100 achieved the best performance on the validation set and no further significant improvement was observed when increasing the number of hidden nodes. The window sizes for training the word embedding and sequence labeling were fixed at 11 and 5, respectively. All DNN parameters were updated using the standard stochastic gradient descent through back propagation.
Results
Table 2 shows the performance on the test dataset for the CRFs, the DNN with random initialized word embedding, and the DNN with word embedding derived from the unlabeled corpus. All evaluation scores were based on exact-matching criteria. At the minimal feature setting, the baseline method (CRFs) achieved an F-1 score of 0.9197. The DNN with randomly initialized word embedding achieved an F1-score of 0.9071, which was not better than the baseline performance. The F-1 score of the DNN approach was remarkably improved to 0.9280 by using the word embedding derived from the unlabeled corpus. The best DNN-based NER system outperformed the CRFs by more than 0.8% on F1-score. The Wilcoxon test showed that the best DNN-based NER system outperformed the CRFs with a significant p-value of 1.832e-07. Table 3 shows the performance of the best DNN system for each entity type. The DNN system with word embeddings achieved the highest F1-score of 0.9489 for lab tests and the lowest F1-score of 0.8113 for procedures.
Table 2.
Performances of machine learning methods
| Precision | Recall | F1-score | |
|---|---|---|---|
| CRFs | 0.9265 | 0.9130 | 0.9197 |
| DNN | 0.9007 | 0.9136 | 0.9071 |
| DNN+Embedding | 0.9237 | 0.9321 | 0.9280 |
Table 3.
Performances for each entity type (DNN + embedding)
| Precision | Recall | F1-score | |
|---|---|---|---|
| Problems | 0.9267 | 0.9356 | 0.9311 |
| Procedures | 0.8119 | 0.8107 | 0.8113 |
| Medications | 0.8604 | 0.8366 | 0.8483 |
| Lab Tests | 0.9427 | 0.9553 | 0.9489 |
Discussion
In this study, we examined a novel deep learning method for NER tasks in Chinese clinical text. When only word feature was used, our DNN-based NER system that utilizes word embeddings derived from another unlabeled corpus achieved better performance than the traditional CRF-based NER system, indicating the potential of using DNN for clinical NER in Chinese clinical documents. To the best of our knowledge, this is the first study that investigated deep learning technologies for NER tasks in clinical documents written in Chinese.
We conducted further analysis of the DNN-based NER system and found that the performance improvement was from the semantic information automatically captured by the DNN-based word embeddings. Table 4 shows some examples of semantically related words captured through word embeddings. The nearest neighbors were derived by calculating the cosine similarity using word embeddings. Most of the neighbors in Table 4 are related to the target words, and such semantic relatedness was captured in the embedding space. However, they may not have any relation in the representation space like bag-of-words.
Table 4.
Examples of nearest neighbors captured in word embeddings
| 一 (one) |
左 (left) |
肢 (extremity) |
喉 (larynx) |
|---|---|---|---|
| 三(three) | 右(right) | 颌(jaw) | 颠(temporal) |
| 二(two) | 双(bilateral) | 月市(lung) | 局(local) |
| 半(half) | 两(double) | 臂(arm) | 鼻(nose) |
| 0 (zero) | 上(upper) | 舌(tongue) | 窦(sinus) |
| 两(double) | 并(accompany) | 壁(wall) | 腔(tibia) |
| 数(several) | 有(have) | 述(mentioned) | 睑(eyelid) |
| 有(have) | 0. | 午(noon) | 峡(isthmi) |
| 较(compare) | 前(front) | 显(show) | 脚(foot) |
| 0-0 | 枕(Occipital) | 颈(neck) | 涕(nasal mu- cus) |
| 每(each) | 下(lower) | 臀(buttocks) | 髋(hip) |
As NER is an extensively studied task in NLP, many types of features, including the orthographic information, the prefix and suffix, part-of-speech (POS) tags, syntax from parse trees, and existing knowledge bases, have been proved to be useful. On the same dataset, the state-of-the-art CRFs-based NER system that was optimized through manual feature engineering could achieve the best F1-score of 0.9353 [21], which was higher than the DNN-based NER developed in this study. However, the DNN-based NER approach uses word feature only and it requires a minimal effort on feature engineering. The simplicity of the DNN-based approach not only reduces human time on feature engineering, but also makes the implementation of NLP systems easier. For example, obtaining syntax features by parsing sentences could be time consuming and sometimes not feasible in certain real-time applications. However, by using DNN-based approaches, an NER system could still achieve reasonable performance even without running parsers to get the syntax features. Moreover, we believe that the unsupervised feature learning from large unlabeled corpora would significantly reduce NLP researchers’ time on task-specific feature engineering, thus enabling efficient development of NER systems for different tasks in various domains.
In the future, we plan to investigate different approaches to further improve the performance of DNN-based NER systems for Chinese clinical text. A straightforward approach is to increase the size of the unlabeled clinical corpus, as previous work has shown that the DNN model benefited from a larger unlabeled corpus. For example, Collobert et al. used the entire English Wikipedia and the Reuters corpus to derive word embeddings. Another appealing research direction is to integrate other linguistic or domain-specific features with the DNN model built on word embeddings to further improve the performance. A previous study by Wang et. al. [31] revealed that low dimension continuous space representation works significantly better in DNN than in the traditional systems (e.g., CRFs). However, high dimension discrete feature representation works better in the traditional systems than in the DNNs. Incorporating the high dimension discrete features into the deep neural network remains a challenge. Recent research from Ma et al. [32] showed that the performance of DNN based POS tagging system can be improved by combining a dense embedding feature based neural network with a discrete feature based neural network using a shared output layer. We will further investigate new methods that could combine the two types of feature representations in the future.
Conclusion
In this study, we investigated deep neural network for NER from Chinese clinical text. Our results showed that DNN outperformed CRFs at the minimal feature setting, achieving the highest F1-score of 0.9280. Further analysis showed that the performance improvement was from the semantic information automatically captured by the DNN-based word embeddings, indicating the usefulness of unsupervised feature learning.
Acknowledgments
This study was supported by grant from the NLM R01LM010681-05 and the National Natural Science Foundation of China 81171426.
Footnotes
References
- [1].Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- [2].Grishman R, Sundheim B. Message Understanding Conference-6: A Brief History. pp. 466–471.
- [3].Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994 Mar-Apr;1(no. 2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- [5].Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010 May-Jun;17(no. 3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Savova Gk Fau - Masanz JJ, Masanz Jj Fau - Ogren PV, Ogren Pv Fau - Zheng J, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. no. 1527-974X (Electronic), 20100907 DCOM- 20101115. [DOI] [PMC free article] [PubMed]
- [7].Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010 Sep-Oct;17(no. 5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Uzuner O, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011 Sep-Oct;18(no. 5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF eHealth Evaluation Lab 2013. In: Forner P, Müller H, Paredes R, et al., editors. Information Access Evaluation. Multilinguality, Multimodality, and Visualization. Springer; Berlin Heidelberg: 2013. pp. 212–231. Lecture Notes in Computer Science. [Google Scholar]
- [10].Pradhan S, Elhadad N, South BR, et al. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J Am Med Inform Assoc. 2014 Aug 21; doi: 10.1136/amiajnl-2013-002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Jan-Feb;17(no. 1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].de Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc. 2011 Sep-Oct;18(no. 5):557–62. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc. 2011 Sep-Oct;18(no. 5):601–6. doi: 10.1136/amiajnl-2011-000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assoc. 2011 Sep-Oct;18(no. 5):580–7. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. 2001. [Google Scholar]
- [16].Tsochantaridis I, Joachims T, Hofmann T, et al. Large margin methods for structured and interdependent output variables. pp. 1453–1484. [Google Scholar]
- [17].Tang B, Cao H, Wu Y, et al. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med Inform Decis Mak. 2013;13(Suppl 1):S1. doi: 10.1186/1472-6947-13-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang S.-k., Li S.-z., Chen T.-s. Recognition of Chinese Medicine Named Entity Based on Condition Random Field. J Xiamen University (Natural Science) 2009;48:349–64. [Google Scholar]
- [19].Wang Y, Liu Y, Yu Z, et al. A preliminary work on symptom name recognition from free-text clinical records of traditional chinese medicine using conditional random fields and reasonable features. Proceedings of the 2012 Workshop on Biomedical Natural Language Processing. 2012:223–230. [Google Scholar]
- [20].Xu Y, Wang Y, Liu T, et al. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. J Am Med Inform Assoc. 2014 Feb;21(no. e1):e84–92. doi: 10.1136/amiajnl-2013-001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lei J, Tang B, Lu X, et al. A comprehensive study of named entity recognition in Chinese clinical text. J Am Med Inform Assoc. 2014 Sep-Oct;21(no. 5):808–14. doi: 10.1136/amiajnl-2013-002381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Turian J, Ratinov L, Bengio Y. Word representations: a simple and general method for semi-supervised learning. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics; Uppsala, Sweden. 2010.pp. 384–394. [Google Scholar]
- [23].Collobert R, Weston J, et al. Natural Language Processing (Almost) from Scratch. J. Mach. Learn. Res. 2011;12:2493–2537. [Google Scholar]
- [24].Socher R, Chen D, Manning CD, et al. Reasoning With Neural Tensor Networks for Knowledge Base Completion. Advances in Neural Information Processing Systems. 2013;26:926–934. [Google Scholar]
- [25].Jones N. Computer science: The learning machines. Nature. 2014 Jan 9;505(no. 7482):146–8. doi: 10.1038/505146a. [DOI] [PubMed] [Google Scholar]
- [26].Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(no. 5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- [27].Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012:1097–1105. [Google Scholar]
- [28].Hinton G, Deng L, Yu D, et al. Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. Signal Processing Magazine, IEEE. 2012;29(no. 6):82–97. [Google Scholar]
- [29].Deselaers T, Hasan S, Bender O, et al. A deep learning approach to machine transliteration. Proceedings of the Fourth Workshop on Statistical Machine Translation.2009. pp. 233–241. [Google Scholar]
- [30].Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781. 2013 [Google Scholar]
- [31].Wang M, Manning CD. Effect of Non-linear Deep Architecture in Sequence Labeling. Proceedings of the Sixth International Joint Conference on Natural Language Processing.2013. pp. 1285–1291. [Google Scholar]
- [32].Ma J, Zhang Y, Zhu J. Tagging The Web: Building A Robust Web Tagger with Neural Network. Association for Computational Linguistics (ACL) 2014:144–154. [Google Scholar]


