SUMMARY
The Hippo/YAP signaling pathway is a crucial regulator of tissue growth, stem cell activity and tumorigenesis. However, the mechanism by which YAP controls transcription remains to be fully elucidated. Here, we utilize global chromatin occupancy analyses to demonstrate that robust YAP binding is restricted to a relatively small number of distal regulatory elements in the genome. YAP-occupancy defines a subset of enhancers and super-enhancers with the highest transcriptional outputs. YAP modulates transcription from these elements predominantly by regulating promoter-proximal Polymerase II (PolII) pause release. Mechanistically, YAP interacts and recruits the Mediator complex to enhancers, allowing the recruitment of the CDK9 elongating kinase. Genetic and chemical perturbation experiments demonstrate the requirement for Mediator and CDK9 in YAP-driven phenotypes of overgrowth and tumorigenesis. Our results here uncover the molecular mechanisms employed by YAP to exert its growth and oncogenic functions, and suggest strategies for intervention.
INTRODUCTION
At the core of the Hippo pathway, a network of kinases controls the subcellular localization of the transcriptional co-activator YAP, and its paralogue TAZ (Ramos and Camargo, 2012). YAP/TAZ can translocate into the nucleus to activate gene expression by associating with a number of DNA-binding transcription factors, particularly of the TEAD family (Zhao et al., 2008). Ablation of Hippo signaling or forced expression of constitutively active YAP results in increased organ size, expansion of tissue progenitor compartments, and ultimately, occurrence of cancers (Benhamouche et al., 2010; Camargo et al., 2007; Dong et al., 2007; Zhou et al., 2009). The remarkable effects of Hippo/YAP on organ growth and its potential to crosstalk with multiple oncogenic signaling cascades(Barry et al., 2013; Kapoor et al., 2014), make this pathway a very attractive target for cancer therapeutics. However, despite the tremendous importance of YAP/TAZ in developmental and disease biology, major gaps in our mechanistic understanding of the transcriptional function of YAP remain. For instance, it is still unclear where YAP, TAZ and TEADs bind in the genome of cancer cells, as is the identity of the transcriptional complexes that are recruited by these factors to drive gene transcription.
The transcriptional state of a specific cell type is determined by the wiring of transcription factor networks that occupy a variety of genomic elements dispersed throughout the non-coding area of the genome. Such elements are able to impede (named insulators)(Wendt et al., 2008), or to drive transcription (named enhancers) of distal genes by recruiting a number of cell type specific transcription factors(Calo and Wysocka, 2013). Enhancers are known to be the major determinants of cell-type and cancer-type specific gene expression program(Akhtar-Zaidi et al., 2012; Heintzman et al., 2009). More recently, a very restricted subset of enhancers, named “super-enhancers” has been shown to drive higher transcription rate of genes that define cell identity(Hnisz et al., 2013), and of key oncogenic drivers in cancer cells (Loven et al., 2013).
Mediator is a large multi-subunit complex, which binds to enhancer elements and, together with the structural complex cohesin, is involved in bringing distal elements in close proximity to target promoters(Kagey et al., 2010). Mediator is able to regulate both basal transcription driven by RNA Polymerase II as well as recruitment of CDK9 to boost transcriptional elongation by releasing Pol II promoter pausing (Malik and Roeder, 2010). Such function, coupled to the binding of Mediator to a variety of cell-type specific transcription factors allows Mediator to integrate multiple signaling cues to deliver appropriate transcriptional activation (Malik and Roeder, 2010).
Here, we utilize multiple genomic technologies and biochemistry to provide a mechanistic insight into YAP/TAZ-driven transcription. We demonstrate, that in cancer cells, YAP/TAZ occupy a very restricted number of TEAD positive enhancers and super-enhancers that drive very high transcriptional activity. We find that YAP/TAZ predominantly control transcription of their targets by modulating elongation of paused PolII at promoter regions. Additionally, we show that YAP/TAZ recruit the Mediator complex in order to promote CDK9-dependent transcriptional activity at target sites. Further, we provide evidence demonstrating that the YAP-Mediator interaction and CDK9 activity are necessary for YAP-induced liver proliferation and cancer growth. Our data provide a molecular mechanism behind YAP-driven cell growth and tumorigenesis, and highlights transcriptional control as a potential therapeutic strategy for YAP-driven cancers.
RESULTS
YAP predominantly binds to a restricted number of putative enhancer elements
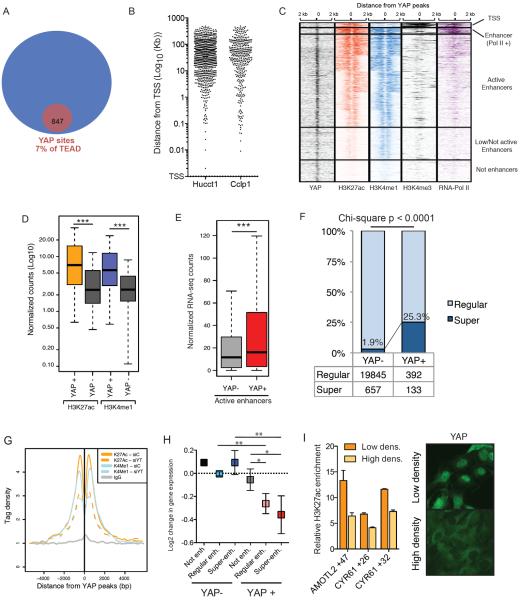
In order to characterize the molecular functions of YAP in transcriptional regulation, we sought to analyze its genome-wide occupancy by ChIP-seq. We chose to study liver cancer cell lines given the spectacular effects of YAP on liver growth and the general dependency of liver tumors on YAP activity(Camargo et al., 2007; Dong et al., 2007). We optimized and performed ChIP-seq for YAP using antibodies against the endogenous protein (Fig. S1A), and also against an epitope-tagged protein expressed at sub-endogenous levels (Fig. S1A, B). This analysis was done in two human cholangiocarcinoma lines (HuCCT1 and Cclp1). We also performed ChIP-seq for the two predominantly expressed members of the TEAD family (TEAD1 and TEAD4) (Fig. S1C). Our strategy (see Methods) identified 847 and 389 high-confidence binding sites for YAP in HuCCT1 and Cclp1 cells, respectively (Fig. S1D–S1G). Unexpectedly, such binding sites represented only a very small fraction (7% and 4%, respectively) of TEAD occupancy in the genome (Fig. 1A, S1H–I). We also optimized ChIP-Seq for TAZ, which revealed redundant patterns of occupancy with YAP (Fig. S1J). Thus, robust YAP/TAZ occupancy is associated only with a very small subset of TEAD binding sites (Fig. 1A and S1I), in contrast to the widespread co-binding previously reported (Zhao et al., 2008).
Figure 1. YAP binds to potent enhancers.
A) Venn diagram of the common TEAD1/4 and YAP peaks in HuCCT1 cells. B) Scatter plot of the distance distribution of YAP peaks from the closest TSS in the indicated cell lines. C) Heatmap for the ChIP-seq signal of the indicated antibodies +/−2Kb from the center of YAP peaks. Clustering results from K-means method. D) Box plots of the normalized counts of H3K27ac and H3K4me1 signal at YAP+ enhancer peaks or YAP− enhancers. *** p<0.0001. E) Boxplot indicating expression levels of genes associated to YAP+ or YAP− enhancers following RNAseq in HUCCT1 cells. F) Histogram indicating the fraction of YAP− or YAP+ enhancers catalogued as regular o super- enhancers in HuCCT1 cells. G) Line plots depicting H3K27Ac and H3K3me1 average signal in YAP+ enhancers upon siYT treatment. H) Log2 changes in gene expression between siControl / siYT treated cells for genes associated with different genomic elements 48h post transfection. Error bars represent 95% CI. I) Left panel, ChIP-qPCR for H3K27ac in selected YREs in cells at low and high density. Error bars = SD. Right panel, immunofluorescence for YAP in these conditions. See also Figure S1.
YAP transcriptional functions have been previously associated to the binding of TEADs around the promoter of target genes in embryonic stem cells(Lian et al., 2010). Similarly, studies in Drosophila have demonstrated promoter-enriched binding for the YAP orthologue, Yorkie(Oh et al., 2013). Yet surprisingly, annotation of YAP target loci revealed that YAP binds predominantly 20 Kilobases (Kb) away from the closest transcriptional start site (TSS) (Fig. 1B), suggesting instead binding to distal regulatory regions. Indeed, clustering following ChIP-seq analysis for histone marks, revealed that the majority of YAP binding sites consisted of enhancer elements, as defined by typical enhancer-associated histone post-translational modifications (H3K27ac+, H3K4me1+ and H3K4me3−) (Fig. 1C). A few of these regions were cloned into luciferase reporter vectors and their YAP-dependent enhancer activity validated in transfection assays (Fig. S2A). Thus YAP/TAZ binding is restricted to distal regulatory elements with enhancer features.
YAP binding defines a subset of highly active enhancers and super-enhancers
We next asked whether the defined YAP bound (referred as YAP+) enhancer regions would have distinct features than YAP− enhancers similarly defined by double H3K27ac+ and H3K4me1+ presence across the genome. Strikingly, YAP+ putative enhancer regions displayed a significantly higher density of H3K27ac and H3K4me1 modification versus the average signal at active enhancers not occupied by YAP (Fig. 1D). Higher density of such post-translational modifications in histones is typically associated with more robust enhancer activity. To further support this idea, we used a proximity algorithm to assign enhancers to their targets genes and performed RNAseq to determine their expression levels. As predicted, this analysis revealed that genes associated with YAP+ enhancers, as a whole, were expressed at significantly higher levels than genes linked to active YAP− enhancers (Fig. 1E). Thus, our data indicate that YAP binding defines a subset of highly active enhancers that drive potent expression of their target genes. We will refer to these enhancers as YAP-bound regulatory elements (YREs) hereafter.
The properties of the YREs are highly reminiscent of the recently described super-enhancers (Hnisz et al., 2013). We thereby compiled a list of super-enhancers in HUCCT1 cells based on H3K27ac presence across the genome (Hnisz et al., 2013). We find that approximately 25% of YREs are catalogued as super-enhancers in comparison to only 2% of YAP− enhancers (Fig. 1F). On the other hand, these YAP+ super-enhancers represent approximately 17% of all super-enhancers in the cell, indicating that YAP/TAZ binding defines a distinct subset of these elements (Fig. 1F). Indeed, YAP occupancy was associated with higher transcriptional outputs when compared to YAP− super enhancers (Fig. S2B).
We next assessed whether the presence of YAP/TAZ was required for YRE activity. To do so, we performed multiple ChIP-Seq analyses and RNAseq following acute YAP/TAZ knockdown (siYT) in HUCCT1 cells. We combined YAP and TAZ manipulation given the highly overlapping genomic binding patterns of both co-activators (Fig S1J). Silencing of YAP/TAZ, indeed, resulted in reductions of H3K27ac+ and H3K4me1+ presence at YREs (Fig. 1G, S2C–D). Furthermore, siYT also led to a concomitant loss of expression of genes associated with both regular and super-enhancer YREs (Fig. 1H). Importantly, genes associated with YAP− enhancers did not exhibit changes in expression levels after siYT (Fig. 1H). We next tested whether YREs would be dynamic in their response to upstream Hippo signaling. Indeed, H3K27ac+ presence at YREs was reduced at high cell densities (Fig. 1H), a condition associated with Hippo signaling-driven YAP inactivation (Fig. 1I) and reduction of target gene expression. Together, our results indicate that YAP/TAZ binding defines a set of highly active and dynamic enhancer elements in the genome, including a subset of super-enhancer driving the highest transcriptional outputs, and that YAP/TAZ play a critical role in sustaining the transcriptional program driven by these elements.
YAP controls RNA Polymerase II pause-release of YRE-associated genes
Given the peculiar pattern of YAP binding at a restricted number of enhancer elements, we next investigated the mechanisms by which YAP/TAZ could regulate transcription from YREs. First, we tested the association between YAP/TAZ distal binding sites and their computationally assigned promoters. We performed Circular Chromosome Conformation Capture (4C-seq) (van de Werken et al., 2012), using 21 different YREs as anchors in HuCCT1 cells. We generally observed that YREs assigned to target genes interacted with their respective TSS and other enhancer regions, indicating that YREs were indeed sites of long-range chromatin interactions (Fig. S2D and data not shown). However, upon siYT, we observed minor, or no differences in the interaction frequency score of chromatin (Fig. S2D), concluding that YAP/TAZ are dispensable for normal chromatin looping.
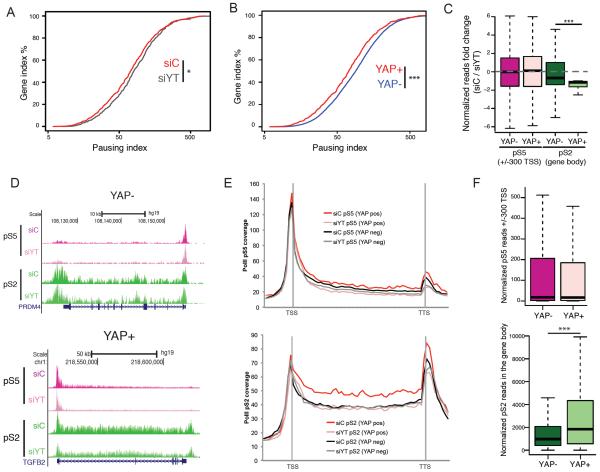
Next, we examined the role of YAP/TAZ in regulating PolII recruitment and/or elongation, two major regulatory steps in the transcriptional cascade(Wade and Struhl, 2008). To do this, we determined the patterns of PolII occupancy in genes associated with YREs following siYT. While we observed reduction of PolII density at the promoters of some YAP target genes, the most drastic effect was the widespread loss of PolII presence at gene bodies (Fig. S3A), indicating impaired transcriptional elongation. We utilized these genomic data to calculate the PolII pausing index (PI), otherwise known as the traveling ratio(Adelman and Lis, 2012). This analysis revealed a significant increase in the PI, indicative of impaired elongation, at YRE-associated genes following siYT (Fig. 2A). Further supporting a key role for YAP in elongation, we show that YRE-associated genes display significantly lower pausing than genes regulated by non-YAP bound enhancers at steady state (Fig. 2B).
Figure 2. YAP controls RNA Polymerase II promoter pause-release.
A–B) Line graph representing the pausing index (PI) for YAP positive genes upon YAP/TAZ silencing (siYT) (A); and YAP+ enhancer genes vs. YAP− (B). C) Box plot representing the fold change of RNA PolII pS5 at the promoter (pink shades) or pS2 in the gene body (green shades) in YAP+ and YAP− enhancer genes upon siYT treatment. D) ChIP-seq tracks representing signal for RNA PolII pS5 or pS2 around a representative YAP− gene and a YAP+ gene. E) Metagene profile of RNAPolII pS5 (upper) and pS2 (lower) for YAP+ and YAP− super-enhancer genes in cells treated with siC and siYT. F) Box plot showing the occupancy of PolII pS5 at the promoter (pink shades) or pS2 in the gene body (green shades) in YAP+ and YAP− enhancer genes. See also Figure S2.
Proximal PolII pausing is an important and widespread mechanism to regulate elongation (Adelman and Lis, 2012). Recruitment of the P-TEFb complex to paused promoters is one of the rate-limiting steps for transcriptional pause release(Adelman and Lis, 2012; Wade and Struhl, 2008). The core component of P-TEFb, CDK9, catalyzes PolII Serine 2 phosphorylation, a mark that is associated with elongating, productive Polymerase II throughout the gene body of active genes. If YAP/TAZ regulate proximal pause release, then siYT would be predicted to cause a reduction in the levels of Ser2-phosphorylated PolII, but should not affect the Ser5-PolII phosphorylation (a marker of initiation)(Adelman and Lis, 2012). Using ChIP-Seq analysis, we found a significant change in Ser2 PolII-density in the gene body of YRE-associated genes, whereas Ser5 at the promoter remained unaffected (Fig. 2C–E). Importantly, no changes were evident in non-YAP target genes. Additionally, we, also show that YRE-driven genes have significantly higher Ser2 PolII in their gene bodies compared to non-YAP target genes (Fig. 2E, 2F). Our results demonstrate that YAP, by binding to distal elements, confers higher transcriptional rate to its targets by promoting RNA Polymerase II promoter release and thereby its elongation.
YAP recruits Mediator complex at YRE to drive transcriptional activity
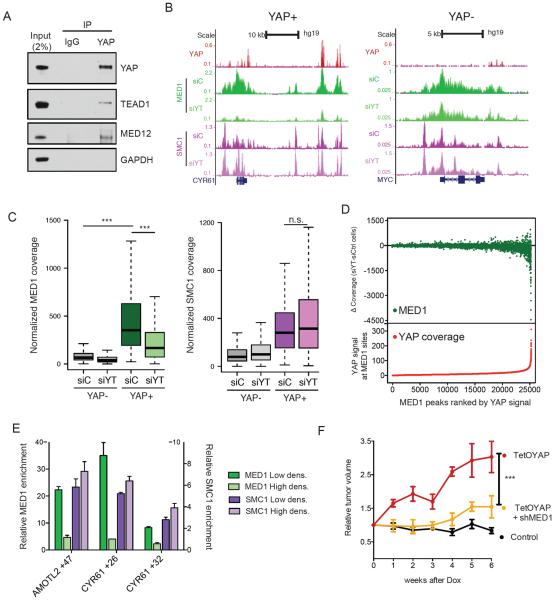
To provide insight into the molecular mechanisms employed by YAP to control elongation we performed IP-Mass spectrometry studies. This analysis revealed the enrichment for different subunits of the Mediator complex (Fig. S3B). Mediator is a large protein complex known to integrate signals from transcription factors in order to control multiple aspects of transcriptional activation (Malik and Roeder, 2010). Biochemical experiments validated the endogenous interaction between YAP and Mediator subunit (Fig. 3A) and demonstrated that these interactions occurred onto chromatin (Fig. S3C–D), and could occur independent of TEAD binding (Fig. S3E). Next, we performed ChIP-seq for the MED1 subunit in HUCCT1 cells and evaluated co-occupancy with YAP across the genome. We found that >87% of YAP sites overlapped with MED1 bound regions (Fig. 3B and S3F), further supporting an important functional interaction. Considering that there are more than 20,000 MED1 binding peaks in the genome, YAP/TAZ occupancy represents a very small subset of MED1 sites, which have some of the highest MED1 density (Fig. 3C), even when using as a comparison a size-matched dataset of YAP− enhancers displaying equal levels of H3K27ac (Fig S3G).
Figure 3. YAP recruits Mediator to regulate transcription.
A) Validation co-IP between YAP-TEAD1 and MED12. B) ChIP-seq tracks of a YAP+ and YAP− locus for YAP, MED1 and SMC1 in HuCCT1 cells treated with siC or siYT. C) Boxplot representing normalized MED1 (left) or SMC1 (right) coverage in YAP+ or YAP− enhancers in siC- or siYT-treated cells. D) Scatter plot correlating YAP occupancy to loss of MED1 signal upon YAP/TAZ silencing. On the X-axis MED1 peaks are ranked according to YAP signal. Y-axis in the top panel represents MED1 signal changes between siC and siYT cells. Y-axis in the lower panel represents the counts of YAP signal in each MED1 peak. E) ChIP-qPCR for MED1 and SMC1 at selected YREs in H69 cells at low and high density. Error bars = SD. F) Tumor volume of xenograft HuCCT1 bearing TetOYAP and/or shMED1 constructs relative to volume measured before the administration of Doxycycline (n= 5 mice, error bars = SEM). See also Figure S3.
We next asked whether YAP is required for Mediator occupancy. Notably, siYT leads to a dramatic decrease of MED1 signal specifically around YREs (Fig. 3B,C, and S3H, J), with the degree of MED1 loss directly correlating to the density of YAP binding at such sites (Fig. 3D). YAP/TAZ are selectively required for Mediator binding, as siYT does not affect the occupancy of Cohesin, a known binding partner of Mediator involved in establishing chromosome looping (Kagey et al., 2010) (Fig. 3B, C and S3H). Mediator binding at YREs was dynamic and highly responsive to cell density cues, in contrast to Cohesin occupancy, which remained unchanged at different confluences (Fig. 3E and S6J, K). These experiments demonstrate that YAP is necessary and sufficient to induce Mediator recruitment to target sites, and reinforce our observations that YAP presence is important for enhancer activity, and not for enhancer organization per se.
Our data point to Mediator recruitment by YAP as an essential step for transcriptional activation of YAP target genes. We thereby functionally tested Mediator function downstream of YAP by silencing Mediator subunits in cells overexpressing a doxycycline (Dox) inducible constitutively active YAPS127A allele. In mouse xenografts, silencing of Mediator results in decreased growth (Fig. 3F, S4A), as well as attenuation of the activation of YRE associated genes (Fig. S4B–D). Collectively, our data demonstrate that the Mediator complex is a functionally important downstream transducer of the YAP transcriptional program.
CDK9 activity mediates YAP-driven transcriptional elongation
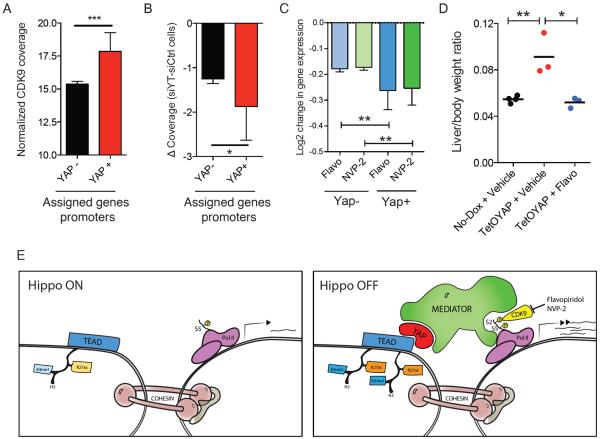
It has recently been demonstrated that Mediator, through its head module can act as a regulator of elongation by recruiting P-TEFb (Donner et al., 2010; Wang et al., 2005). Thus, we posited, that YAP/TAZ promotes pause release by recruiting P-TEFb through Mediator. Two pieces of evidence support this idea: first, YRE-associated genes display higher levels of CDK9 occupancy around their promoter (Fig. 4A) and second, YAP target genes display preferential loss of CDK9 upon siYT (Fig. 4B). To explore the functional role of CDK9 downstream of YAP, we used Flavopiridol (FP, a pan CDK inhibitor with selectivity towards CDK9) along with the and highly specific CDK9 inhibitor NVP-2 (Lu et al., 2015). These compounds are indeed able to rescue, in a dose-dependent manner, the activation of YAP target genes driven by a Dox-inducible YAPS127A transgene (Fig. S4E). Transcriptional profiling of cells treated for 3 hours with these two compounds demonstrate that YRE-associated genes are particularly sensitive to inhibition of CDK9 (Fig. 4C and S4F). This mechanism was corroborated in an animal model, as one-week FP administration results in a full rescue of YAPS127A-driven increase in liver size and target gene expression (Fig. 4D and S4G). In summary, these data demonstrate that YAP recruits the Mediator complex to YREs in order to drive exceptionally high transcriptional activity via CDK9 recruitment to mediate transcriptional pause release.
Figure 4. YAP-Mediator regulates transcriptional elongation via CDK9 recruitment.
A) Normalized CDK9 coverage around the promoter of YAP negative- and YRE-associated genes. Error bars = 95% CI B) Fold change of CDK9 occupancy upon siYT treatment around the promoter of YAP negative- and YRE-associated genes. Error bars = 95% CI C) Log2 (FC) of expression of YAP− vs. YAP+ genes in HuCCT1 cells treated with CDK9 inhibitors Flavopiridol and NVP-2. Error bars = 95% CI. D) Liver/Body weight ratio of mice overexpressing YAP for 1 week treated with vehicle or Flavopiridol. E) Model for YAP/Mediator-driven transcriptional elongation in active Hippo signaling (left) or inactive Hippo signaling (right). See also Figure S4.
Discussion
The Hippo/YAP signaling pathway has emerged as a tremendously important regulator of stem cell biology and tumorigenesis. The mechanism by which YAP/TAZ control gene expression has remained an important open question in the field. Our data provide evidence demonstrating that YAP/TAZ function is predominantly restricted to the control of elongation of a relatively small number of target genes. This has important implications for our conceptual understanding of Hippo signaling. Our results imply that even when Hippo signaling is on (cytoplasmic YAP/TAZ) their target loci still carry transcriptionally engaged, but proximally paused RNAPII (Fig. 4E). Loss of Hippo-mediated regulation, and consequent YAP/TAZ nuclear relocation, allows pause release via Mediator. We hypothesize that pausing allows for a mechanism to induce rapid gene expression in response to physiological growth or regeneration cues, in a manner analogous to the regulation of immediate early genes and inflammatory responses (Adelman et al., 2009; Donner et al., 2010). Our idea fits with the observation that knockout of YAP in the adult results in the absence of phenotypes in many tissues at steady-state (Barry et al., 2013; Chen et al., 2014; Zhang et al., 2014) whereas YAP is fully required for injury or oncogene-driven responses in those same tissues(Barry et al., 2013; Chen et al., 2014; Zhang et al., 2014). Our data also suggest that most YAP/TAZ targets in a cell would need to have engaged PolII, so that cell-type basal transcription would determine the observed tissue specificity in YAP/TAZ targets.
The overall pattern of YAP binding in the genome of liver cancer cells is significantly distinct from what has been recently described in Drosophila(Oh et al., 2013; Oh et al., 2014), where YAP has been found enriched at promoter regions(Oh et al., 2013). Our results are not specific to the liver cancer cell lines used as TEAD binding has also been found to be highly enriched at enhancer regions in two other cellular contexts (Beyer et al., 2013; Cebola et al., 2015). We believe that such differences might not be completely surprising, given some intrinsic differences between human and drosophila genomes(Wilson and Odom, 2009). Moreover, in terms of transcriptional effectors, the Hippo pathway significantly differs in the mammalian system compared to Drosophila. Indeed, 4 TEAD members exist in mammals whereas just one (Scalloped), in the fly. Such evolutionary expansion could be due to an increase the in complexity of transcriptional regulation in mammals, possibly via the control of different sets of tissue specific enhancer elements.
Our analysis identifies a relatively small number of YAP binding sites (Fig 1), which represent only a fraction of the TEAD binding regions. These peaks represent robust and `high-confidence' YAP-binding sites determined after application of a very stringent analysis pipeline encompassing multiple negative controls. While the number of YAP+ peaks found in our analysis are significantly less than what has been described previously, recent work in cardiomyocytes has identified ~1300 YAP peaks, which is in the range of the approximately 850 sites detected by our methodology(Lin et al., 2015). However, it is important to consider that since YAP does not bind DNA directly, its affinity and interaction frequency for chromatin is likely to be lower than a DNA-binding transcription factor, such as TEAD. Thus, it is possible that our analysis might miss a number of sites that may have low levels of YAP bound. Still, one important insight regarding the validity of our peak finding strategy can be obtained from the analysis of functional consequences of YAP/TAZ knockdown. If many other functionally relevant YAP+ sites would exist outside of the ~850 high-confidence peaks that we describe, then siYT should result in loss of MED binding in those extra sites as well. Importantly, our data demonstrate that most of MED1 loss occurs in the few hundred genes with highest YAP binding (Fig. 3D). Reduction in MED1 binding outside of this subset of genes is negligible or barely above noise. These data indicate that, at least in regards to MED binding, additional YAP sites that we may have missed would represent sites of low functional relevance.
Mechanistically, our results point to recruitment of Mediator as the crucial step in YAP-driven transcription. The interaction of YAP with Mediator has been described previously in human embryonic stem cells (Varelas et al., 2008). Additionally, Yorkie also forms a complex with Mediator in Drosophila, suggesting the conservation of this interaction(Oh et al., 2013). Yorkie has been shown to recruit a histone methyltransferase complex via Ncoa6 (Oh et al., 2014), a subunit of the Thritorax-related (Trr) methyltransferase complex. While we did not observe peptides for any Trr complex members in our mass spectrometry analysis, it has been previously reported that Mediator can exist in complex with mammalian Ncoa6 (Ko et al., 2000), thus YAP could still be recruiting mammalian histone methyltransferase activity to its target sites. The fact that we observe a significant loss of H3K4me1 upon siYT also suggests that this might be the case. Furthermore, knockdown of Ncoa6 in H69 cells resulted in the suppression of expression of 2 out 5 YAP target genes tested (Fig. S4H). Thus, in this cellular context, Ncoa6 seems to be functionally important for the expression of a subset of YAP target genes. Future work should address whether recruitment of histone methyltransferase activity, and other chromatin remodeling activity (Skibinski et al., 2014), occurs indirectly via Mediator or is independently regulated by YAP/TAZ.
Finally, our results significantly add to the emerging theme of enhancer misregulation in cancer and disease(Akhtar-Zaidi et al., 2012; Chapuy et al., 2013; Loven et al., 2013). In contrast to other regulators of enhancer activity, which bind most expressed genes within a cell(Akhtar-Zaidi et al., 2012; Chapuy et al., 2013; Loven et al., 2013), YAP/TAZ binding is predominantly restricted to enhancers associated with a only few hundred genes that are critical for growth, making YAP/TAZ ideal targets for cancer therapeutics. Still, our data demonstrate that therapies based on elongation inhibitors could be of use for YAP-driven tumors.
EXPERIMENTAL PROCEDURES
Cell lines and transfections
HuCCT1 (parental and Flag-HA-YAP5SA expressing) cells were maintained in RPMI containing 10% FBS, 1X Hepes, 1X L-glutamine. Cclp1 (parental and Flag-Bio-YAP5SA expressing) were maintained in DMEM containing 10% FBS. shRNAs, siRNAs and compounds employed are detailed in extended experimental procedures.
Mouse models
Tetracycline-inducible YAPS127A expression mice were previously described (Camargo et al., 2007). Viral injections and xenograft assays are detailed in supplemental experimental procedures.
ChIP-seq and RNA-seq
Chromatin Immunoprecipitation was performed essentially as previously described (Galli et al., 2012) and detailed in supplemental experimental procedures.
Libraries for ChIP-sequencing were generated by using NEBNext Ultra DNA Library Prep Kit for Illumina (NEB) and barcoded added using NEBNext® Multiplex Oligos for Illumina (Index Primers Set 1) (NEB) according to manufacturer's recommendation.
RNA-seq libraries were generated using TruSeq RNA Sample Prep Kit v2 (Illumina) according to manufacturer's recommendation. All the High-throughput sequencing experiments were run on a Hi-seq2000 (Illumina) sequencer at the Center for Cancer Computational Biology (Dana Farber Cancer Institute, Boston, USA). Bioinformatic analyses are detailed in supplemental experimental procedure.
Sequencing data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE62275.
4C-sequencing
4C templates were prepared as described previously23. DpnII digestion was used as the first restriction enzyme to generate high resolution 3C template, which was further trimmed with Csp6I, NlaIII or BfaI. 4C primers were design following the general consideration as described23. The primers carried additional 5′ overhangs composed of adaptor sequences for Illumina single-read sequencing. Samples were sequenced on a Hi-seq 2000 machine (Illumina).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Camargo lab for insightful discussion and support. This study was supported by grants from the National Institutes of Health (AR064036 and DK099559 to FDC). FDC was a Pew Scholar in the Biomedical Sciences. GGG was supported by an American-Italian Cancer Foundation Post-Doctoral Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews. Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes & development. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell reports. 2013;5:1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Molecular cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cebola I, Rodriguez-Segui SA, Cho CH, Bessa J, Rovira M, Luengo M, Chhatriwala M, Berry A, Ponsa-Cobas J, Maestro MA, et al. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nature cell biology. 2015;17:615–626. doi: 10.1038/ncb3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang N, Gray RS, Li H, Ewald AJ, Zahnow CA, Pan D. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes & development. 2014;28:432–437. doi: 10.1101/gad.233676.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature structural & molecular biology. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli GG, Honnens de Lichtenberg K, Carrara M, Hans W, Wuelling M, Mentz B, Multhaupt HA, Fog CK, Jensen KT, Rappsilber J, et al. Prdm5 regulates collagen gene transcription by association with RNA polymerase II in developing bone. PLoS genetics. 2012;8:e1002711. doi: 10.1371/journal.pgen.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L, Cardona GR, Chin WW. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes & development. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhou P, von Gise A, Gu F, Ma Q, Chen J, Guo H, van Gorp PR, Wang DZ, Pu WT. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circulation research. 2015;116:35–45. doi: 10.1161/CIRCRESAHA.115.304457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xue Y, Yu GK, Arias C, Lin J, Fong S, Faure M, Weisburd B, Ji X, Mercier A, et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. eLife. 2015;4 doi: 10.7554/eLife.06535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature reviews. Genetics. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell reports. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, White KP, Mann RS, Irvine KD. Yorkie Promotes Transcription by Recruiting a Histone Methyltransferase Complex. Cell reports. 2014;8:449–459. doi: 10.1016/j.celrep.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends in cell biology. 2012;22:339–346. doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski A, Breindel JL, Prat A, Galvan P, Smith E, Rolfs A, Gupta PB, Labaer J, Kuperwasser C. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell reports. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nature methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nature cell biology. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Current opinion in genetics & development. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Molecular cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Odom DT. Evolution of transcriptional control in mammals. Current opinion in genetics & development. 2009;19:579–585. doi: 10.1016/j.gde.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Zhang W, Nandakumar N, Shi Y, Manzano M, Smith A, Graham G, Gupta S, Vietsch EE, Laughlin SZ, Wadhwa M, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Science signaling. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & development. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.