Abstract
The small GTP-binding proteins Arl2 and Arl3, which are close homologs, share a number of interacting partners and act as displacement factors for prenylated and myristoylated cargo. Nevertheless, both proteins have distinct biological functions. Whereas Arl3 is considered a ciliary protein, Arl2 has been reported to be involved in tubulin folding, mitochondrial function, and Ras signaling. How these different roles are attained by the two homolog proteins is not fully understood. Recently, we showed that the N-terminal amphipathic helix of Arl3, but not that of Arl2, regulates the release of myristoylated ciliary proteins from the GDI-like solubilizing factor UNC119a/b. In the biophysical study presented here, both proteins are shown to exhibit a preferential localization and clustering in liquid-disordered domains of phase-separated membranes. However, the membrane interaction behavior differs significantly between both proteins with regard to their nucleotide loading. Whereas Arl3 and other Arf proteins with an N-terminal amphipathic helix require GTP loading for the interaction with membranes, Arl2 binds to membranes in a nucleotide-independent manner. In contrast to Arl2, the N-terminal helix of Arl3 increases the binding affinity to UNC119a. Furthermore, UNC119a impedes membrane binding of Arl3, but not of Arl2. Taken together, these results suggest an interplay among the nucleotide status of Arl3, the location of the N-terminal helix, membrane fluidity and binding, and the release of lipid modified cargos from carriers such as UNC119a. Since a specific Arl3-GEF is postulated to reside inside cilia, the N-terminal helix of Arl3•GTP would be available for allosteric regulation of UNC119a cargo release only inside cilia.
Introduction
ADP-ribosylation factor-like (Arl) proteins belong to the Arf subfamily of small GTP-binding proteins (for a review on Arl proteins, see Gillingham and Munro (1)). As expected from their high sequence identity (50%), the small G-proteins Arl2 and Arl3 share several effector-type interacting partners, including Bart (2), the GDI-like solubilizing factors UNC119a and UNC119b (3, 4, 5), and the delta subunit of type 6 phosphodiesterase (PDEδ) (6, 7, 8). Despite their homology and shared downstream effectors, Arl2 and Arl3 seem to have nonredundant interactions and cellular functions. The GTPase-activating protein (GAP) RP2 has been shown to be specific for Arl3 (9), whereas Arl2 is apparently downregulated by the specific GAP ELMOD3 (10). Arl3 is found exclusively in ciliated organisms and considered as a ciliary protein, with Arl3 knockout mice showing phenotypes similar to ciliopathies, including renal, pancreatic, and retinal defects (11, 12). Arl2, on the other hand, has been shown to be involved in the tubulin-folding pathway (13). Furthermore, knockdown by small interfering RNA and/or the introduction of GTPase-negative mutants of Arl2/3 have been shown to have different effects on mammalian cell lines (14, 15). In addition, Arl2 has been implicated in the regulation of mitochondrial function (15).
Arl proteins are structurally related to Arf proteins, which exhibit a unique and common structural feature: a two-residue shift in the β2 and β3 strands (termed the interswitch toggle) of the GDP-bound state relative to other GDP-bound small G-proteins (16). In the GDP-bound state, the N-terminal amphipathic helix is located in a hydrophobic pocket on the protein surface and caps the interswitch. Upon binding to GTP, the two β strands connecting the nucleotide-sensitive switch 1 and 2 regions (i.e., the β2 and β3 strands) undergo a two-residue register shift, leading to an exposed conformation of this interswitch in the GTP-loaded state and thus an exposed N-terminal helix, because the interswitch toggle requires the displacement of the N-terminal amphipathic helix from the surface of the protein (16, 17). Hence, this amphipathic helix, which carries an N-terminal myristoyl group in several Arf proteins, is only exposed in Arf•GTP and is believed to bind to membranes (18), thereby connecting the membrane binding capacity of Arf to its nucleotide status. Therefore, the N-terminal helix can be considered as part of the switch regions. Arl2 and Arl3 also possess a short amphipathic N-terminal helix and have been shown to undergo the same type of structural transition (Fig. 1). However, despite the presence of a conserved glycine at position 2 of the NH2-terminus (the N-myristoylation site in Arf proteins), they have not been shown to be myristoylated (1, 19). Thus, if membrane binding of Arl2 and Arl3 takes place, it is expected to be mediated only by the amphipathic helix, as in the case of the Arf-related protein Sar1, where the amphipathic helix has been shown to be sufficient for membrane binding (20, 21).
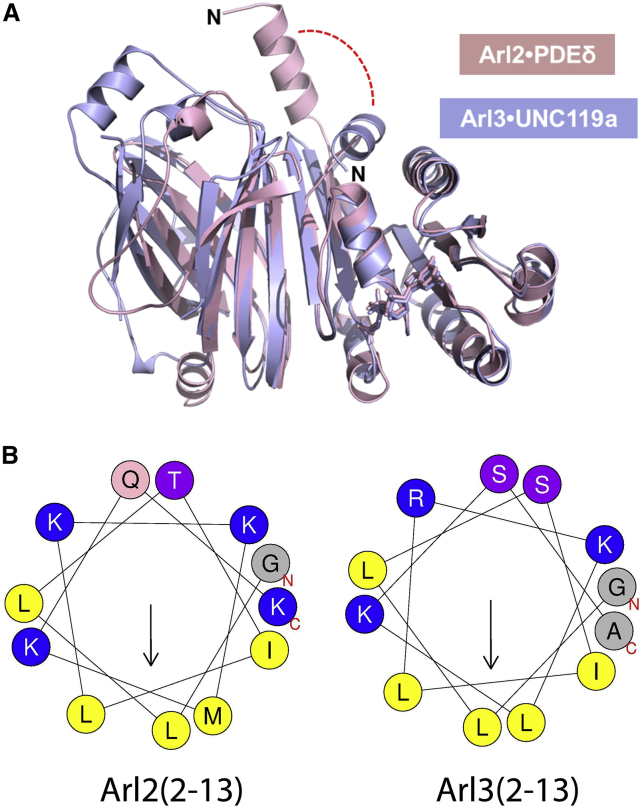
Figure 1.
(A) Ribbon representation of the overall structure of full-length Arl2 and Arl3 illustrating the conformational change of the N-terminal helix with regard to the different interaction of the N-terminus of Arl2 and Arl3 with effectors such as PDEδ and UNC119a (PDB codes: 1KSJ and 4GOJ, respectively). (B) Helical wheel projection of the N-terminal amphipathic helix of Arl2 and Arl3 (amino acid residues 2–13). Positively charged residues (K and R), glutamine (Q), serine (S) and threonine (T), as well as hydrophobic residues (L, M, and I) and alanine (A) and glycine (G) are indicated in color accordingly. The arrow corresponds to the hydrophobic moment. The helical wheel projection was generated using the program HELIQUEST (23). To see this figure in color, go online.
Arl2 and Arl3 serve as displacement factors for lipid-modified proteins bound to the GDI-like solubilizing factors UNC119a, UNC119b, and PDEδ (4, 7, 22). Recently, we showed structurally and biochemically that Arl3, but not Arl2, regulates the release of myristoylated ciliary proteins from UNC119a and UNC119b (4). The allosteric displacement is mediated by the N-terminal helix of Arl3, suggesting a difference in the (dynamic) properties of the Arl2 and Arl3 N-terminal helices. Since this amphipathic helix is predicted to mediate Arl2 and Arl3 membrane binding, we hypothesized that Arl2 and Arl3 might have different membrane interaction properties that could account for the differences in their cellular localization and functions. So far, however, information about the targeting of Arl2 and Arl3 to different membrane compartments is scarce.
In this study, we analyzed the role of the N-terminal amphipathic helix of Arl2/3 in membrane binding and its influence on complexation with UNC119a to further investigate the differences between Arl2 and Arl3. For this purpose, we utilized surface plasmon resonance (SPR), infrared reflection absorption spectroscopy (IRRAS), fluorescence-based kinetic analysis, and atomic force microscopy (AFM). The combined data verify the requirement of the N-terminal amphipathic helix for membrane binding of Arl2/3 and their interaction with UNC119a. Surprisingly, and in contrast to what was found for other Arf family members, a nucleotide-independent membrane interaction was detected for Arl2. The results demonstrated the necessity of GTP binding and the N-terminal helix for Arl3 membrane binding. Moreover, the N-terminal helix of Arl3, but not that of Arl2, strongly increased the binding affinity to UNC119a. Finally, a GTP-specific binding of UNC119a to Arl3 was revealed, with binding of UNC119a preventing membrane binding of Arl3 in the UNC119a-complexed state owing to the unavailable amphipathic helix.
Materials and Methods
Materials and sample preparation
The phospholipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (DOPG), 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (DPPG), and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were purchased from Avanti Polar Lipids (Alabaster, AL). Cholesterol (Chol) and 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (Hepes) were obtained from Sigma Aldrich (Deisenhofen, Germany). Magnesium chloride, tris(hydroxymethyl)-aminomethane (Tris), and chloroform were obtained from Merck (Darmstadt, Germany). Bovine serum albumin was obtained from Pierce (Bonn, Germany). Stock solutions of 10 mg mL−1 lipid (DOPC, DOPG, DPPC, DPPG, and Chol) in chloroform/methanol 4:1 for DPPG and in chloroform for all other lipids were prepared and mixed to obtain 1.94 mg of total lipid with the desired composition of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 for AFM and SPR experiments. After removal of the solvent by drying under vacuum overnight, the dry lipids were resuspended in 1 mL of 10 mM Hepes, 5 mM MgCl2, 150 mM NaCl, pH 7.5 for SPR, or 25 mM Tris, 5 mM MgCl2, 150 mM NaCl, pH 7.5 for the AFM experiments to yield a total lipid concentration of 3 mM. Details regarding the formation of large unilamellar vesicles (100 nm in size) by extrusion can be found in Weise et al. (24, 25). The extruded lipid solution was further diluted to a concentration of 0.5 mM for the SPR experiments.
Protein production and purification
UNC119a, C-terminal His-tagged full-length Arl2 and Arl3, and GST-tagged, N-terminal truncated Arl2 and Arl3 were produced and purified as described previously (5, 9, 26). After purification, both proteins were bound to GDP as detected by high-performance liquid chromatography. Exchange for the nonhydrolyzable GTP analog (GppNHp) was carried out as described previously (9).
SPR
SPR experiments were carried out with a Biacore 3000 system (Biacore (now GE Healthcare), Uppsala, Sweden). For protein-membrane interaction studies, the L1 sensor chip (GE Healthcare, Munich, Germany) was used. All measurements were performed at a temperature of 25°C, and samples were cooled at 10°C in the autosampler before the measurements were started. Sample preparation, vesicle immobilization, SPR measurements, regeneration of the chip surface, and analysis of the SPR sensorgrams were carried out as described previously (27) and in Supporting Materials and Methods in the Supporting Material. Briefly, 15 μL of the extruded lipid vesicle solution (0.5 mM) was injected twice at a flow rate of 2 μL/min for vesicle immobilization. This was followed by a stabilization phase with injection of 50 μL of Hepes buffer at a flow rate of 100 μL/min and three further injections of 10 μL of 25 mM NaOH at a flow rate of 5 μL/min. Finally, the lipid surface was stabilized by injecting 40 μL Hepes buffer at a flow rate of 20 μL/min. After baseline stabilization, 40 μL of the protein-containing solution (cArl = 2 μM, cUNC119a = 3 μM) was injected at a flow rate of 20 μL/min and the dissociation was followed for 30 min. For membrane interaction studies with the UNC119a-complexed Arl, both proteins were mixed before injection into the SPR flow cell to yield a final concentration of 2 μM Arl and 3 μM UNC119a. The degree of chip surface coverage with lipids was determined with the use of 0.5 μM of bovine serum albumin and was found to be ≥75% in all cases (cf. Weise et al. (27)). For the curve-fitting procedure, BIAevaluation software 4.1 (Biacore) and Origin 7 (OriginLab, Northampton, MA) were used.
IRRAS
Experiments were carried out on an IRRAS setup consisting of two linked Teflon troughs and a Vertex 70 FT-IR spectrometer connected to an A511 reflection attachment (both Bruker, Mannheim, Germany) with an MCT detector. The measurements, setup, sample preparation, and spectra analysis were performed as described previously (25, 27). The temperature of the subphase was maintained at 20°C ± 0.5°C and time-dependent measurements were performed in the small (reference) trough at constant surface area. The resulting curve of surface pressure versus time is referred to as the π/t isotherm. Both troughs were filled with 25 mM Tris, 5 mM MgCl2, 150 mM NaCl, pD 7.5. Monolayers of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 (mol %) were formed by directly spreading the lipid solution (1 mM) in a mixture of chloroform and methanol (3:1) onto the subphase. Protein adsorption measurements were carried out by injecting the protein solution into the aqueous subphase below the lipid monolayer to yield concentrations of Arl and UNC119a of 200 and 300 nM, respectively.
AFM
Preparation of the supported lipid bilayers and the AFM setup have been described in detail elsewhere (24, 25). Briefly, vesicle fusion on mica was carried out by depositing 35 μL of the extruded lipid vesicle solution together with 35 μL of Tris buffer on freshly cleaved mica (NanoAndMore, Wetzlar, Germany) and incubating the solution in a wet chamber at 70°C for 2 h. For protein-membrane interaction studies, 200 μL of either Arl2/3•GDP, Arl2/3•GppNHp, or ΔArl2/3•GppNHp (2 or 5 μM) in 25 mM Tris, 5 mM MgCl2, 150 mM NaCl, pH 7.5 were injected into the AFM fluid cell at room temperature and allowed to incubate for 1 h. Measurements were performed on a MultiMode scanning probe microscope equipped with a NanoScope IIIa controller (Digital Instruments (now Bruker), Santa Barbara, CA) and a J-Scanner (scan size 125 μm). Images were obtained by applying the tapping mode in liquid with sharp nitride lever probes mounted in a fluid cell (MTFML; both from Veeco (now Bruker)). Tips with nominal force constants of 0.24 N m−1 were used at driving frequencies of ∼9 kHz and drive amplitudes between 170 and 700 mV. The scan frequencies were between 0.75 and 2.0 Hz. Images with a resolution of 512 × 512 pixels were analyzed using the image analysis and processing software NanoScope version 5 (Veeco (now Bruker)) and Origin 7 (OriginLab).
Fluorescence-based kinetic measurements
Kinetic measurements were monitored by means of a stopped-flow apparatus (Applied Photophysics) in the polarization mode. Experiments were performed at 20°C in a buffer containing 25 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 3 mM DTE, using an excitation wavelength of 366 nm and a 420 nm cutoff filter for mantGppNHp-bound Arl proteins. Data were analyzed using the GraFit 5.0 program (Erithracus Software).
Results and Discussion
Membrane interaction of Arl3
Ciliary membranes form specialized compartments of the plasma membrane of eukaryotic cells with peculiar biophysical properties and compositions. They were reported to have different lipid and protein compositions compared with periciliary and cell-body membranes, being highly enriched in cholesterol and sphingolipids (28, 29). This led to the assumption that cilia might be enriched in lipid raft microdomains and thus be more ordered than the bulk plasma membrane (29). In this study, we used an anionic model raft membrane system that consisted of DOPC/DOPG/DPPC/DPPG/Chol at a molar ratio of 20:5:45:5:25 and was segregated into liquid-ordered (lo) and liquid-disordered (ld) domains under ambient conditions, thus mimicking a heterogeneous plasma membrane with different degrees of membrane order (30). In addition, the integration of anionic lipids took the positive net charge of the amphipathic helices of Arl2 and Arl3 into account (+4 and +3, respectively). Phosphatidylglycerol (PG) is widely used as a simplified model of negatively charged phospholipids that mimic the electrostatic effects of monovalent acidic lipids present in mammalian membranes. Previous studies on small GTPases, such as Ras, revealed a membrane interaction behavior that is independent of the heterogeneous membrane composition (31), justifying the use of the low-melting-point lipid DOPC in liquid-phase coexistence model systems, though DOPC is rare in mammalian membranes.
To analyze the membrane binding kinetics of full-length Arl3 in the inactive (Arl3•GDP) and active (Arl3•GppNHp as a nonhydrolyzable GTP analog) states, as well as truncated Arl3 (ΔArl3, i.e., without the N-terminal residues 1–16), we carried out SPR measurements. The resulting sensorgrams (Fig. 2 A) indicate that the maximum surface coverage of the lipid bilayer (which was immobilized on a lipophilic modified dextran matrix of a L1 sensor chip) with the different Arl3 proteins is on the order of 50−300 resonance units (1 RU = 1 pg mm−2). From SPR sensorgrams, one can quantify the membrane interaction of proteins by determining three representative parameters: 1) the initial binding rate (slope) of the association phase, which allows a comparison of the incipient affinities of the proteins to lipid membranes; 2) the average dissociation rate constant, , which quantifies the overall dissociation rate of the protein from the lipid membrane; and 3) the relative amount of quasi-irreversibly bound protein at the end of each dissociation phase, which reflects the ability of the protein to stably insert into the lipid membrane (27). Because subsequent AFM experiments showed a clustering of Arl proteins in heterogeneous membranes (see below), we applied a two-step reaction model to analyze the association and dissociation phases of the protein-membrane interaction process (27) (Supporting Materials and Methods; Fig. S1; Tables S1–S3).
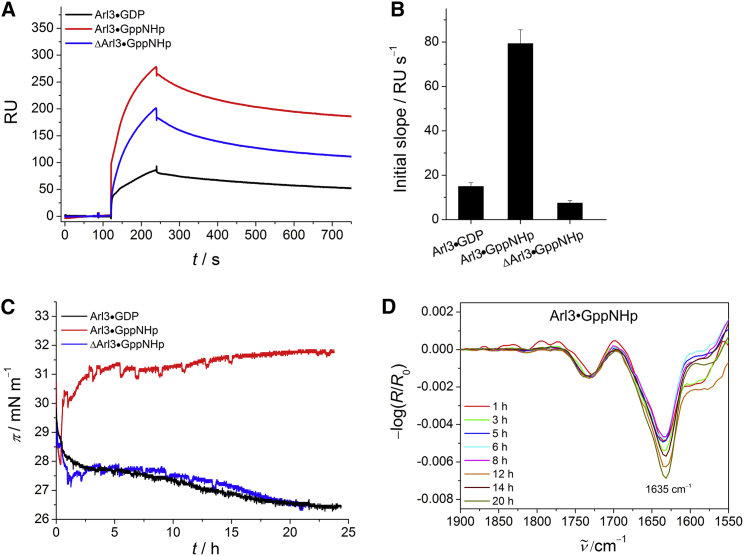
Figure 2.
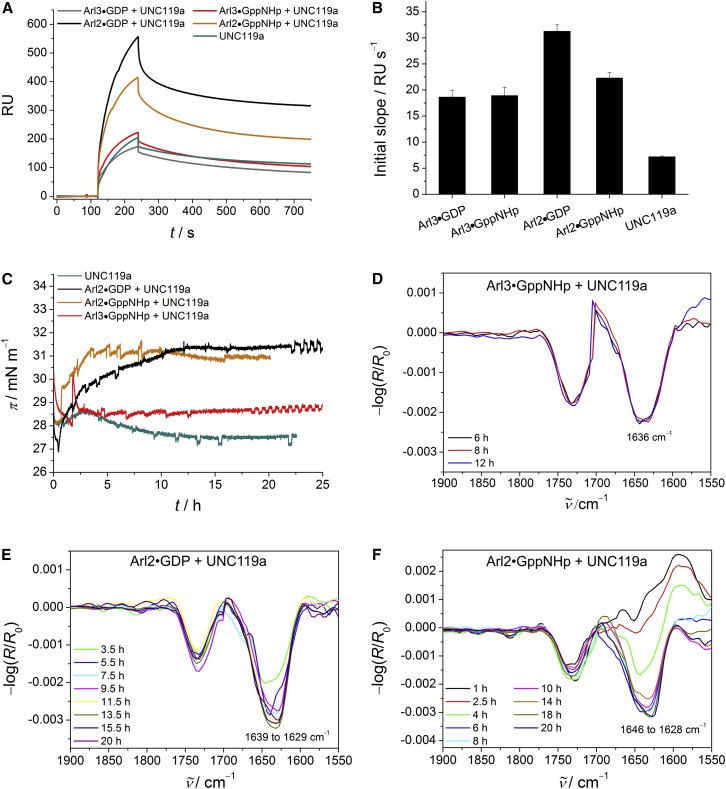
Membrane interaction of GDP- and GppNHp-loaded full-length and GppNHp-bound truncated Arl3. (A) SPR sensorgrams of the binding of Arl3 (c = 2 μM) to anionic model raft membranes composed of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 (mol %). (B) Corresponding SPR data for the initial association process. The error bars represent the standard deviation from three to six measurements. (C) Surface pressure/time isotherms for the membrane interaction of Arl3 with anionic lipid raft monolayers. (D) Corresponding time-dependent IRRA spectra for the amide-I′ region of membrane-bound Arl3•GppNHp. All IRRA spectra were recorded with p-polarized light at a 35° angle of incidence because the use of p-polarized light resulted in larger signals and better signal/noise ratios (cf. Fig. S2). To see this figure in color, go online.
Our analysis of the membrane association of the different proteins reveals that the initial slope of Arl3•GppNHp differs significantly from that of Arl3•GDP and ΔArl3•GppNHp (Fig. 2 B). It indicates a higher binding rate of Arl3•GppNHp, which is most likely due to the exposed N-terminal helix in the active state. The detected incipient membrane affinity of the inactive Arl3 is weak, as would be expected considering that the N-terminal region of Arl3•GDP is located in a hydrophobic pocket on the surface of the G domain. This conclusion is supported by results obtained using the truncated form of Arl3, which also showed a lower membrane binding rate even in the GppNHp-bound state. For comparison, a membrane binding affinity on the order of 105 M−1 was reported for Sar1 (KD = 10.5 μM) (32).
Because of the nonsimple-exponential association and dissociation curves and the observed membrane interaction behavior, analysis of the SPR sensorgrams was quite complex. To facilitate interpretation, we employed IRRA spectroscopy to study the membrane binding of Arl3 in its different states by simultaneously following the IRRA spectra and surface pressure/time (π/t) isotherms. We injected proteins underneath the lipid monolayer at a surface pressure of ∼28–30 mN m−1, which reflects the physiological lipid density found in lipid membranes. The π/t profiles in Fig. 2 C show an effective insertion into the anionic lipid raft monolayer for Arl3•GppNHp only, as indicated by a significant increase (∼4 mN m−1) in surface pressure. No membrane insertion was observed for ΔArl3•GppNHp, supporting the finding that membrane binding of Arl3 in the GTP-bound state is mediated by the N-terminal amphipathic helix. Arl3•GDP also did not display any membrane insertion, confirming that the nucleotide status plays a regulatory role by modulating the membrane interaction of Arl3 through the availability of the amphipathic helix for membrane binding.
The surface pressure data are accompanied by the corresponding IRRA spectra in Fig. 2, where detection of the infrared signature of the protein signifies the presence of membrane-bound protein at the lipid interface. In particular, the amide-I′ band of Arl3•GppNHp showed a maximum around 1635 cm−1 in the IRRA spectra (which is typical for α/β proteins) that remained constant over time, implying a relatively stable conformation/orientation of the membrane-bound Arl3•GppNHp (Fig. 2 D). The observed absence of an amide-I′ band in the IRRA spectra of Arl3•GDP and ΔArl3•GppNHp strongly suggests a very weak membrane binding for these two proteins.
Membrane interaction of Arl2
To compare the membrane binding behavior of Arl2 and Arl3, we carried out analogous SPR measurements for Arl2. The resulting sensorgrams of Arl2 in the inactive (Arl2•GDP), active (Arl2•GppNHp), and truncated (ΔArl2•GppNHp, i.e., missing residues 1–16) states are given in Fig. 3 A. Surprisingly, the analysis reveals similar initial binding rates for Arl2•GDP and Arl2•GppNHp (Fig. 3 B). This is in contrast to Arl3, although the incipient membrane affinities of both Arl2 proteins are considerably lower than that of Arl3•GppNHp (cf. Fig. 2 B).
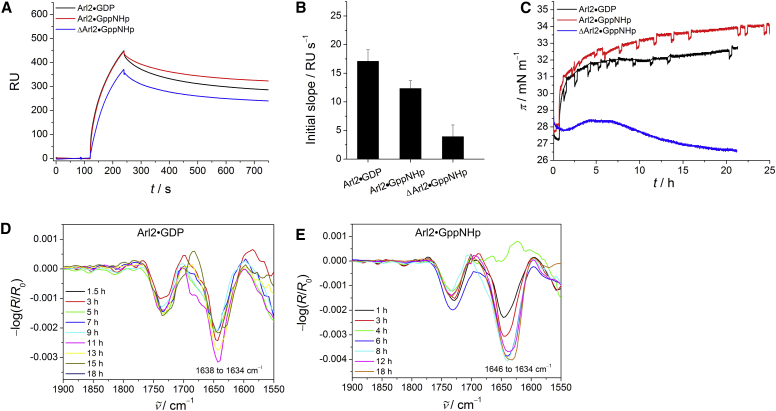
Figure 3.
Membrane interaction of GDP- and GppNHp-loaded full-length and GppNHp-bound truncated Arl2. (A) SPR sensorgrams of the binding of Arl2 (c = 2 μM) to anionic model raft membranes composed of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 (mol %). (B) Corresponding SPR data for the initial association process. The error bars represent the standard deviation from three to five measurements. (C) Surface pressure/time isotherms for the membrane interaction of Arl2 with anionic lipid raft monolayers. (D and E) Corresponding time-dependent IRRA spectra for the amide-I′ region of membrane-bound Arl2•GDP (D) and Arl2•GppNHp (E). All IRRA spectra were recorded with p-polarized light at a 35° angle of incidence. To see this figure in color, go online.
The SPR results are in agreement with the corresponding IRRAS experiments, where no nucleotide-dependent membrane insertion could be detected for Arl2 (Fig. 3, C–E). The surface pressure/time isotherms show a significant membrane insertion for both nucleotide states, with a slightly larger increase in surface pressure for Arl2•GppNHp (Δπ ≈ 5 mN m−1) compared with Arl2•GDP (Δπ ≈ 4 mN m−1; Fig. 3 C). The finding of a comparable membrane interaction behavior for active and inactive Arl2 is further supported by the detection of an amide-I′ band for both GDP- and GppNHp-loaded Arl2 in the IRRA spectra, contrary to Arl3. The wavenumber for the amide-I′ band of membrane-bound Arl2•GppNHp shows a time-dependent shift from 1646 to 1634 cm−1, implying orientational changes (Fig. 3 E). Arl2•GDP exhibits a small random shift in wavenumber for the time-dependent amide-I′ band. However, this is not significant due to the resolution in IRRAS of ∼4 cm−1 (Fig. 3 D). Hampering of the membrane insertion by truncation of the N-terminal helix in Arl2•GppNHp reveals that membrane binding of active Arl2 is still mainly mediated by the N-terminal amphipathic helix, as indicated by the lack of an amide-I′ band in the IRRA spectra of ΔArl2•GppNHp and the missing increase in surface pressure (Fig. 3 C). Taken together, these results show that although the G domains and in particular the N-terminal helices of Arl2 and Arl3 are similar, they behave rather differently in their nucleotide-dependent interaction with membranes, which seems to indicate a different dynamic behavior rather than a different structure of the N-terminus. Thus, the N-terminal helix of Arl2 might even be flexible and exposed in the GDP-loaded state to mediate an interaction with the membrane.
Partitioning of Arl2/3 in heterogeneous membranes
By using AFM, we were able to gain detailed insights into the partitioning behavior of Arl3 and Arl2 in membranes of varying degrees of order. AFM images of the protein-free anionic lipid raft model membrane system indicate phase separation into ld and lo membrane domains under ambient conditions (cf. Fig. S3). The coexisting ld/lo phases can be clearly distinguished by a height difference of ∼1 nm (30). After incubation of the membrane with Arl3•GDP, only a few protein-enriched domains could be detected in a disturbed ld phase, even at a higher protein concentration (5 μM; Fig. 4). A comparable membrane interaction behavior was observed for ΔArl3•GppNHp, i.e., only a few proteins were visible in a thinned ld phase and there were almost no protein-enriched domains. In contrast to the relatively weak membrane interaction of inactive and truncated Arl3, a strong clustering of Arl3•GppNHp (c = 2 μM) was detected in a perturbed ld phase, as indicated by the large amount of huge protein-enriched domains in the AFM image (Fig. 4, white areas). In addition, the pronounced insertion of Arl3•GppNHp into the membrane leads to a significant disturbance and thinning of the ld phase. Insertion of positively charged amphipathic helices into membranes often leads to membrane thinning, which can be due to hydrophobic mismatch, interfacial localization of the basic residues that would result in an increase of the lipid headgroup distance, or a pushing down of the lipid headgroups in the vicinity of the membrane-inserted protein.
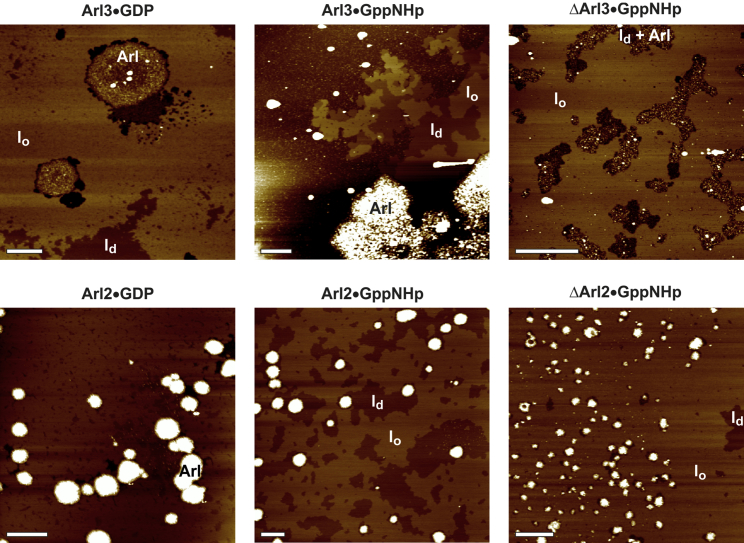
Figure 4.
Membrane partitioning of full-length and truncated Arl2/3 in the different nucleotide-bound states. All AFM images showed a defect-free, continuous lipid bilayer on mica with isolated liquid-disordered (ld) domains in a liquid-ordered (lo) membrane matrix at room temperature before injection of proteins (cf. Fig. S3). Representative AFM images after the addition of Arl2/3•GDP, Arl2/3•GppNHp, and ΔArl2/3•GppNHp to a membrane consisting of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 (mol %) are shown. The overall height of the vertical color scale corresponds to 6 nm for Arl3•GDP, ΔArl3•GppNHp, and ΔArl2•GppNHp, and 12 nm for Arl3•GppNHp, Arl2•GppNHp, and Arl2•GDP. The scale bar represents 1 μm for all images. To see this figure in color, go online.
A possible explanation for the strong protein clustering observed for Arl3•GppNHp is that the N-terminal amphipathic helix is only exposed in the active state of Arl3. Hence, insertion of the positively charged helix (+3) into the negatively charged membrane (as indicated by IRRAS) could lead to an accumulation of anionic lipids at the protein-binding site. Sequestering of acidic lipids by membrane-bound basic peptides is known to result in attractive interactions between membrane-bound proteins. A comparable lipid-sorting mechanism that is controlled by electrostatic interactions has been proposed for the clustering of the small GTPase K-Ras4B in ld domains of heterogeneous membranes (25). Even though the N-terminal helix is not available for membrane binding in ΔArl3•GppNHp and Arl3•GDP (as shown by IRRAS), membrane adsorption can still occur through interactions with the Arl protein surface, although it is much less pronounced (as seen in AFM and SPR).
When the membrane partitioning of Arl3 is compared with that of Arl2, it becomes apparent that the membrane interaction of Arl2•GDP and Arl2•GppNHp is much more pronounced than that of Arl3•GDP. This is evident from the larger amount and size of protein-enriched domains in the AFM images (Fig. 4, white areas) even at a protein concentration of 2 μM. However, just like Arl3, Arl2 partitions into the ld phase, which leads to a disturbance of the membrane when large protein clusters are formed. Deletion of the N-terminus of Arl2•GppNHp results in a diminished membrane interaction, as indicated by a significantly lower amount of protein-enriched domains that are also much smaller in size.
Taken together, the data indicate the same kind of membrane partitioning for all Arl3 and Arl2 proteins, with the strongest membrane binding and clustering occurring for active Arl3, in accordance with the SPR and IRRAS results. In addition, the data also agree in showing a nucleotide-dependent membrane interaction for Arl3, but not Arl2, and in emphasizing the significance of the N-terminal amphipathic helix for the membrane binding process. Moreover, the data reveal that Arl2 and Arl3 partition preferentially into less-ordered domains of heterogeneous, anionic membranes. This is in accord with a previous report showing that the N-terminal helix of Arf1•GTP does not favor ordered lipid domains (33), strengthening the assumption that Arl3•GTP may not be able to bind to more ordered ciliary membranes.
Role of the N-terminal helix in the interaction of Arl2/3 with UNC119a
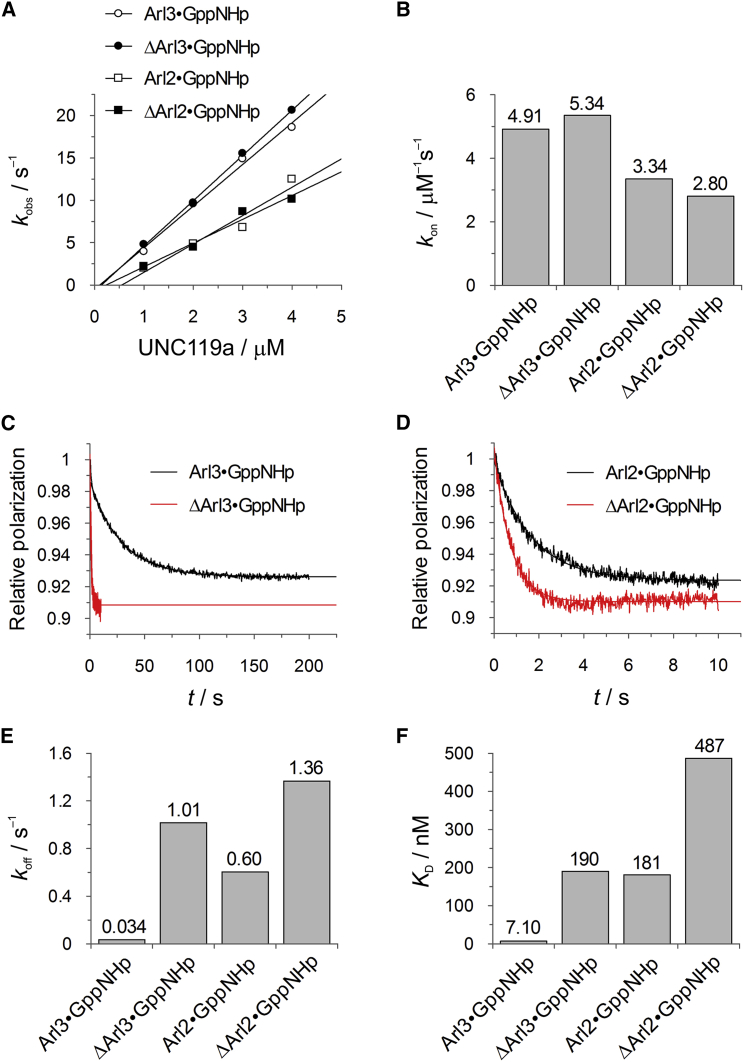
Arl proteins serve as GDI-like displacement factors for cargo bound to the shuttle factors PDEδ and UNC119. To ascertain whether the N-termini of Arl2 and Arl3 have an effect on the interaction with UNC119a, and if so, how it affects the membrane interaction, we first determined the affinities of full-length and truncated Arl2/3 for UNC119a using kinetic measurements (Fig. 5). We measured the association rate constants of the interactions via stopped-flow experiments using pseudo-first-order conditions. The second-order rate constants, kon, obtained from these data are on the order of 2.8–5.3 × 106 M−1 s−1, are somewhat higher for Arl3 than for Arl2, and are not dependent on the presence of the N-terminus (Fig. 5, A and B). The dissociation rate constants, koff, are very similar for full-length and truncated Arl2, as well as for truncated Arl3, on the order of 1 s−1. However, full-length Arl3 shows a dramatic difference, with a koff of 0.034 s−1 (Fig. 5, C–E). Using the kinetic rate constants to determine the equilibrium dissociation constants (Fig. 5 F), we find that binding of full-length Arl3 to UNC119a (7 nM) is ∼25-fold higher than that of full-length Arl2 (0.18 μM). The N-terminus is a major determinant of this higher affinity and its deletion decreases the affinity by 27-fold (0.19 μM), whereas it decreases Arl2’s affinity by only 3-fold (0.48 μM). The affinities determined here are different from those determined earlier by an equilibrium method (5), which is less accurate for such high-affinity interactions. The results achieved here are similar to those obtained for the interaction of Arl2/3 with PDEδ, where we also found a higher affinity for Arl3 and a strong dependence on the presence of the N-terminus for Arl3 only (E.K.F. and A.W., unpublished data).
Figure 5.
Influence of the Arl3 N-terminal helix on UNC119a binding. (A) Stopped-flow fluorescence polarization kinetic measurements of the association of 0.2 μM mantGppNHp-loaded Arl proteins with increasing concentrations of UNC119a. The pseudo-first-order rate constants (kobs) thus obtained are plotted against the concentration of UNC119a. (B) Bar charts of the second-order association rate constants (kon) determined from the data given in (A). (C and D) In stopped-flow fluorescence polarization kinetic experiments, complexes of 2 μM UNC119a with 0.2 μM of mantGppNHp-loaded Arl proteins were mixed with a 200-fold excess of unlabeled Arl proteins to determine koff as indicated. (E) Bar charts of the dissociation rate constants (koff) determined in (C) and (D). (F) Equilibrium dissociation constants (KD) of Arl protein complexes with UNC119a as determined from the kinetic constants in (A)–(E) using koff/kon. To see this figure in color, go online.
Influence of UNC119a on the membrane interaction of Arl2 and Arl3
Recently, we showed that the N-terminal helix of Arl3, but not that of Arl2, mediates the allosteric regulation of cargo binding to UNC119a (4). By solving the x-ray structure of the UNC119-Arl3•GppNHp complex, we showed that the N-terminal helix is not detached, as in other Arf/Arl effector complexes (e.g., the Arl2-PDEδ complex (5)), but is retained on the G domain (Fig. 1) (4). Hence, the N-terminal helix seems to be involved in the interaction between Arl3 and UNC119a, but not in the interaction between Arl2 and UNC119a. Based on these findings, we would expect UNC119a binding to Arl3 or Arl2 to have different effects on the interaction of Arl2/3 with membranes. To test this assumption, we performed SPR and IRRAS experiments as described above for Arl2 and Arl3 in the presence of UNC119a. To achieve complex formation of Arl3 or Arl2 and UNC119a, we premixed Arl2/3 and UNC119a in bulk solution and then injected this solution across the lipid bilayer surface in the SPR flow cell or underneath the lipid monolayer in the IRRAS experiments.
As indicated by the SPR data, UNC119a significantly reduced the initial membrane association rate of Arl3•GppNHp, but not that of Arl3•GDP (Figs. 2 B and 6, A and B). This would be in agreement with a GTP-specific binding of UNC119a to Arl3, with binding of UNC119a preventing membrane binding of Arl3 in the complexed state owing to the fixed position of the amphipathic helix. This is in line with the finding that the amount of quasi-irreversibly bound protein and thus stable membrane binding were significantly reduced by UNC119a for Arl3•GppNHp only (cf. Tables S2 and S3). On the other hand, no substantial nucleotide-dependent effect of UNC119a on the membrane association of Arl2 could be detected. In both the GDP- and GTP-bound states, the membrane association rate was slightly increased by the presence of UNC119a (Figs. 2 B and 6 B). For Arl2•GppNHp, this supports a previous structural analysis indicating that in the Arl2-effector complexes, the N-terminal helix is pointing into solution and thus would be available for membrane insertion (4).
Figure 6.
Membrane interaction of GDP- and GppNHp-loaded Arl2/3 in complex with UNC119a. (A) SPR sensorgrams of the binding of UNC119a-complexed Arl2/3 (cArl2/3 = 2 μM, cUNC119a = 3 μM), as well as UNC119a alone, to anionic model raft membranes composed of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 (mol %). Side-by-side comparisons of the SPR curves are shown in Fig. S5. (B) Corresponding SPR data for the initial association process. The error bars represent the standard deviation from three to nine measurements. (C) Surface pressure/time isotherms for the membrane interaction of Arl2/3 in complex with UNC119a (2:3 molar ratio), as well as UNC119a alone, with anionic lipid raft monolayers. (D–F) Corresponding time-dependent IRRA spectra for the amide-I′ region of Arl3•GppNHp (D), Arl2•GDP (E), and Arl2•GppNHp (F) in complex with UNC119a in the presence of anionic lipid raft monolayers. All IRRA spectra were recorded with p-polarized light at a 35° angle of incidence. To see this figure in color, go online.
In corresponding IRRAS experiments, UNC119a itself did not show an insertion into the membrane. However, it seemed to weakly interact with and perturb the membrane, possibly via lipid headgroup interactions. The absence of an amide-I′ band strongly suggests a rather weak membrane binding for UNC119a (cf. Fig. S4). As expected, the membrane binding capacity of the N-terminal helix of Arl3•GppNHp was highly compromised in complex with UNC119a, leading to a reduced membrane insertion of Arl3•GppNHp in complex with UNC119a (∼75% reduction, Δπ ∼0.9 mN m−1; Figs. 2 C and 6 C). Furthermore, the measured amide-I′ band intensity of UNC119a-complexed Arl3•GppNHp was 4-fold lower, indicating a lower amount of membrane-bound protein (Fig. 6 D). These results confirm the importance of the Arl3 N-terminal helix in regulating cargo release from UNC119a and in binding to membranes. This is in sharp contrast to the behavior of Arl2, where the N-terminal helix is involved in binding to membranes independently of the nucleotide state of the protein, but is not competent to allosterically regulate ciliary cargo release from UNC119a (4). Complex formation with UNC119a did not significantly alter the membrane binding behavior of Arl2 proteins in the different nucleotide-bound states (Fig. 6 C), suggesting that the N-terminal helix is not involved in the interaction between Arl2 and UNC119a. Finally, the appearance of the amide-I′ band shoulder centered around 1628/29 cm−1 (which is typical for proteins with predominantly β-sheet structures, such as UNC119a) with time for both complexes suggests the presence of UNC119a at the membrane along with Arl2•GppNHp/GDP (Fig. 6, E and F), also pointing toward different and mutually exclusive signals for an interaction of Arl2 with UNC119a and membranes. Although the interaction of Arl2 with UNC119a is GTP-specific in solution (5), the combined data of this study show that the membrane interaction of Arl2 in the presence of UNC119a is not influenced by the nature of the nucleotide bound to Arl2. This is clearly different from Arl3, which has a higher affinity to UNC119a. In turn, this affinity is highly dependent on the N-terminus, where the presence of UNC119a has a strong influence on the interaction of Arl3•GppNHp with membranes.
Conclusions
The membrane binding of Arl2 and Arl3 is expected to be mediated only by the N-terminal helix since they lack a myristoyl anchor at the N-terminal glycine. Thus, the net positive charge of the amphipathic helices of Arl2 and Arl3 is thought to promote their interaction with anionic lipid membranes. However, control experiments in which Arl3•GppNHp interacted with a zwitterionic model raft membrane revealed that membrane insertion of the N-terminal helix of Arl3•GppNHp occurred independently of the membrane composition and presence of anionic lipids (cf. Fig. S6). Moreover, each basic residue probably contributes only ∼0.5 kcal/mol to the binding free energy, possibly due to a compensation for the helix net charge reduction by a closer localization of the basic residues to the anionic membrane (34). Consequently, the different positive net charges of +4 and +3 for the helices of Arl2 and Arl3, respectively, are not supposed to significantly affect the binding free energy and membrane interaction behavior. For the Arf-related protein Sar1, it has been shown that the amphipathic helix is indeed sufficient for membrane binding (20, 21). The results presented here show that the N-terminal amphipathic helix is essential and sufficient for proper membrane binding of both Arl2 and Arl3. However, their membrane interaction behavior differs significantly with regard to nucleotide loading. Unlike Arl3 and other Arf proteins, Arl2 binds to membranes in a nucleotide-independent manner, whereas the membrane interaction of Arl3 is regulated by its nucleotide status. We propose that these differences are due to the availability of the N-terminal amphipathic helix for membrane insertion. Whereas the N-terminal helix of Arl2 seems to be flexible and sufficiently exposed in both nucleotide states to mediate an interaction with the membrane, the N-terminal helix of Arl3 is only exposed in the GTP-loaded state, preventing proper membrane binding of Arl3•GDP (Fig. 7).
Figure 7.
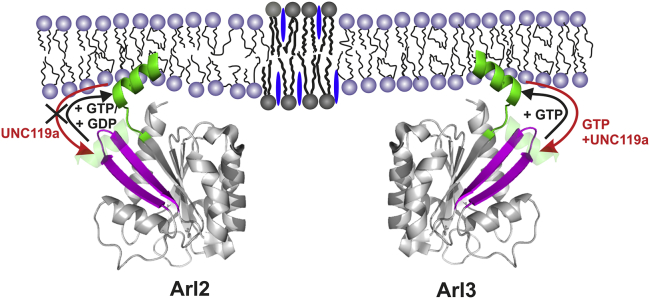
Schematic representation of the membrane interaction of Arl2 and Arl3, and the influence of UNC119a. To see this figure in color, go online.
The interaction of Arl2/3 with GDI-like solubilizing factors has already been studied in solution, but data obtained in the presence of membranes are rather scarce. Here, we show that binding of the effector UNC119a, which is GTP dependent in solution for both Arl2 and Arl3 (5), is much tighter for Arl3 than for Arl2. The fact that the N-terminal helix is almost exclusively responsible for the higher affinity seems to suggest that its position in Arl2•GTP is different from that in the Arl3•GTP-UNC119 complex structure (4), and it may be detached from the core protein, as observed with PDEδ (Fig. 1). Since Arl2 binding to membranes is independent of the nucleotide state of Arl2, we hypothesize that the N-terminal helix is flexible in both states and thus is able to interact with membranes. Correspondingly, we found that complex formation with UNC119a did not significantly alter the membrane binding behavior of Arl2 in both nucleotide-bound states, suggesting that the N-terminal helix is not involved in the interaction between Arl2 and UNC119a (Fig. 7). Moreover, this confirms the previous notion that the N-terminal helix is pointing into solution and thus would be available for membrane insertion in the Arl2•GppNHp-UNC119a complex. This might also be a reason for the inability of Arl2•GTP to allosterically regulate cargo release from UNC119a. In contrast, we show that UNC119a selectively impedes membrane binding of Arl3•GppNHp, since the N-terminal helix of Arl3•GppNHp is no longer available for membrane insertion in the UNC119a-complexed state. In turn, this implies that membrane-bound Arl3•GTP is not able to bind UNC119a/b.
Arl2 and Arl3 regulate the release of cargo from the shuttle factors PDEδ, UNC119a, and UNC119b. However, a distinction has to be made in the cell between cargo destined for cilia and cargo for the rest of the cell. Examples of the former would include myristoylated NPHP3 and farnesylated INPP5E, which are localized exclusively in cilia (11, 35, 36). Since one can assume that Arl2 and Arl3 travel freely across the functional barrier of the transition zone, a mechanism is required to retain Arl3 in the ciliary compartment. Because we and others have postulated the presence of an Arl3-specific GEF in cilia, and it has been shown that RP2, the specific Arl3-GAP, is located outside, such a GEF would specifically activate Arl3 and allow its N-terminal helix to interact with the membrane (4, 5, 11). Recently, it has also been shown that proteins interacting with the NH2-terminus of Arl3 may function as displacing factors as an alternative to membranes (37, 38).
Author Contributions
S.K., S.A.I., A.W., R.W., and K.W. conceived and designed experiments. S.A.I. and E.K.F. synthesized and purified proteins. S.K., E.K.F., S.M., and K.W. performed experiments and analyzed data. S.K., S.A.I., A.W., and K.W. wrote the article.
Acknowledgments
This research was supported by the Deutsche Forschungsgemeinschaft (DFG, SFB 642) and the Max Planck Society (IMPRS Chemical and Molecular Biology).
Editor: Heiko Heerklotz.
Footnotes
Shobhna Kapoor and Eyad K. Fansa contributed equally to this work.
Supporting Materials and Methods, six figures, and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00868-1.
Contributor Information
Alfred Wittinghofer, Email: alfred.wittinghofer@mpi-dortmund.mpg.de.
Katrin Weise, Email: katrin.weise@tu-dortmund.de.
Supporting Citations
References (39, 40, 41, 42, 43) appear in the Supporting Material.
Supporting Material
References
- 1.Gillingham A.K., Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 2.Sharer J.D., Kahn R.A. The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J. Biol. Chem. 1999;274:27553–27561. doi: 10.1074/jbc.274.39.27553. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi A., Kubota S., Inana G. Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 2003;534:26–32. doi: 10.1016/s0014-5793(02)03766-3. [DOI] [PubMed] [Google Scholar]
- 4.Ismail S.A., Chen Y.X., Wittinghofer A. Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J. 2012;31:4085–4094. doi: 10.1038/emboj.2012.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veltel S., Kravchenko A., Wittinghofer A. Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 2008;582:2501–2507. doi: 10.1016/j.febslet.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 6.Hanzal-Bayer M., Renault L., Hillig R.C. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ismail S.A., Chen Y.X., Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 8.Linari M., Hanzal-Bayer M., Becker J. The delta subunit of rod specific cyclic GMP phosphodiesterase, PDE delta, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS Lett. 1999;458:55–59. doi: 10.1016/s0014-5793(99)01117-5. [DOI] [PubMed] [Google Scholar]
- 9.Veltel S., Gasper R., Wittinghofer A. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat. Struct. Mol. Biol. 2008;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- 10.Jaworek T.J., Richard E.M., Riazuddin S. An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans. PLoS Genet. 2013;9:e1003774. doi: 10.1371/journal.pgen.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright K.J., Baye L.M., Jackson P.K. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grayson C., Bartolini F., Cheetham M.E. Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum. Mol. Genet. 2002;11:3065–3074. doi: 10.1093/hmg/11.24.3065. [DOI] [PubMed] [Google Scholar]
- 13.Bhamidipati A., Lewis S.A., Cowan N.J. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou C., Cunningham L., Kahn R.A. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell. 2006;17:2476–2487. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman L.E., Zhou C.J., Kahn R.A. The ARL2 GTPase is required for mitochondrial morphology, motility, and maintenance of ATP levels. PLoS One. 2014;9:e99270. doi: 10.1371/journal.pone.0099270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqualato S., Renault L., Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasqualato S., Ménétrey J., Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonny B., Beraud-Dufour S., Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- 19.Bologna G., Yvon C., Veuthey A.L. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 20.Lee M.C., Orci L., Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Jin H., White S.R., Nachury M.V. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wätzlich D., Vetter I., Ismail S. The interplay between RPGR, PDEδ and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep. 2013;14:465–472. doi: 10.1038/embor.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier R., Douguet D., Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 24.Weise K., Triola G., Winter R. Influence of the lipidation motif on the partitioning and association of N-Ras in model membrane subdomains. J. Am. Chem. Soc. 2009;131:1557–1564. doi: 10.1021/ja808691r. [DOI] [PubMed] [Google Scholar]
- 25.Weise K., Kapoor S., Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- 26.Kühnel K., Veltel S., Wittinghofer A. Crystal structure of the human retinitis pigmentosa 2 protein and its interaction with Arl3. Structure. 2006;14:367–378. doi: 10.1016/j.str.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Weise K., Kapoor S., Winter R. Dissociation of the K-Ras4B/PDEδ complex upon contact with lipid membranes: membrane delivery instead of extraction. J. Am. Chem. Soc. 2012;134:11503–11510. doi: 10.1021/ja305518h. [DOI] [PubMed] [Google Scholar]
- 28.Kaneshiro E.S. Lipids of ciliary and flagellar membranes. In: Bloodgood R.A., editor. Ciliary and Flagellar Membranes. Plenum Press; New York: 1990. [Google Scholar]
- 29.Tyler K.M., Fridberg A., Engman D.M. Flagellar membrane localization via association with lipid rafts. J. Cell Sci. 2009;122:859–866. doi: 10.1242/jcs.037721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapoor S., Werkmüller A., Winter R. Temperature-pressure phase diagram of a heterogeneous anionic model biomembrane system: results from a combined calorimetry, spectroscopy and microscopy study. Biochim. Biophys. Acta. 2011;1808:1187–1195. doi: 10.1016/j.bbamem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Vogel A., Nikolaus J., Huster D. Interaction of the human N-Ras protein with lipid raft model membranes of varying degrees of complexity. Biol. Chem. 2014;395:779–789. doi: 10.1515/hsz-2013-0294. [DOI] [PubMed] [Google Scholar]
- 32.Loftus A.F., Hsieh V.L., Parthasarathy R. Modulation of membrane rigidity by the human vesicle trafficking proteins Sar1A and Sar1B. Biochem. Biophys. Res. Commun. 2012;426:585–589. doi: 10.1016/j.bbrc.2012.08.131. [DOI] [PubMed] [Google Scholar]
- 33.Manneville J.B., Casella J.F., Goud B. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc. Natl. Acad. Sci. USA. 2008;105:16946–16951. doi: 10.1073/pnas.0807102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Gambhir A., McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J. Biol. Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 35.Thomas S., Wright K.J., Attié-Bitach T. A homozygous PDE6D mutation in Joubert syndrome impairs targeting of farnesylated INPP5E protein to the primary cilium. Hum. Mutat. 2014;35:137–146. doi: 10.1002/humu.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacoby M., Cox J.J., Schurmans S. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009;41:1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- 37.Behnia R., Panic B., Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- 38.Setty S.R., Strochlic T.I., Burd C.G. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- 39.Gohlke A., Triola G., Winter R. Influence of the lipid anchor motif of N-ras on the interaction with lipid membranes: a surface plasmon resonance study. Biophys. J. 2010;98:2226–2235. doi: 10.1016/j.bpj.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper M.A., Hansson A., Williams D.H. A vesicle capture sensor chip for kinetic analysis of interactions with membrane-bound receptors. Anal. Biochem. 2000;277:196–205. doi: 10.1006/abio.1999.4389. [DOI] [PubMed] [Google Scholar]
- 41.Besenicar M., Macek P., Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem. Phys. Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Mozsolits H., Thomas W.G., Aguilar M.I. Surface plasmon resonance spectroscopy in the study of membrane-mediated cell signalling. J. Pept. Sci. 2003;9:77–89. doi: 10.1002/psc.439. [DOI] [PubMed] [Google Scholar]
- 43.Mendelsohn R., Mao G., Flach C.R. Infrared reflection-absorption spectroscopy: principles and applications to lipid-protein interaction in Langmuir films. Biochim. Biophys. Acta. 2010;1798:788–800. doi: 10.1016/j.bbamem.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.