Abstract
Recent genetic analyses of large populations have revealed that somatic mutations in hematopoietic cells leading to clonal expansion are commonly acquired during human aging. Clonally restricted hematopoiesis is associated with an increased risk of subsequent diagnosis of myeloid or lymphoid neoplasia and increased all-cause mortality. Although myelodysplastic syndromes (MDS) are defined by cytopenias, dysplastic morphology of blood and marrow cells, and clonal hematopoiesis, most individuals who acquire clonal hematopoiesis during aging will never develop MDS. Therefore, acquisition of somatic mutations that drive clonal expansion in the absence of cytopenias and dysplastic hematopoiesis can be considered clonal hematopoiesis of indeterminate potential (CHIP), analogous to monoclonal gammopathy of undetermined significance and monoclonal B-cell lymphocytosis, which are precursor states for hematologic neoplasms but are usually benign and do not progress. Because mutations are frequently observed in healthy older persons, detection of an MDS-associated somatic mutation in a cytopenic patient without other evidence of MDS may cause diagnostic uncertainty. Here we discuss the nature and prevalence of CHIP, distinction of this state from MDS, and current areas of uncertainty regarding diagnostic criteria for myeloid malignancies.
Introduction
The advent of inexpensive high-throughput genome sequencing platforms has facilitated discovery of genetic lesions that drive the pathogenesis of many human disorders, including myelodysplastic syndromes (MDS).1,2 Once viewed as a biologically obscure group of preleukemic disorders, defined primarily by peripheral blood cytopenias and dysplastic cellular morphology, commonly (but not invariably) associated with clonal karyotypic abnormalities, MDS has now been associated with recurrent somatic point mutations in >40 different genes.3 Biological organization of the proteins encoded by these genes into cellular pathways for pre-mRNA splicing, epigenetic regulation, and chromatin conformation has provided additional insight into disease mechanisms and opened promising new lines of investigation into MDS pathogenesis and treatment strategies.
At the same time, recent large-scale sequencing studies have also revealed that acquisition of clonally restricted somatic mutations in MDS-associated genes in hematopoietic cells is not limited to individuals with MDS or related myeloid neoplasms.4,5 These mutations can be detected in people with normal blood counts and without any apparent disease, and their presence confers an increased risk of subsequent hematological malignancy diagnosis, as well as higher all-cause mortality.5-8 Conversely, some patients have persistent blood cytopenias for which no explanation is apparent, so-called idiopathic cytopenias of undetermined significance (ICUS), yet have unremarkable marrow morphology and lack a known MDS-associated somatic mutation or karyotypic abnormality.9,10 Patients with ICUS have no definitive evidence of a specific disorder and can only be monitored expectantly; some individuals with ICUS will subsequently be diagnosed with MDS or acute myeloid leukemia (AML).11
The rapidly emerging developments in MDS molecular biology warrant reconsideration of the definition of MDS, including reassessment of minimal diagnostic criteria. We propose the term clonal hematopoiesis of indeterminate potential (CHIP) to describe individuals with a hematologic malignancy-associated somatic mutation in blood or marrow, but without other diagnostic criteria for a hematologic malignancy. The rate of progression of CHIP to a hematologic malignancy appears to be similar to the rate of progression of other known clonal pre-malignant disorders, such as the transition of monoclonal gammopathy of undetermined significance (MGUS) to multiple myeloma. Insights into the genetic basis of these disorders, and the ongoing introduction of sequencing studies into routine clinical practice, will continue to refine distinctions between CHIP, ICUS, and MDS.
Somatic mutations in MDS
Before 2005, the only acquired point mutations recurrently associated with MDS were TP53, NRAS, KRAS, RUNX1, and ATRX; each of these lesions is present in <10% of cases.12 A few other mutations such as FLT3, KIT, or NPM1 were known to be acquired occasionally as MDS progresses to AML. In the last decade, recurrent mutations in dozens of genes, encoding proteins of diverse function, have been linked to MDS, and ≥1 mutation is detectable in almost all cases.13 Unlike myeloproliferative neoplasms, in which a mutation in 1 of just 3 genes (JAK2, CALR, and MPL) that constitutively activate hematopoietic growth factor receptor signaling is present in >85% of cases, no single mutation class is dominant in MDS. Mutations in SF3B1 and TET2, each mutated in 20% to 25% of patients with MDS, are the most commonly described abnormalities to date.14
Detection of an MDS-associated mutation can provide additional diagnostic support in cases for which the clinical presentation and morphology are ambiguous. In MDS cases with established diagnoses, certain specific mutations and the overall mutation burden predict risk of leukemic progression and death, independent of clinicopathological risk stratification tools such as the International Prognostic Scoring System.15-18 Furthermore, the presence of specific mutations (eg, TET2 and perhaps DNMT3A) predicts a higher likelihood of response to hypomethylating agent therapy, whereas other mutations (eg, TP53) predict higher relapse risk and inferior survival after allogeneic stem cell transplantation.19-21 Some acquired mutations in MDS are associated with specific clinicopathological presentations, such as the strong association of SF3B1 with ring sideroblasts,22 ATRX with acquired α thalassemia,23 RUNX1 with thrombocytopenia,16 and TP53 with a complex karyotype, therapy-related disease, and dysgranulopoiesis.24 MDS/myeloproliferative neoplasm overlap diseases also have unique genetic signatures compared with pure MDS without proliferative features, including enrichment for mutations in SRSF2, CBL, and ASXL1 in chronic myelomonocytic leukemia,25 the association of SETBP1 with atypical chronic myeloid leukemia,26,27 and coexistence of JAK2 (or, less commonly, MPL or CALR) with SF3B1 or other splicing mutations in refractory anemia with ring sideroblasts and marked thrombocytosis (RARS-T).28,29
Somatic mutations and their consequences in populations of apparently healthy persons
Hematopoietic progenitor and stem cells, like stem cells in other tissues, accumulate somatic mutations throughout life, most of which are nonpathogenic passengers without functional consequence or potential to contribute to clonal expansion.30,31 On average, 1.3 ± 0.2 somatic exonic mutations are acquired per hematopoietic stem cell per decade.31
Analysis of inactivation patterns of polymorphic X-linked genes such as glucose-6-phosphate dehydrogenase (G6PD) or the androgen receptor (AR/HUMARA) provided early evidence of the clonal nature of myeloid neoplasms.32,33 It has long been recognized via similar techniques that age-related clonal skewing of hematopoietic cells is also a relatively common finding after the age of 55 to 65 years, and although this phenomenon can in rare cases result in late presentation of an X-linked disorder in a female (eg, initial presentation of X-linked congenital sideroblastic anemia caused by germ-line ALAS2 mutation in an octagenarian34), age-related clonal skewing is infrequently associated with disease.35-38
As sensitive assays including polymerase chain reaction and fluorescent in situ hybridization became widely available in the 1990s, hematologic neoplasm-associated genetic abnormalities were described in the blood of some healthy people, especially older adults. Recurrent abnormalities observed in the absence of disease include low-level oncogenic BCL2 translocations in patients without evidence of lymphoma and low-allele-burden BCR-ABL fusions that never evolve into chronic myeloid leukemia.39,40 When these reports first appeared, it was unclear why such changes did not inevitably lead to disease. Speculation focused on whether mutation-bearing cells were eliminated by an adaptive immune response, mutations occurred in cells lacking self-renewal properties, or additional cooperating mutations were required to cause disease. The allele fraction for these mutations was also low compared with more recently described mutations that contribute to clonal expansion in older individuals.
In 2012, 2 analyses of genome-wide association study (GWAS) cohorts described copy number changes at chromosomal loci associated with hematological neoplasia, such as 20q, 5q, 11q, and 17p, in 2% of apparently healthy people >70 years of age.41,42 In these studies, acquired clonal mosaicism predicted an increased risk of subsequent diagnosis of a neoplasm, indicating that such changes can represent disease-initiating events in some cases.42 Additionally, TET2 mutations were described in some older women with age-related hematopoietic clonal skewing but normal blood counts, and DNMT3A point mutations identical to those observed in MDS were reported in nonneoplastic blood cells from other patient cohorts.4,5
In the last few months, 3 studies have further expanded our understanding of the premalignant genetic changes that may serve as initiating events in hematologic neoplasms by promoting clonal expansion (Figure 1), illuminating the frequency of variant alleles in older people and clinical associations with those alleles.6-8 The Washington University genomics group analyzed The Cancer Genome Atlas blood sequencing data from 2728 patients with nonhematologic malignancies and identified blood-specific clonal mutations in >2% of all samples and in 5% to 6% of people >70 years of age.6 Similarly, exome sequencing study of 12 380 Swedish patients without hematologic malignancy and with medical follow-up ranging from 2 to 7 years demonstrated clonal hematopoiesis in 10% of individuals >65 years of age.8 In the third analysis, investigators pooled whole exome sequencing datasets derived from 17 182 people in 22 GWAS cohorts focused on risk factors for diabetes mellitus; detectable somatic mutations were rare in study subjects <40 years of age but were present in 9.6% of 2299 people aged 70 to 79, 11.7% of 317 people aged 80 to 89, and 18.4% of 103 people aged ≥90 years.7 These overall mutation rates, as well as increased mutation frequency with aging, parallel epidemiologic patterns observed with other clonal states known to be precursors to hematological neoplasms, such as MGUS (a precursor state for multiple myeloma, light-chain amyloidosis, and some lymphoid neoplasms) and monoclonal B-cell lymphocytosis (MBL, a precursor for chronic lymphocytic leukemia and other B-cell lymphomas).43-45
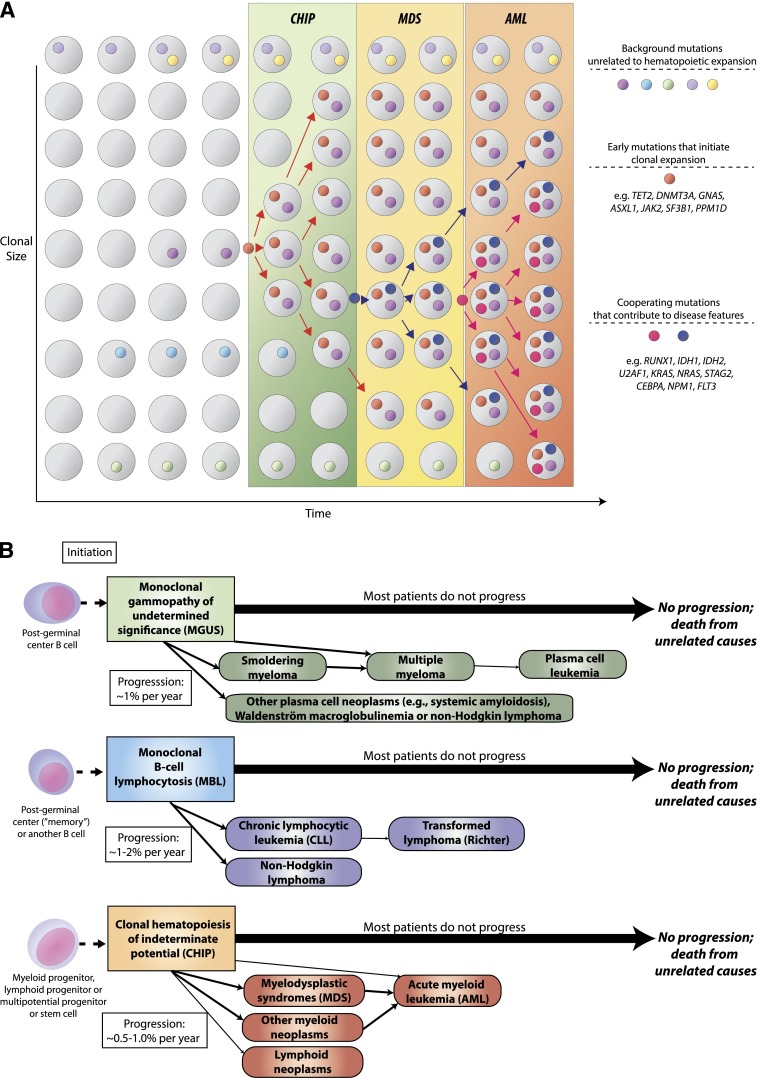
Figure 1.
CHIP as a precursor state for hematological neoplasms. (A) A model for evolution from normal hematopoiesis to CHIP and then, in some cases, to MDS or AML. (B) Comparison of evolution patterns of MGUS, MBL, and CHIP. Hematopoietic progenitor or stem cells commonly acquire mutations throughout the human lifespan; most of these are passenger mutations that have no consequence for hematopoiesis. Certain mutations, however, confer a survival advantage to the mutated cell and its progeny and allow clonal expansion. Serial acquisition of mutations in an expanded clone can lead to a disease phenotype and ultimately morbidity and mortality. Although this article primarily discusses CHIP in the context of its distinction from MDS, CHIP can also directly progress to AML without an intervening MDS stage, and CHIP can progress to other conditions such as myeloproliferative neoplasms or lymphoid neoplasms. Just as with MGUS and MBL, the majority of patients with CHIP will never develop an overt neoplasm, and patients will eventually die of unrelated causes.
More than 80% of the observed mutations in the The Cancer Genome Atlas dataset were in 19 genes previously associated with leukemia or lymphoma, including ASXL1, TP53, BCORL1, GNAS, and SF3B1, as well as the previously noted DNMT3A, TET2, and JAK2. In the Swedish cohort, the most common genes identified as mutated were the hematologic malignancy-associated DNMT3A, ASXL1, and TET2, as well as PPM1D, which encodes a phosphatase sometimes found to be mutated in blood cells in patients with brainstem gliomas, breast cancer, and other nonhematopoietic neoplasms.7 Individuals with clonal mutations had an increased risk of a subsequent hematological malignancy diagnosis (hazard ratio [HR], 12) and death (HR, 1.4) compared with age-matched persons without mutations. Variants in DNMT3A, TET2, and ASXL1 were also the most commonly identified sequence abnormalities in the diabetes mellitus GWAS study, and the presence of clonal mutations was associated with increased risk of subsequent hematologic malignancy diagnosis (HR, 11 during a median follow-up of 95 months) and all-cause mortality (HR, 1.4), as well as development of coronary heart disease (HR, 2.0) and ischemic stroke (HR, 2.6). Increased mortality was synergistic with elevated red cell distribution width, which might be a marker of disordered erythropoiesis because of the expanded clone.
Current understanding of the multistep pathogenesis of cancer suggests that individuals with clonal mutations may already be partway along the path to evolution of frank malignancy. However, although a higher rate of subsequent cancer diagnosis in patients with mutations is not surprising, mutations by themselves do not currently define a diagnosis of MDS/AML.
Current minimal diagnostic criteria for MDS and limitations
Minimal diagnostic criteria for MDS published by the World Health Organization (WHO; 2008 classification, currently undergoing a revision to be released in 2016) require the presence of blood cytopenias and exclusion of reactive or other nonhematopoietic causes of those cytopenias.46 In addition, ≥1 of the following diagnostic features must be present to diagnose MDS: excess blasts (≥5%) with a myeloid phenotype (but <20% blasts, which would qualify as AML); >10% dysplastic cells in at ≥1 of the 3 myeloid lineages (erythroid, granulocytic, megakaryocytic) or ≥15% ring sideroblasts as a proportion of erythroid precursors; or evidence of clonality as manifested by an abnormal MDS-associated karyotype.46 If the latter group of cytogenetically abnormal cases does not meet blast or dysplasia criteria for MDS diagnosis, they are diagnosed as “MDS, unclassifiable” and have a natural history similar to MDS.47 Overreporting of unclassifiable MDS in population-based registries such as the US Surveillance, Epidemiology and End Results program illustrates the challenges in diagnosing and classifying MDS.48
Although karyotyping is an important part of the evaluation of a patient with suspected MDS, the authors of the WHO classification recognized that not all clonal markers detectable by conventional cytogenetic assays have equivalent diagnostic importance. Three karyotypes that can be seen in other non-MDS situations are specifically excluded as MDS-qualifying anomalies by the WHO: isolated loss of the Y chromosome, which is present in 5% to 10% of healthy older men49; trisomy 8, which is frequently associated with aplastic anemia and predicts a higher likelihood of response to immunosuppressive therapy50; and del(20q), which has been recurrently identified in some patients with cytopenias but without clear evidence of MDS.51-53 Others have proposed that additional karyotypic abnormalities, such as isolated trisomy 15, also lack diagnostic significance.54
Because patients presenting with idiopathic cytopenias may have nondiagnostic marrow morphology and a normal karyotype, additional diagnostic information from mutational analysis could be clinically useful. Many academic hematopathology groups and commercial pathology laboratories have recently incorporated mutation assay platforms into diagnostic testing algorithms for patients with suspected hematologic malignancies, so somatic mutation data are increasingly provided to clinicians, sometimes accompanied by interpretation of findings. Unlike current morphologic and cytogenetic diagnosis of MDS, which requires marrow aspiration, MDS-associated mutations can usually be detected in a blood sample; therefore, it is tempting to consider detection of an MDS-associated somatic mutation by itself as sufficient to diagnose MDS.
However, the presence of MDS-associated mutations in older individuals without evidence of disease suggests caution is indicated in rendering a diagnosis of MDS or another myeloid neoplasm primarily on the basis of a somatic mutation, especially when only a single mutation is present and if it is one of those commonly seen in healthy people, such as DNMT3A, TET2, or ASXL1. Although the likelihood that such an identified mutation is clinically relevant is higher in patients who are undergoing evaluation for cytopenia than in the general population, it is still possible that such a patient might be among the ∼10% of older adults who have clonal mutations and that the cytopenias could be from a nonclonal cause.
An ad hoc working group proposal for MDS minimal diagnostic criteria largely followed WHO lines, but also included co-criteria for diagnosis that might be useful in difficult cases, such as decreased circulating colony-forming cells, abnormal flow cytometric immunophenotype, aberrant gene expression pattern, or the presence of an MDS-associated somatic mutation.10,55 Questions can be raised about the diagnostic specificity and sensitivity of all these criteria, as well as the WHO morphologic criteria for MDS, which originate from the French-American-British Cooperative Group Classification56 and are based on convention rather than biology. For instance, the 15% threshold for ring sideroblasts used by the French-American-British Cooperative Group and subsequently by the WHO arose from a small study performed in the early 1980s where there appeared to be a break in the distribution of ring sideroblast proportion between 10% and 20%.57,58 Now it is known that the presence of an SF3B1 mutation is almost invariably associated with some ring sideroblasts, but these often represent <15% of erythroid precursors; this observation may warrant revision of the definition of RARS.59 Cogent arguments also exist that any blast excess defines a disease akin to AML and that the current 20% blast threshold is either too high, or, alternatively, that the threshold is too low, because AML with ≥30% blasts is more aggressive than AML with 20% to 29% blasts.60,61 Finally, the WHO 10% dysplasia threshold is equally arbitrary, especially given interobserver differences in dysplasia assessment62,63; thresholds other than 10% for certain types of dysplasia may improve ability of morphologic criteria to distinguish MDS from non-MDS conditions.64
Historically, MDS diagnosis has relied heavily on detection of morphologic dysplasia. However, this reliance is problematic. Not all that is dysplastic is MDS.65 A broad range of pathologies can cause cell morphology changes that may be mistaken for MDS, including viral infections (eg, HIV infection), alcohol abuse, exposure to cytotoxic agents (eg, azathioprine, methotrexate), or nutritional deficiencies (eg, copper, folate, cobalamin). Diagnosticians reviewing a marrow sample for dysplasia may erroneously diagnose MDS in a patient with a cytopenia caused by a non-MDS condition. Conversely, even when significant dysplasia is present, an incomplete history may prevent a pathologist from confidently diagnosing MDS, because of concern that a non-MDS condition that has not been disclosed to the pathologist might be causing dysplasia mimicking MDS.
In addition to the above known causes of dysplasia, a proportion of persons, especially those >50 years of age, have dysplastic changes on marrow aspiration in the absence of cytopenias or a known cause for dysplasia.66 The term idiopathic dysplasia of undetermined significance (IDUS) has been proposed to describe these changes.67 It is not known whether IDUS is associated with a higher incidence of subsequent MDS diagnosis or the frequency with which IDUS has a nonhematopoietic cause. In contrast, patients with cytopenias and MDS-associated somatic mutations but normal morphology can have clinical behavior and outcome similar to MDS, analogous to cytopenic patients with chromosomal abnormalities but without dysplasia, who have a risk of AML progression and death from cytopenias.11,51 Although light microscopy offers advantages of low cost and widespread availability, the fact that genetic abnormalities in the absence of abnormal cell morphology predict clinical evolution similar to MDS argues that mutations should be considered together with microscopy in supporting MDS diagnosis.
Proposed criteria for CHIP
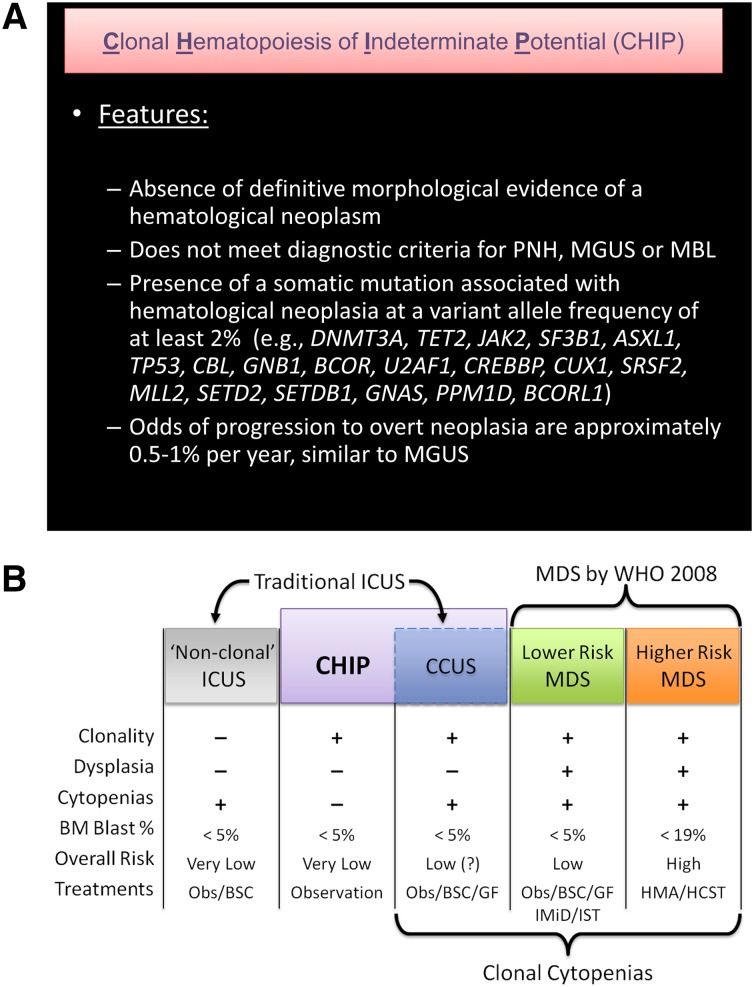
We propose that CHIP be used to describe patients who have detectable somatic clonal mutations in genes recurrently mutated in hematologic malignancies (Figure 2A) but who lack a known hematologic malignancy or other clonal disorder. Under this definition, CHIP would encompass cytopenic patients with concurrent cancer-associated mutations who do not meet diagnostic criteria for MDS, as well as those with normal peripheral blood counts. CHIP would not include clearly described clonal conditions such as paroxysmal nocturnal hemoglobinuria, MBL, or MGUS. We also propose that as a working definition, the mutant allele fraction must be ≥2% in the peripheral blood, because with deep enough sequencing, a mutation can be found in every individual, and current outcomes data are based on a minimum variant allele fraction of >2% in peripheral blood.7,68 It is currently unknown if variants present below this threshold also carry increased risk of adverse outcomes, and this threshold may need to be revised. A copy number variant resulting from a chromosomal rearrangement involving a chromosomal region where hematologic neoplasia-associated genes are encoded is also consistent with CHIP.
Figure 2.
Definition of CHIP and its distinction from MDS and non-clonal cytopenic states. (A) A proposed definition of CHIP. A mutation that is commonly associated with clonal expansion of hematopoietic cells in older persons should be present, whereas criteria for other diagnoses should not be met. Evidence of mildly disordered erythropoiesis such as an elevated red cell distribution width or mean corpuscular volume can be compatible with CHIP rather than MDS, and occasional dysplastic cells might be seen, as is common in the general population with careful scrutiny of blood and marrow. CHIP is associated with an increased risk of all-cause mortality and subsequent diagnosis of hematological malignancy. The 19 genes most commonly mutated in healthy older adults in sequencing studies to date are listed. The roster of CHIP-associated mutations will likely change in the future, with some genes being removed and others being added. As a working definition, we propose a variant allele frequency of 2% in order to be considered CHIP (since extremely deep sequencing will detect mutations in almost every person), but this may need to be revised with further population analyses. PNH, paroxysmal nocturnal hemoglobinuria. (B) The spectrum of clonal hematopoiesis, ICUS, and MDS. ICUS is a broad category that includes a heterogeneous group of individuals, some of whom have benign (nonclonal) hematopoiesis. Other patients with ICUS may have CHIP, differing only from lower risk MDS by their lack of dysplasia and, currently, an undetermined disease risk. CHIP can also include patients with clonal hematopoiesis and nonmalignant causes of cytopenias (eg, immune cytopenias, liver disease, or nutritional deficiencies) that would not be considered to have ICUS because of the presence of a clone, but may have a distinct natural history. Obs, observation; BM, bone marrow; CCUS, clonal cytopenias of undetermined significance; HMA, hypomethylating agent (eg, azacitidine); GF, hematopoietic growth factor (eg, epoetin); IMiD, immunomodulatory drug (eg, lenalidomide); IST, immunosuppressive therapy; BSC, best supportive care; HSCT, hematopoietic stem cell transplant.
CHIP is distinct from MDS because CHIP is associated with a much longer survival, normal blood counts in most cases, and low rate of progression to AML. Individuals with CHIP have an increased risk of disease progression to hematologic neoplasia compared with individuals without detectable mutations, and this risk appears to be proportional to the size of the somatic clone; however, the rate of progression appears to be only 0.5% to 1% per year, similar to MBL and MGUS. Although the annual rate of progression of CHIP, MBL, and MGUS to overt neoplasia is comparable, MBL and MGUS represent expansions of lineage-committed cells, whereas CHIP involves hematopoietic stem cells or less mature progenitor cells, and thus CHIP is a precursor state for a broader range of hematologic neoplasms (Figure 1B).
Although a growing body of evidence confirms that individuals with a first “hit” leading to CHIP are at increased risk of developing overt malignancy and that CHIP is also associated with increased mortality from nonneoplastic causes, the optimal clinical management of these individuals is uncertain. In the absence of any available intervention to prevent progression, surveillance similar to what is currently recommended for MGUS could be considered.4,8,69 In the near term, most individuals who will be screened for somatic mutations will be those with hematologic abnormalities that do not meet criteria for current diagnostic entities. Future studies will be needed to delineate whether certain subgroups with CHIP have a higher risk of progression to malignancy or other adverse outcomes. Such subgroups could include those with an unexplained cytopenia (see below), lymphocytosis, leukocytosis, persistent eosinophilia, or persistent monocytosis that do not meet criteria for recognized entities. The specific genetic mutations, number of mutations, and mutant allele fraction may also influence the risk of progression and could further refine diagnostic criteria. It is likely that a substantial number of people without hematologic abnormalities who have genetic sequencing performed for personal, research, or medical reasons (eg, on a blood-contaminated solid tumor biopsy) will incidentally be found to have CHIP. In certain settings, such as following cytotoxic therapy for a nonhematologic disorder, CHIP may have more ominous implications.70
Proposed diagnostic criteria for MDS
To diagnose MDS, it is important to rule out other disorders that can mimic MDS. Testing algorithms such as those advocated by the National Comprehensive Cancer Network and the European LeukemiaNet are helpful for this purpose.71,72 Given the large percentage of individuals with CHIP, a somatic mutation is not sufficient to diagnose MDS, even in the context of prior therapy for another malignancy with radiation or cytotoxic drugs or a history of another marrow failure disorder such as aplastic anemia.73 Like dysplasia or MDS-associated cytogenetic abnormalities, somatic mutations must have a known association with MDS and be accompanied by clinically meaningful cytopenias to consider an MDS diagnosis.
Patients with cytopenias who lack MDS-defining features but in whom no other cause for cytopenias is evident are said to have ICUS. The ICUS label includes a highly heterogeneous population, and ICUS may resolve spontaneously or may ultimately be determined to be caused by nonmyeloid neoplasia or a nonneoplastic condition. Good natural history studies of ICUS are lacking.9,11,74 In the largest study to date, ICUS evolved to MDS in a minority of cases; further conclusions were limited by a lack of long-term follow-up.11
A critical question is whether patients with cytopenias and MDS-associated somatic mutations, but lacking MDS-defining dysplasia, blast increase, or karyotype criteria, should be diagnosed with MDS. If such patients were to be included in the MDS category, this would represent a major redefinition of MDS that would increase its incidence. Adding additional complexity, some of the mutations that drive MDS and other myeloid malignancies do not, by themselves, cause cellular dysplasia.
Emerging data indicate that clonal cytopenias without morphologic or cytogenetic evidence of MDS are at least as common as bona fide MDS.75 For example, in 1 prospective study that examined patients with unexplained cytopenias for mutations in 21 genes, patients with cytopenias were stratified into 3 groups based on hematopathology evaluation: definite MDS, some evidence of MDS but not fully meeting WHO diagnostic criteria, and cytopenias with no evidence of MDS. Although >80% of the definite MDS patients and 50% of the equivocal cases had typical mutations, so did 22% of the patients lacking any evidence for MDS: a frequency higher than the background rate of CHIP in an age-matched population.75 Genes mutated in the group of cytopenic patients with no evidence of dysplasia included several that have been associated with a poor prognosis in MDS (eg, TP53, RUNX1, and ASXL1). In a retrospective study, mutations were detected in 33% of 250 ICUS cases compared with 83% in a lower-risk MDS control group.76 These data suggest that the incidence of MDS would at least double if patients with unexplained cytopenias and typical somatic mutations, but without MDS-defining morphologic dysplasia, were considered to have MDS. Further study is required to confirm the apparent similarity in disease biology of such cases to bona fide MDS, which would validate their inclusion in MDS despite lack of the dysplasia implied by the disease name.
Because >85% of patients with MDS have somatic mutations detectable in 1 or more of a few dozen recurrently mutated genes,3 mutation analysis has a high negative predictive value. Although the presence of a mutation alone may not mean a patient has MDS, the absence of a mutation is a good predictor of not having MDS, and this may represent a useful role for clinical mutation testing in ambiguous cytopenia cases. Although a few individuals may have pathologically consequential mutations that have not yet been identified or that are not part of widely used testing panels and others might have only copy number abnormalities,77 the majority of MDS patients will have one of the common mutations, and thus an absence of mutations should prompt a more vigorous search for non-MDS causes of cytopenias. Redefinition proposals need to be validated prospectively and fit with current biological understanding and clinical experience. We recognize that testing for somatic mutations is not yet universal (especially in resource-poor settings), but such testing is increasingly available in routine clinical practice and should ultimately reduce the number of diagnostically ambiguous cases.
Is CHIP or MDS cancer?
For many years there has been ambiguity about whether MDS can be described as cancer.78,79 This semantic distinction influences patient self-perception and has practical consequences for regulatory jurisdiction, health care resource allocation, cancer-specific indemnity policy payouts, and research funding eligibility.
MDS is a clonal disorder, but clonality is not the sole defining characteristic of neoplasia. The concept of cancer is somewhat ill defined and includes a variety of factors in addition to a specific biology, such as a natural history component and compromise of normal tissue function.80,81 Although MDS is classified as cancer by the WHO and is treated by oncologists in many settings, and MDS shares some biological features with leukemia or other overt neoplasms, there are other features of MDS that are not typical of cancer, such as response to immunosuppressive therapy in some cases and stability for more than a decade in others.82 Given the similar survival of the 4 International Prognostic Scoring System risk groups of MDS to the 4 stages of non–small-cell lung cancer, the operating classification of MDS as cancer seems appropriate from a natural history standpoint. If cancer were to be defined primarily by burden of disease, as has historically been used to distinguish MGUS from multiple myeloma and MBL from CLL, then CHIP, like MDS, would be considered cancer, because in CHIP a large fraction of hematopoietic cells are clonal. The recent redefinition of smoldering myeloma based on distinct outcomes illustrates the potential for future reassessment of the nature of CHIP and MDS as additional data become available.83
Conclusion
Improved understanding of hematopoiesis in aging persons requires revisiting the boundary between health and disease and re-examination of what specific alterations are necessary to label a patient as having MDS. Clonal hematopoiesis in the absence of cytopenias is of indeterminate potential, but conveys health risks, and is likely to become more common as the global population ages. In the future, detection of an initiating mutation might prompt interventions to eliminate a developing dangerous clone and restore normal hematopoiesis, opening up a new field of preventive hematology applicable to CHIP, as well as MGUS, MBL, and other clonal preneoplastic conditions.
Acknowledgment
D.P.S., B.L.E., M.A.S., R.C.L., and R.B. acknowledge support from the Edward P. Evans Foundation.
Footnotes
There is an Inside Blood Commentary on this article in this issue.
Authorship
Contribution: D.P.S. wrote the initial draft and prepared the figures; and all authors revised and contributed to the manuscript.
Conflict-of-interest disclosure: D.P.S., R.B., and B.L.E. served as consultants for Genoptix. R.B. and B.L.E. have technology licensed to Genoptix. The remaining authors declare no competing financial interests.
Correspondence: David P. Steensma, Dana-Farber Cancer Institute and Brigham and Women's Hospital, 450 Brookline Ave, Boston, MA 02215; e-mail: david_steensma@dfci.harvard.edu.
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bejar R, Steensma DP. Recent developments in myelodysplastic syndromes. Blood. 2014;124(18):2793–2803. doi: 10.1182/blood-2014-04-522136. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Malcovati L, et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shlush LI, Zandi S, Mitchell A, et al. HALT Pan-Leukemia Gene Panel Consortium. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovese G, Kähler AK, Rose SA, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. doi: 10.1056/NEJMoa1409405. 2014;371(26)2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valent P, Bain BJ, Bennett JM, et al. Idiopathic cytopenia of undetermined significance (ICUS) and idiopathic dysplasia of uncertain significance (IDUS), and their distinction from low risk MDS. Leuk Res. 2012;36(1):1–5. doi: 10.1016/j.leukres.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Valent P, Horny HP. Minimal diagnostic criteria for myelodysplastic syndromes and separation from ICUS and IDUS: update and open questions. Eur J Clin Invest. 2009;39(7):548–553. doi: 10.1111/j.1365-2362.2009.02151.x. [DOI] [PubMed] [Google Scholar]
- 11.Hanson CA, Hoyer JD, Zakko L, Hodnefield JM, Steensma DP. Is idiopathic cytopenia of undetermined significance (ICUS) a valid clinical concept? A longitudinal clinicopathological study. In: Proceedings from the 10th International Symposium on Myelodysplastic Syndromes, May 6-9, 2009, Patras, Greece. Abstract P092. [Google Scholar]
- 12.Look AT. Molecular pathogenesis of MDS. Hematology Am Soc Hematol Educ Program. 2005;2005:156–160. doi: 10.1182/asheducation-2005.1.156. [DOI] [PubMed] [Google Scholar]
- 13.Kulasekararaj AG, Mohamedali AM, Mufti GJ. Recent advances in understanding the molecular pathogenesis of myelodysplastic syndromes. Br J Haematol. 2013;162(5):587–605. doi: 10.1111/bjh.12435. [DOI] [PubMed] [Google Scholar]
- 14.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejar R, Stevenson KE, Caughey BA, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejar R. Clinical and genetic predictors of prognosis in myelodysplastic syndromes. Haematologica. 2014;99(6):956–964. doi: 10.3324/haematol.2013.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. [PubMed] [Google Scholar]
- 19.Traina F, Visconte V, Elson P, et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28(1):78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- 20.Bejar R, Lord A, Stevenson K, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bejar R, Stevenson KE, Caughey B, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32(25):2691–2698. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaemmanuil E, Cazzola M, Boultwood J, et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steensma DP, Higgs DR, Fisher CA, Gibbons RJ. Acquired somatic ATRX mutations in myelodysplastic syndrome associated with alpha thalassemia (ATMDS) convey a more severe hematologic phenotype than germline ATRX mutations. Blood. 2004;103(6):2019–2026. doi: 10.1182/blood-2003-09-3360. [DOI] [PubMed] [Google Scholar]
- 24.Lai JL, Preudhomme C, Zandecki M, et al. Myelodysplastic syndromes and acute myeloid leukemia with 17p deletion. An entity characterized by specific dysgranulopoïesis and a high incidence of P53 mutations. Leukemia. 1995;9(3):370–381. [PubMed] [Google Scholar]
- 25.Wassie EA, Itzykson R, Lasho TL, et al. Molecular and prognostic correlates of cytogenetic abnormalities in chronic myelomonocytic leukemia: a Mayo Clinic-French Consortium Study. Am J Hematol. 2014;89(12):1111–1115. doi: 10.1002/ajh.23846. [DOI] [PubMed] [Google Scholar]
- 26.Maxson JE, Gotlib J, Pollyea DA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368(19):1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SA, Hasserjian RP, Fox PS, et al. Atypical chronic myeloid leukemia is clinically distinct from unclassifiable myelodysplastic/myeloproliferative neoplasms. Blood. 2014;123(17):2645–2651. doi: 10.1182/blood-2014-02-553800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malcovati L, Cazzola M. Refractory anemia with ring sideroblasts. Best Pract Res Clin Haematol. 2013;26(4):377–385. doi: 10.1016/j.beha.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Jeromin S, Haferlach T, Weissmann S, et al. Refractory anemia with ring sideroblasts and marked thrombocytosis (RARS-T) cases harbor mutations in SF3B1 or other spliceosome genes accompanied by JAK2V617F and ASXL1 mutations. Haematologica. doi: 10.3324/haematol.2014.119032. 2015;100(4):3125-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barr RD, Fialkow PJ. Clonal origin of chronic myelocytic leukemia. N Engl J Med. 1973;289(6):307–309. doi: 10.1056/NEJM197308092890608. [DOI] [PubMed] [Google Scholar]
- 33.Adamson JW, Fialkow PJ, Murphy S, Prchal JF, Steinmann L. Polycythemia vera: stem-cell and probable clonal origin of the disease. N Engl J Med. 1976;295(17):913–916. doi: 10.1056/NEJM197610212951702. [DOI] [PubMed] [Google Scholar]
- 34.Cotter PD, May A, Fitzsimons EJ, et al. Late-onset X-linked sideroblastic anemia. Missense mutations in the erythroid delta-aminolevulinate synthase (ALAS2) gene in two pyridoxine-responsive patients initially diagnosed with acquired refractory anemia and ringed sideroblasts. J Clin Invest. 1995;96(4):2090–2096. doi: 10.1172/JCI118258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gale RE. Evaluation of clonality in myeloid stem-cell disorders. Semin Hematol. 1999;36(4):361–372. [PubMed] [Google Scholar]
- 36.Knudsen GP, Pedersen J, Klingenberg O, Lygren I, Ørstavik KH. Increased skewing of X chromosome inactivation with age in both blood and buccal cells. Cytogenet Genome Res. 2007;116(1-2):24–28. doi: 10.1159/000097414. [DOI] [PubMed] [Google Scholar]
- 37.Champion KM, Gilbert JG, Asimakopoulos FA, Hinshelwood S, Green AR. Clonal haemopoiesis in normal elderly women: implications for the myeloproliferative disorders and myelodysplastic syndromes. Br J Haematol. 1997;97(4):920–926. doi: 10.1046/j.1365-2141.1997.1933010.x. [DOI] [PubMed] [Google Scholar]
- 38.Hatakeyama C, Anderson CL, Beever CL, Peñaherrera MS, Brown CJ, Robinson WP. The dynamics of X-inactivation skewing as women age. Clin Genet. 2004;66(4):327–332. doi: 10.1111/j.1399-0004.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 39.Biernaux C, Loos M, Sels A, Huez G, Stryckmans P. Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood. 1995;86(8):3118–3122. [PubMed] [Google Scholar]
- 40.Limpens J, Stad R, Vos C, et al. Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals. Blood. 1995;85(9):2528–2536. [PubMed] [Google Scholar]
- 41.Laurie CC, Laurie CA, Rice K, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44(6):642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs KB, Yeager M, Zhou W, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyle RA, Therneau TM, Rajkumar SV, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med. 2006;354(13):1362–1369. doi: 10.1056/NEJMoa054494. [DOI] [PubMed] [Google Scholar]
- 44.Shim YK, Rachel JM, Ghia P, et al. Monoclonal B-cell lymphocytosis in healthy blood donors: an unexpectedly common finding. Blood. 2014;123(9):1319–1326. doi: 10.1182/blood-2013-08-523704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shanafelt TD, Ghia P, Lanasa MC, Landgren O, Rawstron AC. Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia. 2010;24(3):512–520. doi: 10.1038/leu.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 47.Hwang Y, Huh J, Mun Y, Seong C, Chung W. Characteristics of myelodysplastic syndrome, unclassifiable by WHO classification 2008. Ann Hematol. 2011;90(4):469–471. doi: 10.1007/s00277-010-1018-x. [DOI] [PubMed] [Google Scholar]
- 48.Ma X, Does M, Raza A, Mayne ST. Myelodysplastic syndromes: incidence and survival in the United States. Cancer. 2007;109(8):1536–1542. doi: 10.1002/cncr.22570. [DOI] [PubMed] [Google Scholar]
- 49.Pierre RV, Hoagland HC. 45,X cell lines in adult men: loss of Y chromosome, a normal aging phenomenon? Mayo Clin Proc. 1971;46(1):52–55. [PubMed] [Google Scholar]
- 50.Maciejewski JP, Risitano A, Sloand EM, Nunez O, Young NS. Distinct clinical outcomes for cytogenetic abnormalities evolving from aplastic anemia. Blood. 2002;99(9):3129–3135. doi: 10.1182/blood.v99.9.3129. [DOI] [PubMed] [Google Scholar]
- 51.Steensma DP, Dewald GW, Hodnefield JM, Tefferi A, Hanson CA. Clonal cytogenetic abnormalities in bone marrow specimens without clear morphologic evidence of dysplasia: a form fruste of myelodysplasia? Leuk Res. 2003;27(3):235–242. doi: 10.1016/s0145-2126(02)00161-3. [DOI] [PubMed] [Google Scholar]
- 52.Soupir CP, Vergilio JA, Kelly E, Dal Cin P, Kuter D, Hasserjian RP. Identification of del(20q) in a subset of patients diagnosed with idiopathic thrombocytopenic purpura. Br J Haematol. 2009;144(5):800–802. doi: 10.1111/j.1365-2141.2008.07508.x. [DOI] [PubMed] [Google Scholar]
- 53.Braun T, de Botton S, Taksin AL, et al. Characteristics and outcome of myelodysplastic syndromes (MDS) with isolated 20q deletion: a report on 62 cases. Leuk Res. 2011;35(7):863–867. doi: 10.1016/j.leukres.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Hanson CA, Steensma DP, Hodnefield JM, et al. Isolated trisomy 15: a clonal chromosome abnormality in bone marrow with doubtful hematologic significance. Am J Clin Pathol. 2008;129(3):478–485. doi: 10.1309/2VQLDHVJB9YW83KB. [DOI] [PubMed] [Google Scholar]
- 55.Valent P, Horny HP, Bennett JM, et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk Res. 2007;31(6):727–736. doi: 10.1016/j.leukres.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 56.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982;51(2):189–199. [PubMed] [Google Scholar]
- 57.Juneja SK, Imbert M, Sigaux F, Jouault H, Sultan C. Prevalence and distribution of ringed sideroblasts in primary myelodysplastic syndromes. J Clin Pathol. 1983;36(5):566–569. doi: 10.1136/jcp.36.5.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wardrop D, Steensma DP. Is refractory anaemia with ring sideroblasts and thrombocytosis (RARS-T) a necessary or useful diagnostic category? Br J Haematol. 2009;144(6):809–817. doi: 10.1111/j.1365-2141.2008.07526.x. [DOI] [PubMed] [Google Scholar]
- 59.Damm F, Thol F, Kosmider O, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. doi: 10.1038/leu.2011.321. 2012;26(5)1137-1140. [DOI] [PubMed] [Google Scholar]
- 60.Lichtman MA. Does a diagnosis of myelogenous leukemia require 20% marrow myeloblasts, and does <5% marrow myeloblasts represent a remission? The history and ambiguity of arbitrary diagnostic boundaries in the understanding of myelodysplasia. Oncologist. 2013;18(9):973–980. doi: 10.1634/theoncologist.2013-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hasserjian RP, Campigotto F, Klepeis V, et al. De novo acute myeloid leukemia with 20-29% blasts is less aggressive than acute myeloid leukemia with ≥30% blasts in older adults: a Bone Marrow Pathology Group study. Am J Hematol. 2014;89(11):E193–E199. doi: 10.1002/ajh.23808. [DOI] [PubMed] [Google Scholar]
- 62.Ramos F, Fernández-Ferrero S, Suárez D, et al. Myelodysplastic syndrome: a search for minimal diagnostic criteria. Leuk Res. 1999;23(3):283–290. doi: 10.1016/s0145-2126(98)00166-0. [DOI] [PubMed] [Google Scholar]
- 63.Parmentier S, Schetelig J, Lorenz K, et al. Assessment of dysplastic hematopoiesis: lessons from healthy bone marrow donors. Haematologica. 2012;97(5):723–730. doi: 10.3324/haematol.2011.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Della Porta MG, Travaglino E, Boveri E, et al. Minimal morphological criteria for defining bone marrow dysplasia: a basis for clinical implementation of WHO classification of myelodysplastic syndromes. Leukemia. 2015;29(1):66–75. doi: 10.1038/leu.2014.161. [DOI] [PubMed] [Google Scholar]
- 65.Steensma DP. Dysplasia has A differential diagnosis: distinguishing genuine myelodysplastic syndromes (MDS) from mimics, imitators, copycats and impostors. Curr Hematol Malig Rep. 2012;7(4):310–320. doi: 10.1007/s11899-012-0140-3. [DOI] [PubMed] [Google Scholar]
- 66.Bain BJ. The bone marrow aspirate of healthy subjects. Br J Haematol. 1996;94(1):206–209. doi: 10.1046/j.1365-2141.1996.d01-1786.x. [DOI] [PubMed] [Google Scholar]
- 67.Valent P, Jäger E, Mitterbauer-Hohendanner G, et al. Idiopathic bone marrow dysplasia of unknown significance (IDUS): definition, pathogenesis, follow up, and prognosis. Am J Cancer Res. 2011;1(4):531–541. [PMC free article] [PubMed] [Google Scholar]
- 68.McKerrell T, Park N, Moreno T, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10(8):1239-1245. [DOI] [PMC free article] [PubMed]
- 69.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. [DOI] [PMC free article] [PubMed]
- 70.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greenberg PL, Attar E, Bennett JM, et al. Myelodysplastic syndromes: clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2013;11(7):838–874. doi: 10.6004/jnccn.2013.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malcovati L, Hellström-Lindberg E, Bowen D, et al. European Leukemia Net. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood. 2013;122(17):2943–2964. doi: 10.1182/blood-2013-03-492884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lane AA, Odejide O, Kopp N, et al. Low frequency clonal mutations recoverable by deep sequencing in patients with aplastic anemia. Leukemia. 2013;27(4):968–971. doi: 10.1038/leu.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buckstein R, Jang K, Friedlich J, et al. Estimating the prevalence of myelodysplastic syndromes in patients with unexplained cytopenias: a retrospective study of 322 bone marrows. Leuk Res. 2009;33(10):1313–1318. doi: 10.1016/j.leukres.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 75.Hall J, Al Hafidh J, Balmert E, et al. Somatic mutations indicative of clonal hematopoiesis are present in a large fraction of cytopenic patients who lack diagnostic evidence of MDS [abstract].; Blood. 2014;124(21). Abstract 3272. [Google Scholar]
- 76.Kwok B, Reddy P, Lin K, et al. Next-generation sequencing (NGS)-based profiling of idiopathic cytopenia of undetermined significance (ICUS) identifies a subset of patients with genomic similarities to lower-risk myelodysplastic syndrome (MDS) [abstract].; Blood. 2014;124(21). Abstract 166. [Google Scholar]
- 77.Starczynowski DT, Vercauteren S, Telenius A, et al. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood. 2008;112(8):3412–3424. doi: 10.1182/blood-2007-11-122028. [DOI] [PubMed] [Google Scholar]
- 78.Steensma DP. Are myelodysplastic syndromes “cancer”? Unexpected adverse consequences of linguistic ambiguity. Leuk Res. 2006;30(10):1227–1233. doi: 10.1016/j.leukres.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 79.Steensma DP, Komrokji RS, Stone RM, et al. Disparity in perceptions of disease characteristics, treatment effectiveness, and factors influencing treatment adherence between physicians and patients with myelodysplastic syndromes. Cancer. 2014;120(11):1670–1676. doi: 10.1002/cncr.28631. [DOI] [PubMed] [Google Scholar]
- 80.Esserman LJ, Thompson IM, Jr, Reid B. Overdiagnosis and overtreatment in cancer: an opportunity for improvement. JAMA. 2013;310(8):797–798. doi: 10.1001/jama.2013.108415. [DOI] [PubMed] [Google Scholar]
- 81.Wacholder S. Precursors in cancer epidemiology: aligning definition and function. Cancer Epidemiol Biomarkers Prev. 2013;22(4):521–527. doi: 10.1158/1055-9965.EPI-13-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shlush LI, Minden MD. Preleukemia: the normal side of cancer. Curr Opin Hematol. 2015;22(2):77–84. doi: 10.1097/MOH.0000000000000111. [DOI] [PubMed] [Google Scholar]
- 83.Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15(12):e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]