Abstract
Human plasma is a biofluid that is high in information content, making it an excellent candidate for metabolomic studies. 1H NMR has been a popular technique to detect several dozen metabolites in blood plasma. In order for 1H NMR to become an automated, high-throughput method, challenges related to (1) the large signal from lipoproteins and (2) spectral overlap between different metabolites have to be addressed. Here diffusion-weighted 1H NMR is used to separate lipoprotein and metabolite signals based on their large difference in translational diffusion. The metabolite 1H NMR spectrum is then quantified through spectral fitting utilizing full prior knowledge on the metabolite spectral signatures. Extension of the scan time by 3 minutes or 15% per sample allowed the acquisition of a 1H NMR spectrum with high diffusion weighting. The metabolite 1H NMR spectra could reliably be modeled with 28 metabolites. Excellent correlation was found between results obtained with diffusion NMR and ultrafiltration. The combination of minimal sample preparation together with minimal user interaction during processing and quantification provides a metabolomics technique for automated, quantitative 1H NMR of human plasma.
Keywords: Human plasma, 1H NMR, diffusion, spectral quantification, lipoproteins
INTRODUCTION
Human plasma is a biofluid that is high in information content, making it an excellent candidate for metabolomic studies (Psychogios et al. 2011). Besides mass spectrometry (MS)-based methods, 1H NMR has been a popular technique to detect several dozen metabolites in blood plasma. However, in order to become a quantitative and high-throughput method 1H NMR has several challenges to overcome. Firstly, the bulk of signal intensity in a typical 1H NMR spectrum arises from lipoproteins, which complicate the detection and quantification of metabolites. Traditionally, lipoproteins are removed by ultrafiltration (Wevers et al. 1994) or organic solvent precipitation (Nagana Gowda et al. 2014). While robust and straightforward, these methods lengthen the sample preparation period, which could present a bottle neck for high-throughput studies. Diffusion-weighted 1H NMR has previously been used to separate lipoproteins and metabolites based on the large difference in translational diffusion coefficient (de Graaf et al. 2003; Liu et al. 1996). As the separation is performed on intact blood plasma, sample preparation can be minimal. Secondly, spectral overlap between different metabolites often leads to ambiguities in quantification when spectral integration or binning is used. Spectral fitting based on a linear combination of basis set 1H NMR spectra is a technique that can robustly deal with partial spectral overlap and line shape distortions. Spectral fitting has previously been applied to the quantification of 1H NMR brain spectra from rat brain (de Graaf et al. 2011).
Here we present an extension of diffusion-weighted 1H NMR (de Graaf et al. 2003) and combine it with a recently described spectral fitting algorithm (de Graaf et al. 2011). It is anticipated that the combination of minimal sample preparation together with minimal user interaction during processing and quantification will provide a metabolomics technique for automated, quantitative 1H NMR of human plasma.
MATERIALS AND METHODS
Sample collection and preparation
Plasma samples were obtained as part of a long term study on metabolic alterations in childhood obesity (Giannini et al. 2013). Venous blood was drawn into a tube containing heparin, chilled on ice, and centrifuged within an hour. The plasma samples were stored at 193 K for up to several months before analysis.
Two 300 μL samples were taken from each subject (n = 5) and either used directly for 1H NMR spectroscopy or following ultrafiltration. Samples that were used directly were diluted with 300 μL solution containing 250 mM phosphate buffer (pH 7.4), 5 mM formate and 10% D2O. Ultrafiltration was performed using Millipore centrifugal devices (Millipore Microcon YM-10) with a 10,000 molecular weight cutoff. Human plasma (300 μL) was diluted with 300 μL solution containing 250 mM phosphate buffer (pH 7.4), 5 mM formate and 10% D2O and centrifuged at 14,000g and 277 K for up to 3 h. To reduce glycerol (membrane preservative) contamination of the filtrate, all centrifugal devices were washed four times (by 30 min of centrifugation at 10,000g) with 500 μL water immediately prior to usage.
NMR spectroscopy
All experiments were performed on a Bruker 11.74 T magnet (500.13 MHz for 1H) interfaced to an Avance III spectrometer equipped with triple-axis gradients. The MR pulse sequence consisted of an adiabatic double-spin echo with a 0.5 ms adiabatic half passage (tanh/tan modulation with R = bandwidth × pulselength = 150, (Garwood et al. 1991)) pulse for excitation and two 0.5 ms adiabatic full passage pulses (sech/tanh modulation, R = 20, (Silver et al. 1984)) for refocusing. Diffusion-weighting was introduced with bipolar pairs of 4 ms magnetic field gradients on all three axes surrounding the refocusing pulses, giving a maximum b-value of 15 ms/μm2 at an echo-time TE of 17.28 ms. The opposite signs of the first and third magnetic field gradients relative to the second and fourth ensured excellent suppression of eddy current related field perturbations. All samples were measured at 298 K with 96 averages and a repetition time TR of 6,000/3,000 ms for low/high b-value acquisitions. The pulse sequence in Bruker format can be obtained by contacting the corresponding author. Data preprocessing included zero-filling to 32K points, fast Fourier transformation and zero-order phase correction. Prior to spectral fitting manual chemical referencing of formate at 8.444 ppm was performed. The high-quality water suppression achieved with presaturation removed the need for post-acquisition solvent removal.
Spectral fitting
1H NMR spectra were quantified with a spectral fitting algorithm described previously (de Graaf et al. 2011), extended with modifications specific for 1H NMR spectra of plasma as outlined below. In short, the experimental 1H NMR spectrum is approximated by a sum of 1H NMR spectra from pure compounds. The amplitude, line width and frequency shifts are adjusted so that the sum of the pure compound spectra best matches the experimental 1H NMR spectrum. While the amplitude variation is unconstrained, the variations in line width and frequency shifts are typically highly constrained. The spectral fitting algorithm allows different line width and frequency shifts for different multiplets within a given molecule while still retaining a fixed amplitude relation between the multiplets. In addition, the algorithm allows for an arbitrary line shape distortion that is common to all resonances. This is particularly valuable in the presence of line shape distortions due to imperfect magnetic field homogeneity. The algorithm can also accommodate an arbitrary baseline based on a polynomial or cubic spline expansion. In the case of 1H NMR spectra from human plasma the baseline is almost entirely composed of resonances from lipoproteins. Approximating this baseline with a generic polynomial or spline expansion would introduce a large number of unconstrained parameters to the spectral fit, thereby undermining the stability of the algorithm. Previously it was shown (de Graaf et al. 2003) that the lipoprotein baseline can be measured by utilizing the large difference in diffusion coefficient between metabolites and lipoproteins. Small differences in the order of 5-10% between the diffusion-based lipoprotein baseline and the real lipoprotein 1H NMR spectrum were attributed to similarly small differences in diffusion coefficients between the various lipoprotein subclasses. In this study the spectral fitting algorithm was extended to allow the incorporation of the experimentally determined lipoprotein baseline spectrum with the added modification that individual lipoprotein resonance amplitudes were allowed to change by +/− 10% to accommodate the variable diffusion-based suppression. This was achieved by parameterizing the measured lipoprotein 1H NMR spectrum by a limited number (typically < 70 for the entire 0.5 – 4.5 ppm range) of Lorentzian or Gaussian lines. The line width of the individual parameterized lipoprotein resonances was restricted to the range [8 … 40] Hz to minimize their effect on the narrow metabolite resonances with a typical line width of less than 3 Hz. During the spectral fitting, the frequency, phase and line width of the individual parameterized lipoprotein resonances remained constant, whereas the amplitude was allowed to change by up to 10%.
The spectral basis set consisted of 28 previously identified metabolites (Psychogios et al. 2011) and included 2-hydroxy-butyrate (CHEBI:64552), 3-hydroxy-butyrate (CHEBI:37054), acetate (CHEBI:30089), acetoacetate (CHEBI:13705), acetone (CHEBI:15347), alanine (CHEBI:16449), arginine (CHEBI:29016), betaine (CHEBI:22860), carnitine (CHEBI:17126), choline (CHEBI:15354), citrate (CHEBI:53258), creatine (CHEBI:16919), creatinine (CHEBI:16737), ethanol (CHEBI:16236), glucose (CHEBI:17234), glutamate (CHEBI:64243), glutamine (CHEBI:28300), isoleucine (CHEBI:17191), lactate (CHEBI:75228), leucine (CHEBI:25017), lysine (CHEBI:18019), methionine (CHEBI:16811), ornithine (CHEBI:18257), proline (CHEBI:26271), pyruvate (CHEBI:15361), succinate (CHEBI:63675), threonine (CHEBI:16857) and valine (CHEBI:27266). Metabolite identifiers can be found through Chemical Entities of Biological Interest (ChEBI, http://www.ebi.ac.uk/chebi). The spectral basis set was constructed through density matrix simulations in SpinWizard, a home-written program based in Matlab (Matlab 8.0, The Mathworks, Natick, MA, USA). In order to use the same spectral basis set for both filtered and unfiltered human plasma, the ultra-filtered samples were measured with the identical diffusion-weighted MR pulse sequence at low diffusion-weighting (b = 0.01 ms/μm2). The spectral fitting routines may be downloaded from http://mrrc.yale.edu or can be obtained by contacting the corresponding author.
RESULTS AND DISCUSSION
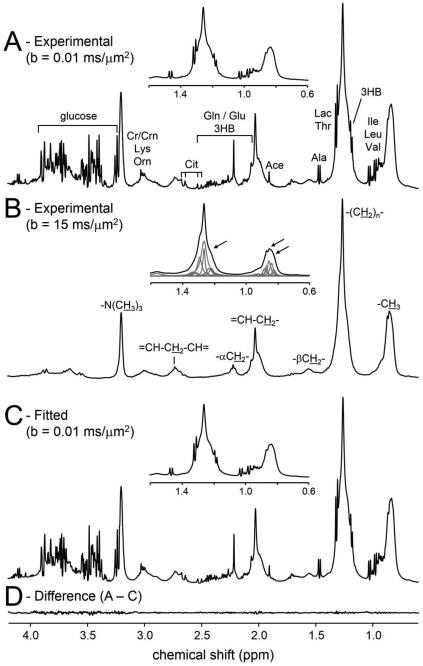
Fig 1A shows a typical 1H NMR spectrum from human plasma with low diffusion sensitization (b = 0.01 ms/μm2) such that both metabolites and lipoproteins are visible. At high diffusion sensitization (b = 15 ms/μm2), only the signals from lipoproteins remain (Fig 1B). Variation in lipoprotein diffusion coefficients (de Graaf et al. 2003) will lead to small (+/− 10%) amplitude variations, such that the lipoprotein spectrum does not exactly match the lipoprotein background signals in Fig 1A. Following parameterization of the lipoprotein spectrum (Fig 1B, inset), the individual lipoprotein signal amplitudes are allowed to vary slightly (+/− 10%) during the spectral fitting algorithm as to provide the best fit (Fig 1C) to the experimental 1H NMR spectrum (Fig 1A). The parameterized lipoprotein line widths, phases and frequency offsets are held constant. Fig 1D shows the residual between the experimental (Fig 1A) and fitted (Fig 1C) 1H NMR spectra.
Figure 1.
1H NMR spectra acquired from human plasma with (A) low diffusion weighting (b = 0.01 ms/μm2) and (B) high diffusion weighting (b = 15 ms/μm2). The insets show the zoomed and scaled spectral regions between 0.8 and 1.5 ppm. The arrows in (B) indicate positions were the lipoprotein spectrum in (B) does not perfectly match the total spectrum in (A). The inset in (B) shows the parameterization of the experimental spectrum into a limited number of Lorentzian lines with fixed frequencies and line widths. (C) Total spectral fit composed of the sum of 28 metabolites and circa 70 lipoprotein resonances. The inset shows that slight amplitude scaling of the lipoprotein resonances provides a perfect match to the experimental spectrum in (A). (D) Difference between the experimental and fitted spectra.
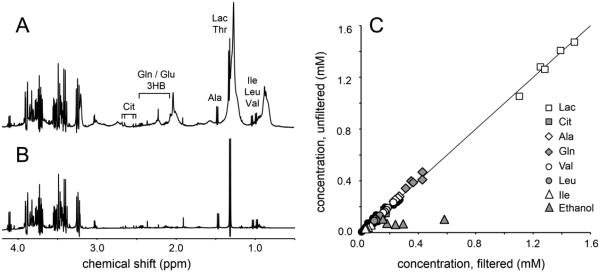
Figs 2A and B show 1H NMR spectra from (A) unfiltered (b = 0.01 ms/um2) and (B) filtered plasma from the same subject. The signal of β-H2 glucose at 3.23 ppm was used to normalize both 1H NMR spectra. Visual inspection quickly reveals that the metabolite levels in both 1H NMR spectra are very similar. This qualitative evaluation is supported by the quantitative correlation plot shown in Fig. 2C. Most metabolite concentrations fall on or near the line of unity with the best linear fit given by Cdiffusion = 0.9983Cfiltration – 0.0005 (R = 0.9981), where C refers to a particular metabolite concentration (in mM). The signals from ethanol represent a contamination accidently introduced by the incomplete drying of reused NMR tubes following a cleaning with water and ethanol. Ethanol was excluded from the correlation coefficient calculation. Supplementary Table S1 holds the numerical values of all data points displayed in Fig. 1.
Figure 2.
1H NMR spectra acquired with low diffusion-weighting (b = 0.01 ms/μm2) on (A) unfiltered and (B) ultra-filtered plasma. (C) Correlation between metabolite concentrations obtained from unfiltered and filtered plasma. For all samples the β-H2 glucose signal at 3.23 ppm was referenced to the glucose concentration measured with a glucometer. The best linear fit is given by Cdiffusion = 0.9983Cfiltration – 0.0005 (R = 0.9981), where C is the concentration in mM. The solid black circles represent all other metabolites not specifically mentioned.
Ultrafiltration is one of the standard techniques to eliminate lipoprotein signals and it has been shown here and elsewhere that ultrafiltration provides reliable and quantitative metabolite concentrations. The primary disadvantage of ultrafiltration is the time, labor and cost involved with the procedure presenting a potential bottleneck for application in high-throughput metabolomics. While the time investment can be reduced by running ultrafiltration samples in parallel, the sample preparation will still be more involved then with the proposed diffusion NMR method. The resonances in 1H NMR spectra obtained from ultra-filtered samples were noticeably sharper than those in non-filtered samples (1.4 ± 0.1 versus 1.8 ± 0.3 Hz), thereby improving the spectral resolution and sensitivity. However, at the same time the T1 relaxation times of ultra-filtered samples appeared significantly longer with fully-relaxed 1H NMR spectra requiring a repetition time TR of 20 s against a fully-relaxed TR of 6 s for unfiltered samples. It can thus be argued that the NMR characteristics between the two methods balance each other out, such that the primary advantage of the presented method comes from the minimal preparation. Recently the use of protein precipitation with methanol was evaluated as an attractive and reliable alternative to ultrafiltration for NMR-based metabolomics (Nagana Gowda et al. 2014). It is interesting to note that the NMR visibility of many metabolites appeared to be enhanced with protein precipitation relative to ultrafiltration, presumably due to the release of the protein-bound metabolite fraction during the precipitation process. This observation thus warrants caution about the NMR-visible and total metabolite concentrations, especially when comparing between different modalities like NMR and MS. On the other hand, the specific features of NMR also provide a window of opportunity to study the dynamics of metabolite-protein interactions.
The spectral fitting routine was able to adequately fit all 1H NMR spectra from filtered and unfiltered plasma in a semi-automated fashion. One of the key features of the algorithm is the ability to vary the line widths and relative chemical shifts of individual multiplets within a given molecule while keeping the relative amplitudes between the multiplets fixed. This feature gives the algorithm some flexibility to deal with small line shape and chemical shift effects due to the chemical environment (ionic strength, pH) or temperature. As the samples were buffered and measured at a constant temperature, the flexibility of the algorithm was sufficient to accommodate the observed effects. However, on samples were the prior knowledge of the basis set does not match the experimental data an unavoidable error in the spectral fit will result. In these cases the generation of a new basis set matching the experimental conditions is required.
Similarly, the presence of unknown signals that are not part of the spectral basis set will lead to errors in the spectral fitting. The presence of large signals not accounted for with the spectral basis set will lead to obvious discrepancies between the experimental and fitted spectra (see Supplemental Figure S3 and Table S21) and can potentially compromise the results for all metabolites (Hofmann et al. 2002; Provencher 1993). Inspection of the difference spectrum immediately reveals the missing compounds missing after which extension of the spectral basis set will remedy the problem. While the 28 metabolites included in the spectral basis set of the current study accounted for > 95% of the spectral signal intensity on all samples, it was obvious that the 1H NMR spectra hold information on many more metabolites below the 10 μM concentration level. The signal-to-noise ratio achievable with the current experimental setup (6 min acquisition) made it difficult to reliably detect compounds below circa 10 μM. Enhanced signal averaging at higher magnetic fields utilizing cold-probe technology could lower the detection threshold to 1 μM or less (Nagana Gowda et al. 2014). It remains to be proven if spectral fitting can adequately deal with the more than four orders of metabolite concentration range and the greatly increased number of detectable compounds (Psychogios et al. 2011). It is likely that under those conditions spectral fitting could benefit from additional NMR methods aimed at spectral simplification, like 2D NMR or spectral editing.
Several previous studies have used diffusion-weighted MRS on plasma to discriminate between low molecular-weight metabolites and high molecular-weight lipoproteins (de Graaf et al. 2003; Liu et al. 1996). Whereas the high diffusion sensitization readily obliterates signals from fast-diffusing metabolites, it also had a small effect on the slow-diffusion lipoproteins. If all lipoprotein subpopulations had the same diffusion coefficient this would simply lead to a global scaling. However, the various lipoprotein fractions can vary in molecular weight and size by an order of magnitude (Ala-Korpela 1995), which gives a circa 1.5-fold spread of lipoprotein diffusion coefficients (0.014 – 0.022 μm2/ms (de Graaf et al. 2003)). At the diffusion-weighting used here (15 ms/μm2) the spread in diffusion translates to a ± 6% variation around the mean signal intensity. Previous studies (de Graaf et al. 2003; Liu et al. 1996) have essentially ignored this relatively small effect by directly subtracting the lipoprotein spectrum from the total NMR spectrum to give a ‘metabolite-only’ NMR spectrum albeit with some minor baseline errors due to the aforementioned effect. In the current study the lipoprotein 1H NMR spectrum obtained at high diffusion-weighting was parameterized in order to take small amplitude variations due to differences in diffusion coefficient into account during the spectral fitting. At first sight the parameterized baseline appears to introduce a lot of variables that could compromise the spectral fit. However, it should be realized that the parameterization was highly constrained. The frequency, line width and phase of the parameterized signals were not allowed to change during the fit, whereas the amplitude was allowed to vary by only 15%, roughly corresponding to the expected lipoprotein amplitude spread due to variations in diffusion coefficients.
It should be emphasized that the parameterization of the lipoprotein spectrum as used here is fundamentally different from previous studies that parameterize part of the lipoprotein spectrum with a basis set of different lipoprotein classes ((Ala-Korpela et al. 1994; Otvos et al. 1991) e.g. VLDL, LDL, HDL) in order to characterize and quantify the various subclasses. In the current study the lipoproteins were simply parameterized to allow sufficient variation during the spectral fitting process. No knowledge on the individual lipoprotein fractions was assumed or obtained during the process. However, as the 1H NMR spectrum acquired under high diffusion weighting is only composed of lipoprotein signal it does provide an ideal platform to separate individual lipoprotein subclasses, provided that the temperature is high enough to ensure complete NMR visibility.
CONCLUDING REMARKS
Here we have described a novel 1H NMR method for the quantitative analysis of human plasma samples that is suitable for automated and high-throughput measurements. The sample preparation is minimal and is limited to mixing the samples with buffer, D2O and an internal chemical shift reference. The NMR acquisition scheme is short, 15 minutes per sample, and entails the measurement of separate lipoprotein and total (metabolite + lipoprotein) 1H NMR spectra based on large differences in diffusion. The NMR spectra analysis does not require any user interaction and has the potential to provide an automated platform for high-throughput data processing of 1H NMR spectra of plasma.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health (NIH) grants K08-DK082618, R03-DK100709 and R01-DK101984.
Footnotes
COMPLIANCE WITH ETHICAL REQUIREMENTS
The study complied with all applicable institutional guidelines and terms of the Declaration of Helsinki of 1975 (as revised in 2008) for investigation of human participants and was approved by the Institutional Review Board of Yale University. Informed consent was obtained from the parents and assent from the children.
REFERENCES
- Ala-Korpela M. 1H NMR spectroscopy of human blood plasma. Prog NMR Spectroscopy. 1995;27:475–554. [Google Scholar]
- Ala-Korpela M, Korhonen A, Keisala J, Horkko S, Korpi P, Ingman LP, Jokisaari J, Savolainen MJ, Kesaniemi YA. 1H NMR-based absolute quantitation of human lipoproteins and their lipid contents directly from plasma. J Lipid Res. 1994;35:2292–2304. [PubMed] [Google Scholar]
- de Graaf RA, Behar KL. Quantitative 1H NMR spectroscopy of blood plasma metabolites. Anal Chem. 2003;75:2100–2104. doi: 10.1021/ac020782+. [DOI] [PubMed] [Google Scholar]
- de Graaf RA, Chowdhury GM, Behar KL. Quantification of high-resolution 1H NMR spectra from rat brain extracts. Anal Chem. 2011;83:216–224. doi: 10.1021/ac102285c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood M, Ke Y. Symmetric pulses to induce arbitrary flip angles with compensation for RF inhomogeneity and resonance offsets. J Magn Reson. 1991;94:511–525. [Google Scholar]
- Giannini C, Feldstein AE, Santoro N, Kim G, Kursawe R, Pierpont B, Caprio S. Circulating levels of FGF-21 in obese youth: associations with liver fat content and markers of liver damage. J Clin Endocrinol Metab. 2013;98:2993–3000. doi: 10.1210/jc.2013-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann L, Slotboom J, Jung B, Maloca P, Boesch C, Kreis R. Quantitative 1H-magnetic resonance spectroscopy of human brain: Influence of composition and parameterization of the basis set in linear combination model-fitting. Magn Reson Med. 2002;48:440–453. doi: 10.1002/mrm.10246. [DOI] [PubMed] [Google Scholar]
- Liu M, Nicholson JK, Lindon JC. High-resolution diffusion and relaxation edited one- and two-dimensional 1H NMR spectroscopy of biological fluids. Anal Chem. 1996;68:3370–6. doi: 10.1021/ac960426p. [DOI] [PubMed] [Google Scholar]
- Nagana Gowda GA, Raftery D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal Chem. 2014;86:5433–5440. doi: 10.1021/ac5005103. doi:10.1021/ac5005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos JD, Jeyarajah EJ, Bennett DW. Quantification of plasma lipoproteins by proton nuclear magnetic resonance spectroscopy. Clin Chem. 1991;37:377–386. [PubMed] [Google Scholar]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver MS, Joseph RI, Hoult DI. Highly selective π/2 and π pulse generation. J Magn Reson. 1984;59:347–351. [Google Scholar]
- Wevers RA, Engelke U, Heerschap A. High-resolution 1H-NMR spectroscopy of blood plasma for metabolic studies. Clin Chem. 1994;40:1245–50. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.