Abstract
Reversible protein phosphorylation is critically important in biology and medicine. Hundreds of thousands of sites of protein phosphorylation have been discovered but our understanding of the functions of the vast majority of these post-translational modifications is lacking. This review describes several chemical and biochemical methods that are under development and in current use to install phospho-amino acids and their mimics site-specifically into proteins. The relative merits of total chemical synthesis, semisynthesis, and nonsense suppression strategies for studying protein phosphorylation are discussed in terms of technical simplicity, scope, and versatility.
Background
The report of enzymatic phosphorylation of the protein casein in 1954 launched six decades of biomedical advances that could not have been imagined by its discoverers Burnett and Kennedy. [1] Over the past 60 years, reversible protein phosphorylation on Ser, Thr, Tyr, His, and Asp has been identified as involved in nearly every important cellular process extending throughout phylogeny. The protein kinases, which catalyze the attachment of the gamma-phosphoryl group from MgATP to protein sidechains, are encoded by more than 500 genes in humans making them one of the largest gene families in the genome.[2] There are over 100 human phosphatases that hydrolyze phosphoprotein modifications, highlighting the delicate balance that appears important for regulating cellular functions.[3] Over 250,000 phosphorylation sites have now been mapped in the biological proteome (http://www.phosphosite.org).
Eukaryotic protein kinases primarily phosphorylate Ser, Thr, and Tyr residues whereas prokaryotic kinases often catalyze the phosphorylation of His residues.[4] These phosphoHis proteins can in turn transphosphorylate Asp residues in transcription factor proteins.[5] In addition, phosphorylation of Arg and Lys are emerging as potentially biologically impactful but are in general poorly understood.[6]
Protein kinases govern cell growth, cell differentiation, cell cycle, and cell migration.[7] They are important in such key processes as angiogenesis[8], hormone response[9], learning and memory[10], metabolism[11], cardiac function[12], and reproductive biology[13]. Dysregulation of protein kinases is a hallmark of cancer and EGFR, Her2/Neu, IGFR1, B-Raf, Abl, Btk, Alk, Ret, and c-Kit are examples of protein kinases that are mutated or show increased expression, bolstering their enzymatic activities.[14-18] Such hyperactive eukaryotic protein kinases can drive the growth of various cancers including lung cancer, breast cancer, melanoma, leukemia, renal cell carcinoma, and gastrointestinal tumors and are established therapeutic targets. There are now more than one dozen clinically approved protein kinase inhibitors on the market for cancer and other diseases and some of these have made a dramatic impact on extending life.[19]
Despite the major discoveries in the protein phosphorylation field, large gaps in our knowledge remain. As is the case with many enzyme reactions, we have only a limited understanding of the catalytic mechanism of protein kinase and phosphatase reactions.[20] For the most part, we do not know which protein kinase(s) is responsible for phosphorylating a particular substrate site and which phosphatase(s) is involved in hydrolytic removal at a particular phospho-site.[20] How kinase and phosphatase enzymes are regulated by partner proteins, lipids, or post-translational modifications is relatively opaque in the majority of cases. Most central to this review, the function of the vast majority of specific phosphorylation events is poorly understood.
Protein phosphorylation functions, sites, and dynamics
At a fundamental chemical level, transfer of a phosphoryl group to an amino acid sidechain adds negative charge, installs strong hydrogen bond acceptor oxygens, and sharply increases the size of the residue.[20] Such phosphoryl group transfer can perturb structure locally in the peptide motif, alter global protein conformation, and confer changes in intermolecular interactions in protein complexes. Specifically, phosphorylation events can modulate enzymatic activity of the phosphoprotein, enhance or inhibit protein binding events, affect protein stability, and influence cellular localization.[20] A number of phosphopeptide binding proteins have been described exemplified by SH2 domain-containing and 14-3-3 proteins that recruit pTyr and pSer/Thr sequences, respectively.[21,22] Most sites of protein phosphorylation have been mapped using high throughput mass spectrometric methods, and they are commonly found in flexible or unstructured regions of proteins.[23] Reversibility is a hallmark of phospho-dependent signaling, although the kinetics of this can be quite variable and depends in part on the actions of cellular phosphatases.[24]
Site-directed mutagenesis and the study of protein phosphorylation
The ability to convert each of the genetically encoded 20 amino acid residues to one of the other 19 is facile using modern methods of site-directed mutagenesis. The attractiveness of this approach is that it can be performed rapidly and applied to the study of phosphorylation sites both in purified recombinant proteins as well as in cell culture and in vivo. The study of multiple phosphorylation sites distributed throughout the protein using site-directed mutagenesis is now routine. Furthermore, using the newer CrispR/Cas9-related technologies, germline editing of the genome is becoming straightforward so a “knock-in” point mutation of a phosphorylation site can be efficiently prepared and analyzed in vivo.[25]
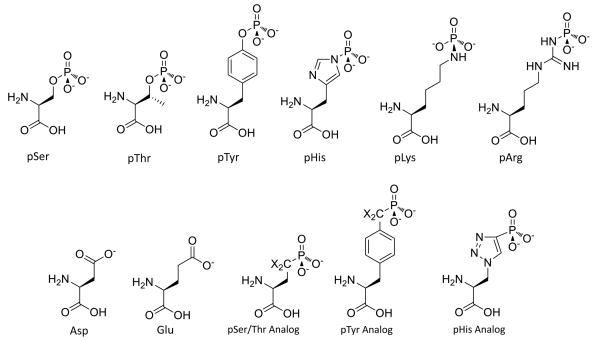
The standard algorithm for phosphorylation site replacements is mutation of a Ser/Thr to Ala, Asp or Glu or Tyr to Phe. The Ala is envisioned to represent a Ser/Thr that cannot be phosphorylated and the Asp/Glu a constitutive phosphoSer/phosphoThr (Figure 1).[26] A hypothesis has been advanced that Asp/Glu residues are in some cases ancestral precursors to sites of phosphorylation. In this theory, Asp/Glu residues have in some instances evolved into phosphorylated Ser or Thr residues, providing for reversible control of phenotype.[27] While powerful, Ala as a non-phosphorylatable Ser mimic has limitations. Loss of hydrogen bond donating/accepting capacity of the Ala methyl or Phe phenyl sidechains versus the hydroxymethyl sidechain of Ser or phenol sidechain of Tyr can impact biochemical function.[28] Moreover, protein Ser residues are subject to alternative PTMs including the commonly observed O-GlcNAc (N-acetyl-glucosamine) modification[29] and more rarely, O-acylation events[30] while Tyr residues can be sulfated or nitrated[31,32]. Thus, phenotypic changes from mutation of Ser to Ala or Tyr to Phe may be due to loss of function unrelated to phosphorylation.
Figure 1.
Structures of Asp, Glu, phosphoamino acids and their phosphonate mimics. X=H or F
Further complicating matters, the mimicry of a phosphoSer/phosphoThr by an Asp of Glu can be quite problematic. Although to a first approximation the Asp/Glu carboxylate can represent a point negative charge associated with phosphate-mediated electrostatic effects, the carboxylate possesses only one negative charge unlike dianionic phosphate monoesters. Moreover, the Asp/Glu residues are smaller and show different geometry versus phosphoSer/phosphoThr. Consequently, the Asp/Glu mimics often fail to capture the function of Ser/Thr phosphorylation, especially in mediating adaptor interactions.[28,33-35] The case of phosphoTyr and phosphoHis mimicry by Asp, Glu or any other standard amino acid is even less productive.[36] Therefore, investigators have sought other approaches to interrogate site-specific phosphorylation effects.
Phosphonate mimics
Protein phosphorylation is reversible and can be very short-lived in cellular systems and lysates.[24] While relatively kinetically stable in physiologic buffers, phosphoSer, phosphoThr, and phosphoTyr are often rapidly removed by phosphatase enzymes with turnover sometimes measured in seconds.[24] PhosphoHis, phosphoAsp, phosphoArg, and phosphoLys residues are intrinsically labile even in enzyme-free buffers due to the strong leaving group characteristics of these sidechains, and these phosphorylations can also be readily cleaved enzymatically.[37,38] Thus, even if one prepared a stoichiometrically, site-specifically phosphorylated protein, functional analysis would be complicated. A number of elegant approaches have been developed for the synthesis of the corresponding phosphonate analogs for peptide and protein incorporation that are not enzymatically labile.[33,39,40] In the case where the oxygen is replaced with a methylene group, the pKa of the phosphate is typically elevated, which can impact molecular recognition. Consequently, the difluoromethylene linker has been used in the case of phosphoSer and phosphoTyr that more closely matches physiologic pKas and ensures dianionic charge states of the phosphonates (Figure 1).[33,39] These have been effectively used in peptides and proteins to illuminate phosphorylation function.[32,33,38,39,41]
Incorporation of phosphoamino acids and analogs into proteins
There are now several methods to incorporate unnatural amino acids and post-translational modifications, including phosphorylation, with precision and complete stoichiometry. These methods include total chemical synthesis, semisynthesis, and unnatural amino acid mutagenesis. Each can be compared regarding technical ease, scope of targets, yield of desired products, and versatility with regard to PTMs. We briefly discuss these methods and their relative strengths and limitations below.
Total chemical synthesis of proteins
For acceptable yields and purities, the solid phase synthesis of peptides in one linear chain is generally limited to about 60 residues. This limitation has been overcome by the use of peptide ligation strategies, primarily based on chemoselective reactions involving a C-terminal thioester in one peptide and an N-Cys residue in a second peptide to be linked.[42] The efficiency of this ‘native chemical ligation’ process is based on initial transthioesterification followed by rearrangement to a standard peptide bond. By ligating multiple synthetic peptide segments, proteins in the range of up to 150 amino acid residues have been constructed using iterations of native chemical ligation. The total chemical synthesis approach offers the virtue of nearly complete control of incorporation of phosphorylated residues and their mimics at any site and at full stoichiometry. In recent years, this native chemical ligation strategy has been expanded to include alternative chemistries for ligation that can obviate the requirement for N-Cys containing peptides.[43,44] However, the need for generation of many peptides, the size limitations, and the typical requirement to fold the protein from a denatured state has perhaps prevented the more widespread adoption of total chemical synthesis in the study of phosphoproteins.
Semisynthesis of proteins
The semisynthesis of proteins can be defined as a hybrid strategy in which a major section of the modified protein is prepared by standard recombinant expression and then subjected to chemical manipulation to afford the semisynthetic protein. Advantages of semisynthesis versus total synthesis are the ease and low cost of production of recombinant proteins and its amenability to constructing larger proteins. Furthermore, the need for the cumbersome and often unreliable removal of denaturants from unstructured polypeptides to produce three-dimensionally normal proteins can often be avoided with semisynthetic strategies since the recombinant piece often emerges from the cell correctly folded. While complete chemical control over the entire protein is lacking with semisynthesis, there are many cases where restricted group alteration suffices for the goal of biological analysis. Below, we describe expressed protein ligation, Cys modification, and Staudinger ligation approaches that have been exploited to study protein phosphorylation.
Expressed protein ligation
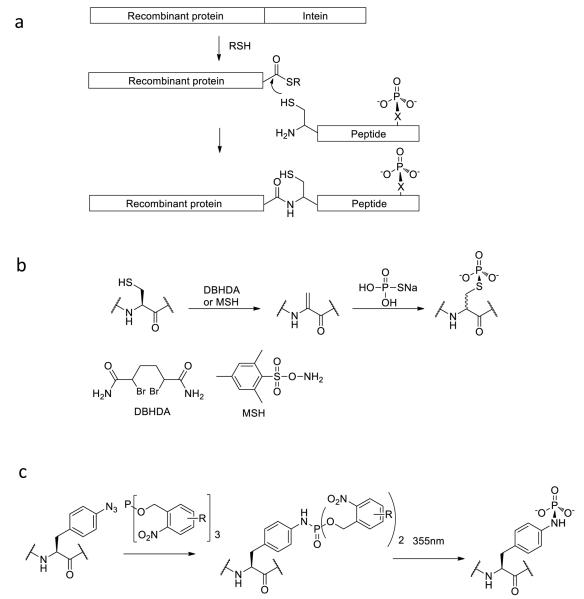
Expressed protein ligation has become a widespread method to study protein post-translational modifications (PTMs) including phosphorylation. The original technique of expressed protein ligation involves the use of recombinant protein thioesters that are then ligated to N-Cys synthetic peptides via the chemoselective ligation reaction described above (Figure 2).[45] This method has become increasingly robust as it has been shown that inteins can be an efficient way to generate recombinant protein thioesters in a variety of different expression conditions. It avoids the somewhat laborious challenge of synthesizing peptide thioesters. However, a variant of expressed protein ligation in which a recombinant protein fragment containing an N-terminal cysteine by virtue of proteolysis is ligated to a synthetic peptide still requires installation of a C-terminal thioester synthetically.[46]
Figure 2.
Strategies for semisynthesis of protein phosphorylation. (a) Expressed protein ligation. An N-Cys peptide containing a phospho-residue is ligated to a recombinant protein fragment containing a C-terminal thioester. (b) Cysteine modification strategy for attaching a thiophosphate to a protein. Sulfur modification with DBHDA or MSH and elimination generates dehydroalanine. (c) Staudinger-phosphite reaction for conversion of azidophenylalanine into phosphotyrosine analog in a protein.
Expressed protein ligation has been broadly applied to study phosphorylation of proteins. Two relatively recent examples are CK2 protein kinase and PTEN lipid phosphatase.[47,48] Using expressed protein ligation, a phosphoThr344-semisynthetic CK2 kinase form was generated and it was shown to exhibit altered protein substrate specificity. By introduction of a PFA (phosphono-difluoromethylene-alanine) non-hydrolyzable analog at the 344 position, it was demonstrated directly that phosphono-modified CK2 resisted proteolytic breakdown in the cell by binding to adaptor proteins. Reciprocal regulation by glycosylation at Ser347 was also analyzed using expressed protein ligation.[47]
In addition, expressed protein ligation facilitated the study of the tumor suppressor PTEN C-tail phosphorylation. The tetra-phosphorylation at Ser380, Thr382, Thr383, and Ser385 of PTEN catalyzed by CK2 kinase was suggested to have an important regulatory role for PTEN. A truncated version of PTEN fused to an intein was expressed in insect cells, and then ligated to a synthetic tetraphosphorylated C-tail peptide, to generate the full-length phosphorylated PTEN. The C-terminal tail phosphorylation was shown to induce a conformational change in PTEN, resulting in reduced phosphatase activity and membrane association.[48]
Expressed protein ligation has been shown to be very useful, but like all methods it has its limitations. Expressed protein ligation is best suited to study phosphorylation sites located at the N- or C-termini of a protein (or protein domain) of interest, to avoid multi-segment ligation. Fortunately, phosphorylation sites are often near protein termini since they are usually located in flexible regions that tend to be at the ends of proteins or protein domains. In some cases, when flexible modified regions are far from the termini, circular permutation strategies can be employed to analyze PTMs.[49] Another potential limitation is the requirement of a cysteine at the ligation junction. Since Cys is one of the least frequently encoded amino acids in proteins, introduction of a Cys by mutation is commonly required for the convenient use of the technique. Careful controls are needed to ensure that Cys mutation per se is not perturbing to structure or function. Some groups have developed alternative ligation chemistries to avoid Cys or refined desulfurization reactions to convert Cys to Ala.[50,51] Another concern is that truncation of a protein for expressed protein ligation can disrupt protein stability and this can lead to low yields or even the need for refolding. The gyrase intein can function in the presence of moderate concentrations of denaturant, partly alleviating this problem, but recovering functional protein after removing denaturing agents is often challenging.[52] Despite these limitations, expressed protein ligation is a relatively attractive method for the study of protein phosphorylation as evidenced by the many cases where it has been successfully employed.
Cysteine modification
Cysteine modification strategies have recently been developed as a potentially useful alternate to expressed protein ligation to study protein phosphorylation.[53-57] With its chemically reactive thiol sidechain, cysteine can be selectively alkylated and ultimately converted to a phosphoSer mimic. This sequence involves mutating the phosphorylated Ser residue to a Cys residue, and replacing the natural Cys residues with Ala or Ser amino acids to ensure site-specificity. Following production of the Ser to Cys recombinant protein, conversion of the Cys residue to a dehydroalanine residue is performed by chemical modification and elimination. O-mesitylenesulfonylhydroxylamine (MSH) and 2,5-dibromohexane diacetamide (DBHDA) have been reported to be effective reagents for cysteine to dehydroalanine conversions (Figure 2). The next step involves reacting the dehydroalanine with sodium thiophosphate, generating a phosphate thioesther adduct by Michael addition. The resulting phospho-cysteine is moderately stable and is a fairly accurate mimic of natural phosphorylated serine or threonine. It has been shown that that phosphoCys mimic can be recognized by a phospho-serine antibody with comparable efficiency to the natural PTM.[54] This method has been applied to the study of Aurora-A[55,57] and p38α[56] protein kinases and their regulation by phosphorylation. The semisynthetic phospho-Cys containing kinases showed similar catalytic activities to the enzymatically phosphorylated enzymes, confirming that the phospho-cysteine nicely recapitulates phospho-serine/phospho-threonine functionally. This Cys modification method facilitates characterization of the individual phosphorylation events in regulation, since site-specific phosphorylation can be achieved.
While in some ways appealing, the Cys modification method for phosphoSer mimicry has several drawbacks. The Cys mutations and chemical transformations can be incompatible with many proteins, challenging to execute, and low yielding. Thiophosphate esters are somewhat chemically labile due to the kinetic instability of the P-S bond in the ester linkage. But perhaps the biggest liability with the Cys modification strategy is the stereochemical scrambling associated with the Michael addition of the thiophosphate to dehydroalanine, predictably resulting in a 1:1 mixture of L- and D-amino acids. It is likely to be quite difficult to chromatographically separate the corresponding epimeric proteins, compromising functional analysis. A current challenge for the Cys modification approach therefore is the need for development of a stereoselective reaction for the protein installation of the thiophosphate by 1,4-addition.
Staudinger ligation
Phosphorylation mimics have been installed by modification of azido amino acid residues using phosphite reagents.[58] In a variant of the Staudinger reaction, the initial iminophosphorane adduct formed between the phosphite and azide undergoes reaction to furnish the N-linked phosphoramide mimic of the PTM (Figure 2). To apply the Staudinger reaction to mimic serine/threonine or tyrosine phosphorylation, initial incorporation of an azidohomoalanine or 4-azidophenylalanine into the protein at the desired site is required. This can in principle be achieved by a protein translation system using amber-stop codon suppression. In their initial work in this area, Serwa et al chose the protein SecB as a model for Staudinger modification.[58] The protein was expressed in a cell-free orthogonal protein translation system to introduce 4-azidophenylalanine at position 156. The 4-azidophenylalanine residue was reacted with a phosphite reagent that contained a 2-nitrobenzyl ester that could undergo light-induced deprotection, and an additional ethylene glycol side chain to improve water solubility. In aqueous solution at pH 8, the modified Staudinger reaction resulted in conversion of the azide to phosphoramidate, and irradiation with 355 nm UV triggered the liberation of the phosphoramidate ester, generating the phosphotyrosine mimic.
More recently, a related chemoselective Staudinger-phosphite reaction has also been applied to generate a site-specifically phosphorylated lysine peptide.[59] The general concept of the reaction is similar to forming the phosphotyrosine mimic but the aliphatic azide is less reactive than the aryl azide. Although this lysine modification has not yet been applied to proteins, the site-specific incorporation of lysine phosphorylation in peptides could be combined with expressed protein ligation to study protein lysine phosphorylation. The requirement for unnatural azide resiues makes this approach technically demanding. Moreover, the overall impact of the phosphoramide mimics of phosphoSer, phosphoThr, and phosphoTyr may be limited by the intrinsic instability of the P-N bonds. But, since phospho-Lys is a natural PTM, the Staudinger approach could be very useful in the biochemical analysis of these functionalities.
Unnatural amino acid mutagenesis
An elegant co-translational approach has been reported that has extended unnatural amino acid mutagenesis to the incorporation of phosphoamino acids into proteins made in bacteria.[60] Investigators engineered a unique tRNACys and a corresponding aminoacyl tRNA synthestase to one that is specific for phosphoserine (SEP) and modified the tRNA to recognize the UAG (amber) codon. In this way, the phosphoserine can be incorporated biosynthetically during ribosome-mediated translation. Other genetic manipulations in this approach designed to increase phosphoprotein yield included deletion of a phosphoserine phosphatase gene and mutating the residues in the amino acid binding pocket of the translation elongation factor EF-Tu in the heterologous host E. coli. Phosphoserine is actually added to the bacterial culture media and the authors note that E. coli expresses a phosphoserine transporter that allows for its intracellular accumulation and utilization in protein synthesis. Using this unnatural amino acid mutagenesis approach, the authors have confirmed the role of Ser218 phosphorylation to activate the kinase activity of the MEK1 protein kinase, and SEP shows more dramatic activation than the glutamate mimic. In a more robust version of the original method in which further selection and screening of the translation machinery was performed, they have also investigated the effects of histone H3 phosphorylation.[61] Overall, this creative approach potentially offers a significant advantage over semisynthesis in that in principle, any residue in a protein can be investigated and the natural PTM installed. However, it remains to be seen how general the phosphoserine mutagenesis strategy will be toward a range of proteins that may not express as well as histones. It is also a concern that, despite the gene deletion of one phosphatase, there are presumably numerous other bacterial phosphatases that could potentially hydrolyze the cellular phosphoprotein during production and isolation. It will also be interesting to see if this method can be extended to other phosphoamino acids and their phosphonate analogs beyond phosphoserine, especially in proteins with multiple PTMs.
Analysis of synthetic phosphoproteins in cellular environments
One general limitation with phosphoproteins produced using the strategies outlined above is that they are produced and/or purified in vitro, so studying their cellular properties requires transporting them into appropriate cell types. For introducing these phosphoproteins into cells, microinjection has been used, but this is technically demanding and has to be done one cell at a time.[47] However, two new methods for protein delivery into live cells offer hope in this regard.[62,63] One involves the use of a dimeric TAT peptide labeled with the fluorophore tetramethylrhodamine (TMR), called dfTAT as an endosomolytic agent.[62] The second is called iTOP (induced transduction by osmocytosis and propanebetaine), which delivers protein through the combination of NaCl hypertonicity-induced micropinocytosis and a small molecule (propanebetaine) as transduction compound.[63] Both methods appear to have the capacity to transport functional proteins into cells with high efficiency and low toxicity, and corresponding cellular effects are observed after certain proteins are delivered. The iTOP method has been shown to be effective for primary cells, and the iTOP-CRISPR/Cas9 system successfully achieves gene editing by delivery of Cas9 protein and the corresponding RNA into cells, which makes it powerful for genetic studies. While it remains to be seen how versatile and robust these cell transporting methods will be, they appear to offer an attractive alternative to microinjection.
Summary
The last two decades has seen the rise of the importance of protein phosphorylation in biomedical research. The success in proteomics studies to identify hundreds of thousands of protein phosphorylation sites has far outstripped our ability to analyze their functional importance. Nevertheless, the increasing range and sophistication of biochemical strategies to obtain proteins containing site-specific phospho-modifications and their close PTM mimics is very exciting. We believe that the further refinement of semisynthetic and nonsense suppression strategies combined with the burgeoning of cellular methods to analyze these modified proteins will allow for a new precision over classical site-directed mutagenesis stratgeies to understand protein phosphorylation. Innovations in cell delivery and synthetic genomes can allow for phospho-modified proteins to be studied in a cellular environment.[64] Complementary approaches to study other important post-translational modifications such as lysine acetylation and Ser glycosylation can allow for combinatorial complexity of protein chemistry to be interrogated.[65-67] Such advances offer promise that can be exploited in molecular medicine and novel therapeutic interventions in the major disorders of cell signaling..
Chen and Cole Highlights.
We discuss synthetic, semisynthetic, and genetic methods to make phosphoproteins.
Acknowledgment
We thank Cole lab members for helpful discussions and the NIH for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burnett G, Kennedy EP. The enzymatic phosphorylation of proteins. J Biol Chem. 1954;211:969–980. [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Besant PG, Attwood PV. Detection and analysis of protein histidine phosphorylation. Mol Cell Biochem. 2009;329:93–106. doi: 10.1007/s11010-009-0117-2. [DOI] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Besant PG, Attwood PV, Piggott MJ. Focus on phosphoarginine and phospholysine. Curr Protein Pept Sci. 2009;10:536–550. doi: 10.2174/138920309789630598. [DOI] [PubMed] [Google Scholar]
- 7.Hunter T. Signaling--2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 8.Holmes K, Roberts OL, Thomas AM, Cross MJ. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal. 2007;19:2003–2012. doi: 10.1016/j.cellsig.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Hubbard SR. The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb Perspect Biol. 2013;5:a008946. doi: 10.1101/cshperspect.a008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudmon A, Schulman H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J. 2002;364:593–611. doi: 10.1042/BJ20020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol. 2015;33:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Zhao W, Meng W, Zhao T, Chen Y, Ahokas RA, Liu H, Sun Y. Platelet-derived growth factor blockade on cardiac remodeling following infarction. Mol Cell Biochem. 2014;397:295–304. doi: 10.1007/s11010-014-2197-x. [DOI] [PubMed] [Google Scholar]
- 13.Rossi P. Transcriptional control of KIT gene expression during germ cell development. Int J Dev Biol. 2013;57:179–184. doi: 10.1387/ijdb.130014pr. [DOI] [PubMed] [Google Scholar]
- 14.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 15.Rossi A, Maione P, Sacco PC, Sgambato A, Casaluce F, Ferrara ML, Palazzolo G, Ciardiello F, Gridelli C. ALK inhibitors and advanced non-small cell lung cancer (review) Int J Oncol. 2014;45:499–508. doi: 10.3892/ijo.2014.2475. [DOI] [PubMed] [Google Scholar]
- 16.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue M, Cao X, Zhong Y, Kuang D, Liu X, Zhao Z, Li H. Insulin-like growth factor-1 receptor (IGF-1R) kinase inhibitors in cancer therapy: advances and perspectives. Curr Pharm Des. 2012;18:2901–2913. doi: 10.2174/138161212800672723. [DOI] [PubMed] [Google Scholar]
- 18.Chu N, Cole PA. Switching immune signals on and off. Elife. 2015;4 doi: 10.7554/eLife.07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Cole PA. Catalytic mechanisms and regulation of protein kinases. Methods Enzymol. 2014;548:1–21. doi: 10.1016/B978-0-12-397918-6.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Curr Opin Cell Biol. 2001;13:131–138. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- 22.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol. 2002;3:177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 23.Via A, Diella F, Gibson TJ, Helmer-Citterich M. From sequence to structural analysis in protein phosphorylation motifs. Front Biosci (Landmark Ed) 2011;16:1261–1275. doi: 10.2741/3787. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 25.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 26.Thorsness PE, Koshland DE., Jr. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987;262:10422–10425. [PubMed] [Google Scholar]
- 27.Pearlman SM, Serber Z, Ferrell JE., Jr. A mechanism for the evolution of phosphorylation sites. Cell. 2011;147:934–946. doi: 10.1016/j.cell.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai H, Ding H, Meng XW, Lee SH, Schneider PA, Kaufmann SH. Contribution of Bcl-2 phosphorylation to Bak binding and drug resistance. Cancer Res. 2013;73:6998–7008. doi: 10.1158/0008-5472.CAN-13-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 30.Barnett BP, Hwang Y, Taylor MS, Kirchner H, Pfluger PT, Bernard V, Lin YY, Bowers EM, Mukherjee C, Song WJ, et al. Glucose and weight control in mice with a designed ghrelin O-acyltransferase inhibitor. Science. 2010;330:1689–1692. doi: 10.1126/science.1196154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feeney MB, Schoneich C. Proteomic approaches to analyze protein tyrosine nitration. Antioxid Redox Signal. 2013;19:1247–1256. doi: 10.1089/ars.2012.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki N. Current status and future prospects for research on tyrosine sulfation. Curr Pharm Biotechnol. 2012;13:2632–2641. doi: 10.2174/138920101314151120122922. [DOI] [PubMed] [Google Scholar]
- 33.Zheng W, Zhang Z, Ganguly S, Weller JL, Klein DC, Cole PA. Cellular stabilization of the melatonin rhythm enzyme induced by nonhydrolyzable phosphonate incorporation. Nat Struct Biol. 2003;10:1054–1057. doi: 10.1038/nsb1005. [DOI] [PubMed] [Google Scholar]
- 34.Littlepage LE, Wu H, Andresson T, Deanehan JK, Amundadottir LT, Ruderman JV. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc Natl Acad Sci U S A. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szczepanowska J, Ramachandran U, Herring CJ, Gruschus JM, Qin J, Korn ED, Brzeska H. Effect of mutating the regulatory phosphoserine and conserved threonine on the activity of the expressed catalytic domain of Acanthamoeba myosin I heavy chain kinase. Proc Natl Acad Sci U S A. 1998;95:4146–4151. doi: 10.1073/pnas.95.8.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia F, Li J, Hickey GW, Tsurumi A, Larson K, Guo D, Yan SJ, Silver-Morse L, Li WX. Raf activation is regulated by tyrosine 510 phosphorylation in Drosophila. PLoS Biol. 2008;6:e128. doi: 10.1371/journal.pbio.0060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kee JM, Muir TW. Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem Biol. 2012;7:44–51. doi: 10.1021/cb200445w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein JA, Cheung YF, Marletta MA, Walsh C. Fluorinated substrate analogues as stereochemical probes of enzymatic reaction mechanisms. Biochemistry. 1978;17:5567–5575. doi: 10.1021/bi00618a037. [DOI] [PubMed] [Google Scholar]
- 39.Lu W, Shen K, Cole PA. Chemical dissection of the effects of tyrosine phosphorylation of SHP-2. Biochemistry. 2003;42:5461–5468. doi: 10.1021/bi0340144. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Shen K, Lu W, Cole PA. The role of C-terminal tyrosine phosphorylation in the regulation of SHP-1 explored via expressed protein ligation. J Biol Chem. 2003;278:4668–4674. doi: 10.1074/jbc.M210028200. [DOI] [PubMed] [Google Scholar]
- 41.Panigrahi K, Eggen M, Maeng JH, Shen Q, Berkowitz DB. The alpha,alpha-difluorinated phosphonate L-pSer-analogue: an accessible chemical tool for studying kinase-dependent signal transduction. Chem Biol. 2009;16:928–936. doi: 10.1016/j.chembiol.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 43.Shang S, Tan Z, Danishefsky SJ. Application of the logic of cysteine-free native chemical ligation to the synthesis of Human Parathyroid Hormone (hPTH) Proc Natl Acad Sci U S A. 2011;108:5986–5989. doi: 10.1073/pnas.1103118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harmand TJ, Murar CE, Bode JW. New chemistries for chemoselective peptide ligations and the total synthesis of proteins. Curr Opin Chem Biol. 2014;22:115–121. doi: 10.1016/j.cbpa.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 45.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT.AP-1.DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 47.Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolduc D, Rahdar M, Tu-Sekine B, Sivakumaren SC, Raben D, Amzel LM, Devreotes P, Gabelli SB, Cole P. Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. Elife. 2013;2:e00691. doi: 10.7554/eLife.00691. •This study is unusual in using insect cell expression for expressed protein ligation.
- 49.Karukurichi KR, Wang L, Uzasci L, Manlandro CM, Wang Q, Cole PA. Analysis of p300/CBP histone acetyltransferase regulation using circular permutation and semisynthesis. J Am Chem Soc. 2010;132:1222–1223. doi: 10.1021/ja909466d. •This study describes how a circularly permuted protein stratgey can be used to install an acetylated loop in the ‘middle’ of the natural protein for functional analysis.
- 50.Rohde H, Seitz O. Ligation-desulfurization: a powerful combination in the synthesis of peptides and glycopeptides. Biopolymers. 2010;94:551–559. doi: 10.1002/bip.21442. [DOI] [PubMed] [Google Scholar]
- 51.Wan Q, Danishefsky SJ. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew Chem Int Ed Engl. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi S, Yamamoto E, Mannen T, Nagamune T, Nagamune T. Protein refolding using chemical refolding additives. Biotechnol J. 2013;8:17–31. doi: 10.1002/biot.201200025. [DOI] [PubMed] [Google Scholar]
- 53.Bernardes GJ, Chalker JM, Errey JC, Davis BG. Facile conversion of cysteine and alkyl cysteines to dehydroalanine on protein surfaces: versatile and switchable access to functionalized proteins. J Am Chem Soc. 2008;130:5052–5053. doi: 10.1021/ja800800p. [DOI] [PubMed] [Google Scholar]
- 54.Chalker JM, Lercher L, Rose NR, Schofield CJ, Davis BG. Conversion of cysteine into dehydroalanine enables access to synthetic histones bearing diverse post-translational modifications. Angew Chem Int Ed Engl. 2012;51:1835–1839. doi: 10.1002/anie.201106432. •This manuscript is noteworthy for the efficient conversion of a cysteine to dehydroalanine as a way to introduce various PTM mimics.
- 55.Rowan FC, Richards M, Bibby RA, Thompson A, Bayliss R, Blagg J. Insights into Aurora-A kinase activation using unnatural amino acids incorporated by chemical modification. ACS Chem Biol. 2013;8:2184–2191. doi: 10.1021/cb400425t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chooi KP, Galan SR, Raj R, McCullagh J, Mohammed S, Jones LH, Davis BG. Synthetic phosphorylation of p38alpha recapitulates protein kinase activity. J Am Chem Soc. 2014;136:1698–1701. doi: 10.1021/ja4095318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowan F, Richards M, Widya M, Bayliss R, Blagg J. Diverse functionalization of Aurora-A kinase at specified surface and buried sites by native chemical modification. PLoS One. 2014;9:e103935. doi: 10.1371/journal.pone.0103935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serwa R, Wilkening I, Del Signore G, Muhlberg M, Claussnitzer I, Weise C, Gerrits M, Hackenberger CP. Chemoselective Staudinger-phosphite reaction of azides for the phosphorylation of proteins. Angew Chem Int Ed Engl. 2009;48:8234–8239. doi: 10.1002/anie.200902118. [DOI] [PubMed] [Google Scholar]
- 59.Bertran-Vicente J, Serwa RA, Schumann M, Schmieder P, Krause E, Hackenberger CP. Site-specifically phosphorylated lysine peptides. J Am Chem Soc. 2014;136:13622–13628. doi: 10.1021/ja507886s. [DOI] [PubMed] [Google Scholar]
- 60.Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S, Oh S, Yang A, Kim J, Soll D, Lee D, Park HS. A facile strategy for selective incorporation of phosphoserine into histones. Angew Chem Int Ed Engl. 2013;52:5771–5775. doi: 10.1002/anie.201300531. •This study describes a relatively high yield of phosphoprotein which permitted functional characterization with histone acetyltransferase.
- 62.Erazo-Oliveras A, Najjar K, Dayani L, Wang TY, Johnson GA, Pellois JP. Protein delivery into live cells by incubation with an endosomolytic agent. Nat Methods. 2014;11:861–867. doi: 10.1038/nmeth.2998. •This paper describes a promising new method to introduce modified proteins into cells.
- 63.D’Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, Lebbink RJ, Rehmann H, Geijsen N. Efficient intracellular delivery of native proteins. Cell. 2015;161:674–690. doi: 10.1016/j.cell.2015.03.028. •This paper describes a promising new method to introduce modified proteins into cells.
- 64.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, Church GM. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 66.Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, et al. Site-specific introduction of an acetyllysine mimic into peptides and proteins by cysteine alkylation. J Am Chem Soc. 2010;132:9986–9987. doi: 10.1021/ja103954u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chalker JM, Bernardes GJ, Davis BG. A “tag-and-modify” approach to site-selective protein modification. Acc Chem Res. 2011;44:730–741. doi: 10.1021/ar200056q. [DOI] [PubMed] [Google Scholar]