Abstract
Hfq facilitates gene regulation by small non-coding RNAs (sRNAs), thereby affecting bacterial attributes such as biofilm formation and virulence. E. coli Hfq recognizes specific U-rich and AAN motifs in sRNAs and target mRNAs, after which an arginine patch on the rim promotes base pairing between their complementary sequences. In the cell, Hfq must discriminate between many similar RNAs. Here, we report that acidic amino acids lining the sRNA binding channel between the inner pore and rim of the Hfq hexamer contribute to the selectivity of Hfq’s chaperone activity. RNase footprinting, in vitro binding and stopped-flow fluorescence annealing assays showed that alanine substitution of D9, E18 or E37 strengthened RNA interactions with the rim of Hfq and increased annealing of non-specific or U-tailed RNA oligomers. Although the mutants were less able than WT Hfq to anneal sRNAs with wild type rpoS mRNA, the D9A mutation bypassed recruitment of Hfq to an (AAN)4 motif in rpoS, both in vitro and in vivo. These results suggest that acidic residues normally modulate access of RNAs to the arginine patch. We propose that this selectivity limits indiscriminate target selection by E. coli Hfq, and enforces binding modes that favor genuine sRNA and mRNA pairs.
Keywords: Hfq, non-coding RNA, RNA chaperone, RNA-protein interactions, molecular beacon, RNA annealing, sRNA, RNase footprinting, Sm protein
Graphical Abstract
INTRODUCTION
In bacteria, small RNAs (sRNAs) regulate gene expression during environmental stress conditions like cold shock, oxidative stress, osmotic shock and quorum sensing 1; 2; 3. Transacting sRNAs activate or repress expression of certain genes by base pairing with the 5′-UTR of the target mRNAs 4, either masking or exposing the Shine-Dalgarno ribosome binding site, or recruiting RNase E for mRNA turnover 5; 6. In Gram-negative bacteria, sRNAs typically act together with the RNA chaperone protein Hfq 6; 7, which accelerates annealing of complementary RNA strands and stabilizes sRNA-mRNA duplexes 8; 9; 10; 11. The observed regulation depends on the specificity of these complexes, as different sRNAs must compete for binding to Hfq and their proper mRNA target.
Bacterial Hfq is a member of the Sm/Lsm super-family that forms a homo-hexamer ring with two distinct single-stranded RNA binding surfaces 7 (Fig. 1a). The proximal face of Hfq binds to U-rich RNA sequences found in sRNAs while the distal face of Hfq binds to AAN motifs found in the 5′-UTR of mRNAs 12; 13; 14. In addition, a patch of highly conserved arginine residues on the rim of Hfq binds U-rich regions of the mRNA and sRNA body 15; 16; 17 and is thought to be responsible for the RNA annealing activity of Hfq 18.
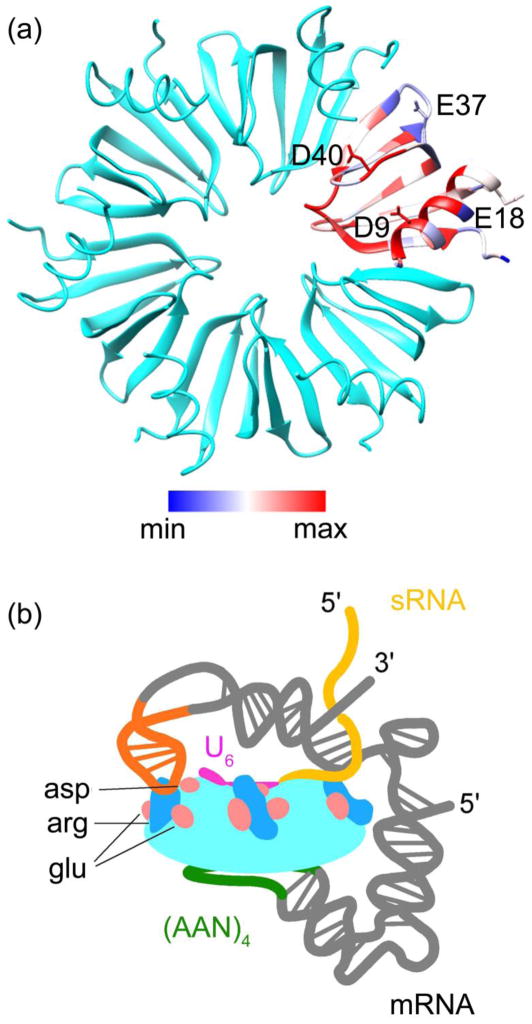
Fig. 1. Conserved acidic residues on the Hfq proximal surface.
(a) Conservation scores of 145 Hfq sequence clusters were rendered from blue (minimum) to magenta (maximum) on one subunit of the hexamer (chain A; 1HK9). Acidic residues (D9, E18, E37 and D40) are shown as sticks. (b) Schematic representation of the sRNA-rpoS-Hfq ternary complex, based on solution data 20. (AAN)4 (green) and U6 (magenta) motifs bind to the distal and proximal faces of Hfq, respectively. The U5 loop in rpoS (orange) interacts with the rim. The complementary regions of the sRNA and mRNA interact with charged residues at the rim, which are needed to initiate base pairing.
In our working model for the Hfq annealing cycle, sRNAs and mRNAs are recruited to the proximal and distal surfaces of Hfq through specific recognition of U and A-rich motifs. When Hfq forms a ternary complex with two complementary RNAs, interactions with the positively charged arginine patch accelerate base pairing between the anti-sense regions 18. In addition, simultaneous interactions with different RNA-binding surfaces of Hfq distort sRNA and mRNA structures, orienting them for anti-sense annealing 15; 19; 20.
The arginine patch in Hfq is surrounded by four conserved acidic residues D9, E18, E37 and D40 in E. coli Hfq (Fig. 1a). Invariant D9 and D40 lie on either side of a channel that sRNAs use as a passage between the inner proximal face RNA binding site and the arginine patch 15; 17, while the moderately conserved E18 and E37 flank the arginine patch on the distal rim of Hfq (Fig. 1a). Although negatively charged residues are not expected to interact with RNA, replacement of the almost universally conserved D9 with alanine impaired the ability of Hfq to support regulation of cirA by OmrA sRNA 21, chiP by ChiX sRNA and sdhCDAB by RyhB sRNA in E. coli 22. These results indicated that D9 is important for sRNA binding to Hfq, or for regulation of these target genes by sRNAs.
Here we show that removing D9 or other acidic residues makes the Hfq rim more accessible to unstructured RNAs, improving the ability of Hfq to act on RNAs lacking a specific Hfq binding site. Further in vitro and in vivo assays confirmed that mutating acidic residues overrides the requirement for AAN motifs which normally recruit Hfq to rpoS mRNA for riboregulation. The D9A mutation was even found to rescue Hfq alleles defective for RNA binding to the rim and distal face. The results presented here suggest that the acidic residues normally act to limit RNA binding to arginine patch, thereby increasing the selectivity of Hfq’s chaperone function.
RESULTS
Conserved acidic residues regulate the accessibility of the rim
Each of the four acidic side chains on the proximal face of Hfq was replaced with alanine (D9A, E18A, E37A and D40A) and the variants tested for RNA binding and annealing activity. The D40A mutant protein could not form a stable hexamer as previously reported 22 and was therefore not analyzed further. Equilibrium binding constants for oligoribonucleotides and sRNAs were measured using fluorescence anisotropy and electrophoretic gel mobility shift assays (EMSA), respectively (Table 1).
Table 1.
Equilibrium binding constant for RNA and Hfq
| Hfq variants | KD1 (μM) | DsrAΔU6 | |
|---|---|---|---|
| D16-FAM | DsrA sRNA | ||
| WT | 0.067 ± 0.005 | 0.22 ± 0.015 | 1.31 ± 0.1 |
| D9A | 0.047 ± 0.002 | 0.46 ± 0.018 | 0.67 ± 0.022 |
| E18A | 0.024 ± 0.004 | 0.38 ± 0.02 | ND |
| E37A | ND | 0.51 ± 0.021 | ND |
Overall dissociation constants per Hfq monomer were measured by fluorescence anisotropy (D16-FAM) or native gel mobility shift (DsrA and DsrAΔU6) at 30 °C in TNK buffer. ND, not done. Values are the average and standard deviation from the mean of three independent experiments. The Kd for D16-FAM and WT Hfq is about half that reported for earlier preparations of the protein 18.
We compared RNAs that primarily bind the rim of Hfq, or that interact with both the inner pore and outer rim of the proximal surface. An unstructured 16 nt RNA derived from the body of DsrA 23, D16, has six internal uridines and is thought to mainly interact with the proximal rim 18. Wild type (WT) E. coli Hfq binds FAM-labeled D16 (D16-FAM) with Kd = 67 nM (Hfq monomer) in TNK buffer at 30°C. The D9A and E18A variants bound D16-FAM more tightly than the WT protein, with dissociation constants of 47 nM and 24 nM, respectively.
Natural sRNAs end in 3′ uridines that tightly bind the inner proximal surface of Hfq, in addition to internal uridines and stem-loops that interact with the rim 24; 25. The D9A and E18A mutations weakened overall binding of DsrA sRNA about twofold compared to WT Hfq (Table 1; Fig. S1). When the 3′ terminal uridines were removed (DsrAΔU6), binding to WT Hfq became almost six-fold weaker (Table I). Thus, the WT Hfq-DsrA complex is stabilized more by interactions between the U-tail and the proximal face than the rim. By contrast, removing the U-tail had a much smaller effect on the overall affinity of DsrA for Hfq:D9A, raising the Kd less than twofold (Table I). As a result, the tail-less sRNA actually bound Hfq:D9A more tightly than it bound WT Hfq. We conclude that the D9A mutation increases RNA binding to the rim, perhaps owing to the increased net positive charge, while weakening the ability of structured sRNAs to simultaneously contact the rim and the inner pore of Hfq.
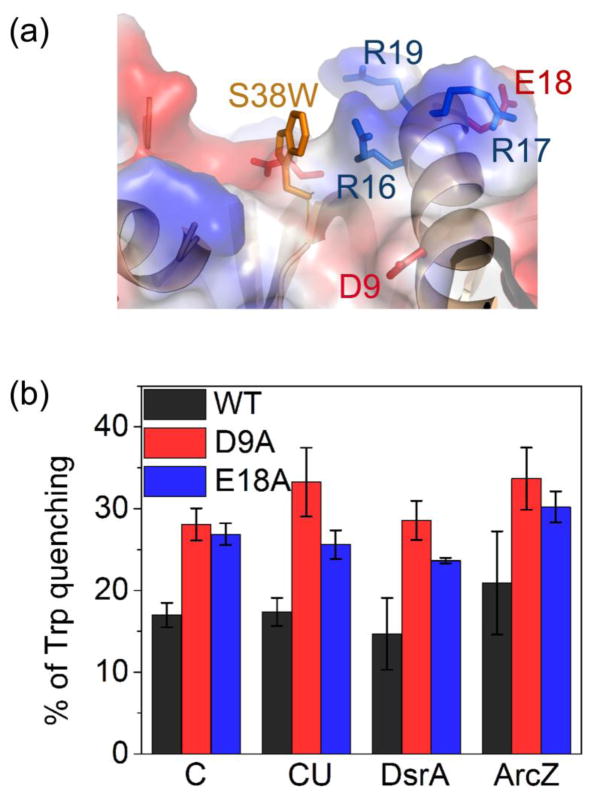
To directly test whether removing an acidic residue strengthens RNA interactions with the rim of Hfq, we monitored the ability of different RNAs to quench the fluorescence of a tryptophan introduced at residue 38 near the arginine patch (Fig. 2a; Fig S2a). This tryptophan substitution reports interactions with the rim of Hfq, as short RNAs that cannot bind the rim do not reduce the emission intensity 18. This mutation does not impair sRNA-mRNA annealing in vitro (Fig. S2b). All of the RNAs tested quenched the tryptophan fluorescence signal of Hfq:S38W by 15–20%. These RNAs were oligo C, an unstructured 16-mer that binds the Hfq rim non-specifically, oligo C-U6 (CU), which interacts with the proximal face and rim, DsrA sRNA and ArcZ56 sRNA. The D9A and E18A mutations resulted in stronger quenching of the tryptophan signal by all of the RNAs tested (Fig. 2b), consistent with stronger binding of RNAs to the rim of Hfq when a negatively charged side chain is removed.
Fig. 2. Acidic side chains reduce RNA binding to the rim of Hfq.
(a) A unique tryptophan at position 38 monitors RNA binding to the rim arginine patch. 18; 42 (b) Percent quenching of tryptophan emission in the presence of 100 nM RNA in TNK buffer at 30 °C, for Hfq variants shown in the key. These variants had similar (±20%) intrinsic fluorescence in the absence of RNA (Fig. S2c). Values were corrected for the inner filter effect (Methods and Fig. S2). Error bars represent the standard deviation from the mean of three independent trials.
Acidic residues contribute to sRNA recognition
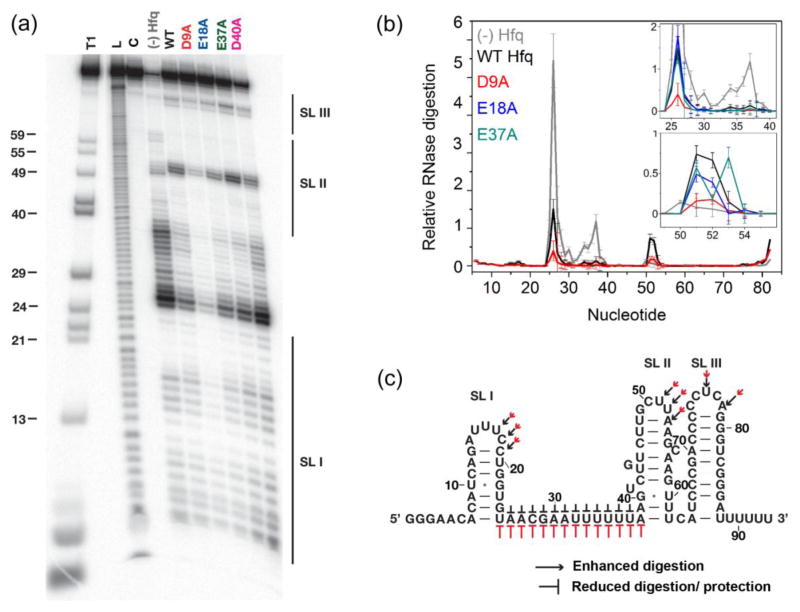
To ask whether the acidic residues influence how Hfq interacts with specific sRNA sequences, partial digestion with RNase If was used to probe the structure of 32P-labeled DsrA with and without Hfq. WT Hfq protects single-stranded nt 25–28 and U’s 32–38 in the body of DsrA while enhancing cleavage of nt 51–53 in loop 2 (black labels in Fig. 3), as reported previously 8; 26. Hfq:D9A protected DsrA much more strongly than WT Hfq (red labels in Fig. 3). The E18A and E37A Hfq mutants protected the same regions of DsrA, but to a degree more like that of WT Hfq (blue and green traces, Fig. 3b inset). Since the body of DsrA is thought to interact with the rim of Hfq 15, the footprinting results also indicate that the D9A mutation especially increases RNA binding to the rim of Hfq, even though the sRNA-Hfq complex is less stable overall owing to weaker interactions with the U6 tail (Table 1). The loss of enhancements in stem-loop I and loops II and III further suggest that Hfq:D9A cannot interact with DsrA as precisely as the WT protein (Fig. 3c).
Fig. 3. Effect of acidic residues on DsrA recognition.
(a) Ribonuclease If footprinting of 5′-32P-labeled DsrA at 30 °C with no Hfq or with 10 μM Hfq monomer as shown. Lanes T1, L and C represent control reactions in urea with RNase T1 (T1), alkaline hydrolysis (L) and no treatment (C). The D40A variant forms unstable hexamers and these data were not analyzed further. (b) Relative RNase If digestion of nucleotides in DsrA. Gray, (−) Hfq; black, WT Hfq; red, D9A; (inset): blue, E18A; dark cyan, E37A. Lines and error bars trace the mean and S.D. of four trials (Methods). (c) Summary of nuclease digestion pattern on the secondary structure of DsrA. Pointed (→) and flat (⊥) arrows indicate enhancement or protection, respectively, while the size indicates the degree of change in the digestion pattern (Black, WT; red, Hfq:D9A).
Acidic residues increase annealing of non-specific and U-rich RNAs
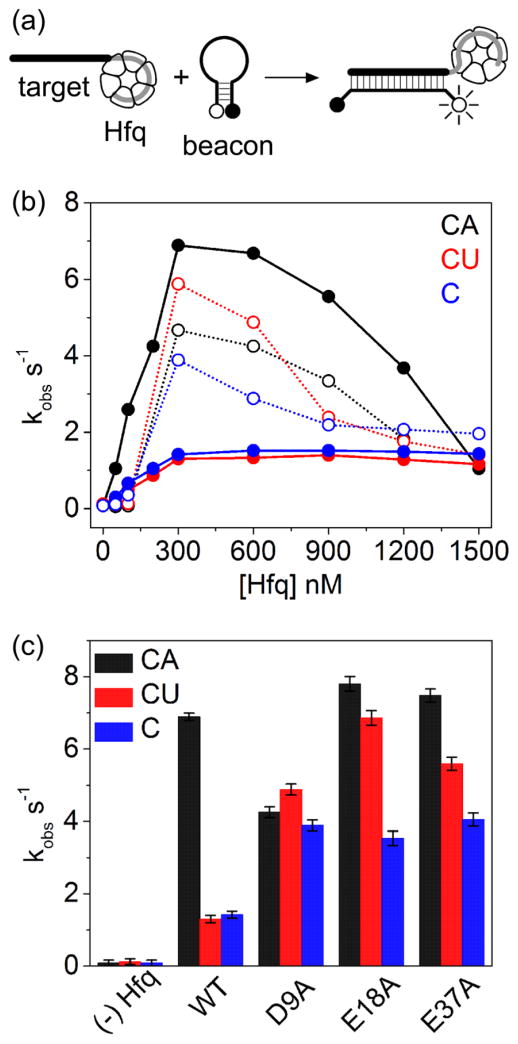
Since the arginine patch on the rim is essential for Hfq’s chaperone activity 18, we asked whether the loss of adjacent acidic residues could affect the annealing of complementary RNA strands. To compare the chaperone activity of each Hfq variant, the annealing kinetics of a molecular beacon and 16 nt target RNAs were measured by stopped-flow fluorescence (Fig. 4a). Wild type Hfq speeds up base pairing between a molecular beacon and unstructured RNAs, such as oligo C, which bind the Hfq rim non-specifically (Fig. S3) 27. This acceleration reached a maximum at 300 nM Hfq, which equaled one Hfq hexamer per RNA pair (Fig. 4b, solid blue). Hfq acts most efficiently, however, when one of the RNA strands contains an A18 tail that binds the distal face of Hfq, increasing the annealing rate 20 to 60 fold (Fig. 4b, solid black) 28.
Fig. 4. Acidic residues help discriminate between A-rich and U-rich RNAs.
(a) Schematic of molecular beacon annealing assay. (b) Observed annealing rate constants (average of five trials) were plotted against Hfq concentration. Annealing rates were measured by stopped-flow fluorescence using 50 nM molecular beacon, 100 nM target RNA and 0–1500 nM Hfq in TNK at 30°C. WT Hfq, filled circles; Hfq:D9A, open circles. (c) Observed annealing rate constants for Hfq variants (300 nM). Black, A18 tail; red, U6 tail; blue, target sequence only. Results of 5 to 6 trials were averaged; errors bars represent the standard deviation of the mean.
When the A-tailed target RNA (oligo CA) was recruited to the distal face of Hfq, the D9A mutation reduced the annealing rate less than twofold (Fig. 4b; compare black open and closed circles). A subtle effect on the A-tailed target was expected because the D9 residue is located on the proximal face of Hfq. Far more surprising was that the D9A mutation greatly improved the annealing rate when a U-tailed target RNA (oligo CU) was recruited to the proximal face of Hfq (Fig. 4b; compare red open and closed circles). Similar increases in annealing rate were observed for oligo C, which lacks a specific Hfq binding site (Fig. 4b; compare blue open and closed circles). Parallel changes in the annealing kinetics were observed for the E18A and E37A variants (Fig. 4c), in that each of these acidic residue Hfq mutations reduced the preference for annealing A-tailed RNA substrates over U-tailed (CU) and untailed (C) substrates (Fig 4c; compare black bar to red and blue bars).
Loss of acidic residues makes sRNA-mRNA annealing less efficient
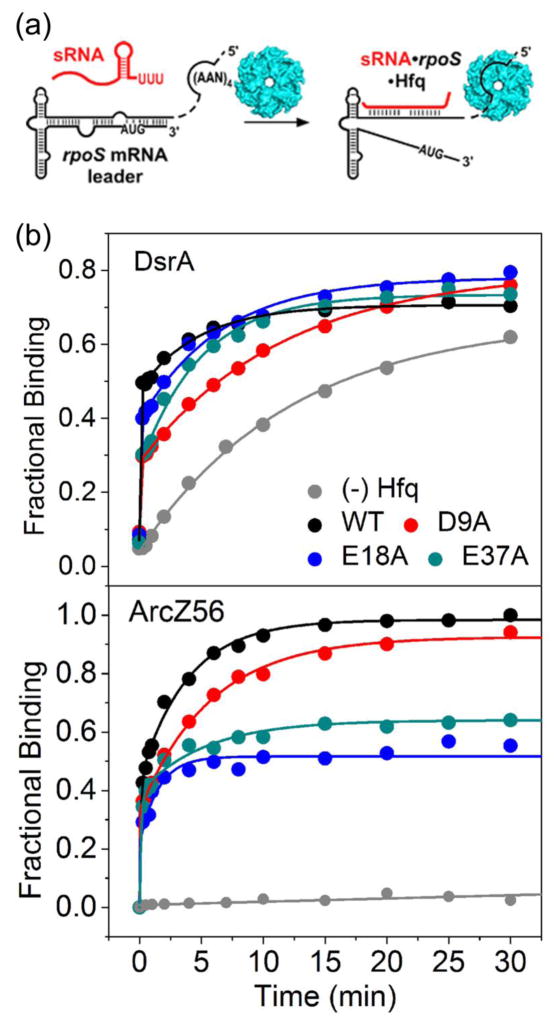
In contrast to unstructured oligonucleotides, natural sRNAs and mRNAs must make specific contacts with the proximal pore and the rim. We next tested whether the acidic residues affect the ability of Hfq to facilitate annealing of DsrA and ArcZ sRNAs to rpoS mRNA in vitro (Fig. 5a). As expected, DsrA and rpoS mRNA base paired with each other about 60 times faster with WT Hfq than without Hfq. All of the Hfq variants could facilitate sRNA annealing to rpoS mRNA, but the reaction became less efficient when an acidic side chain was mutated. For DsrA, the amplitude of the Hfq-dependent burst phase was lowered from 50% for WT Hfq to 33%, 43% and 34% for Hfq:D9A, E18A and E37A, respectively (Fig. 5b).
Fig. 5. Acidic side chains promote sRNA and mRNA annealing.
(a) Annealing kinetics of 32P-labeled rpoS301 and 200 nM DsrA or ArcZ56 sRNA with or without 0.6 μM Hfq was measured by native 6% PAGE. (b) Formation of rpoS-sRNA-Hfq complexes versus time for DsrA (top) and ArcZ56 (bottom). Grey, no Hfq; black, WT Hfq; red, D9A; blue, E18A; dark cyan, E37A.
A 56 nt form of ArcZ sRNA (ArcZ56) requires Hfq to base pair with rpoS mRNA 29; 30, unlike DsrA which slowly base pairs with rpoS in the absence of Hfq (grey lines, Fig. 5a, b). Mutations in the acidic residues did not change the initial burst, but they did diminish the amount of ArcZ56•Hfq•rpoS product formed after 4 minutes, from 80% in the presence of WT Hfq to only 64%, 47% and 55% with D9A, E18A and E37A Hfq, respectively. One explanation for these results is that tighter interactions with the rim make the ArcZ56-rpoS duplex less able to cycle off Hfq after base pairing has occurred, leading to refolding of rpoS and displacement of ArcZ56. Alternatively, the acidic residue mutants may not fold rpoS mRNA correctly for sRNA annealing, reducing the yield of sRNA-mRNA-Hfq complex for both DsrA and ArcZ56.
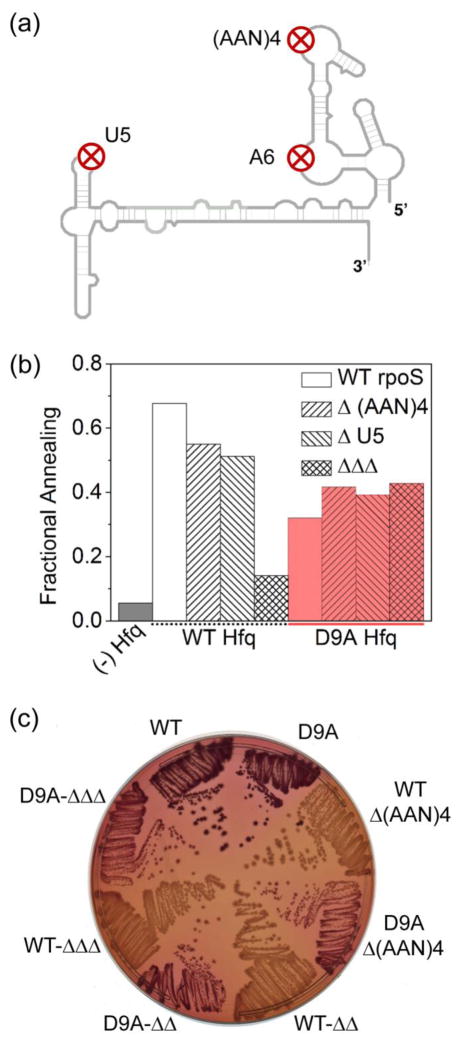
D9A mutation bypasses the requirement for rpoS AAN motifs
Up-regulation of rpoS by sRNAs depends on recruitment of Hfq to (AAN)4, A6 and U5 motifs within the rpoS 5′ UTR (Fig. 6a), which are needed for Hfq to promote sRNA-mRNA base pairing in vivo and in vitro 20; 29; 31. Mutagenesis of either the (AAN)4 or U5 motifs alone, or all three motifs together (ΔΔΔ), lowered the burst of DsrA annealing by WT Hfq from 67% with rpoS301 to 14% with ΔΔΔrpoS301 (Fig. 6b, white and grey bars). In contrast, mutagenesis of canonical Hfq binding motifs in rpoS mRNA had no effect on DsrA annealing by Hfq:D9A (Fig. 6b, red bars).
Fig. 6. D9A mutation bypasses an AAN motif in rpoS mRNA.
(a) Schematic representation of rpoS secondary structure and position of Hfq binding sites. (b) Fraction 32P-labeled rpoS 301RNA bound to DsrA with or without Hfq after 30 s. Mutations in rpoS deleted the upstream AAN motif (Δ(AAN)4), the downstream U5 loop (ΔU5), or both of these plus an A6 sequence (ΔΔΔ). (c) E. coli cells (Table S1) containing a chromosomal copy of WT or mutant rpoS::lacZ fusion, with hfq+ or hfqD9A alleles in the chromosome for each, were grown overnight at 37 °C on MacConkey agar supplemented with 0.0001% arabinose. Strains used, listed clockwise from WT in Figure, were SG30214, SG30236 SG30233, SG30242, SG30277, SG30279, DJS3056, and DJS3059, all described in Table S1. Red colonies indicate up-regulation of lacZ expression by Hfq and chromosomally derived sRNAs (DsrA, ArcZ and RprA). See Fig. S4 and S5 for further data.
We next asked whether D9 affects the requirements for rpoS mRNA regulation by sRNAs in vivo. Over-expression of DsrA and ArcZ sRNAs from an IPTG-inducible promoter increased translation of an rpoS::lacZ fusion in the E. coli chromosome to a similar extent in strains containing wild type or mutant hfq alleles in the bacterial chromosome (Fig. S4). In fact, we observed slightly greater up-regulation of rpoS by ArcZ in strains expressing Hfq:D9A, E18A or E37A compared to WT Hfq. These results were consistent with previous studies, which found that D9A was more important for negative regulation than positive regulation of rpoS, and had little effect on the accumulation of ArcZ and DsrA sRNAs in E. coli 22. Because the rpoS-lacZ mRNA was strongly induced by arabinose in these experiments, however, deficiencies in Hfq binding to the mRNA may have been masked 22.
To test whether the D9A allele also bypassed the need for Hfq to bind the rpoS AAN motif in vivo, we assayed translation of a WT rpoS-lacZ fusion on MacConkey agar plates. In the absence of a plasmid (akin to the vector control in Fig. S4b), rpoS-lacZ expression depends on endogenous levels of DsrA, ArcZ and RprA 30. Deletion of the (AAN)4 motif alone, (AAN)4 and A6 (ΔΔ), or all three motifs (ΔΔΔ) lowered the expression of the rpoS fusion in the presence of WT Hfq (WT sectors in Fig. 6c). However, the deleterious effects of these rpoS mutations were suppressed when Hfq carried the D9A allele (D9A sectors in Fig. 6c), in agreement with the in vitro annealing results (Fig. 6b). The ability of Hfq:D9A to suppress deletion of the rpoS (AAN)4 motif was also seen when mutants in the rim or distal site were combined with a deletion of the (AAN)4 or deletion of all three motifs (Fig. S5). Thus, the D9A mutation not only compensates for weaker rim binding, in agreement with our tryptophan quenching data, but surprisingly bypasses the need to recruit Hfq to the rpoS mRNA via the upstream (AAN)4 motif.
DISCUSSION
E. coli encodes about 100 different sRNAs that regulate up to 700 genes 32; 33. Although not all sRNAs are expressed simultaneously, the Hfq chaperone must still discriminate between sRNAs and other transcripts, while promoting base pairing with a diversity of mRNA targets. This is largely accomplished by specific recognition of U-rich and A-rich sequence motifs present in natural substrates of Hfq. It has been recently recognized, however, that natural RNAs interact with multiple surfaces of Hfq in a manner that likely depends on the RNA secondary structure. These multi-lateral interactions trigger structural rearrangements needed for efficient sRNA-mRNA recognition. For example, single-stranded 3′ uridines anchor sRNAs to the inner pore of the proximal face, while the body of many sRNAs binds the rim of Hfq 15; 18. Hfq restructures the rpoS mRNA leader even more dramatically by recognizing an upstream (AAN)4 motif and the U5 motif downstream of the sRNA binding site 20. These interactions fold the rpoS mRNA 5′-UTR into a compact structure 20, partially melting the inhibitory stem containing the sRNA target sequence 34.
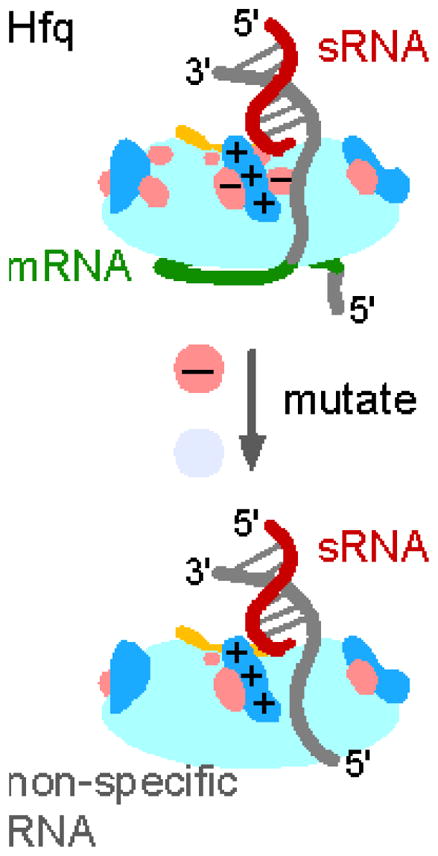
Our results suggest that acidic residues on the proximal face and rim act as “gatekeepers” that increase the selectivity of RNA interactions with arginine patch. Tryptophan quenching data showed that removal of an acidic side chain allows oligonucleotides and sRNAs to bind the rim of Hfq more tightly (Fig. 2). Similarly, Hfq:D9A protected internal residues of DsrA from ribonuclease digestion more strongly than WT Hfq protected them (Fig. 3). Mutations in acidic residues could improve RNA binding to the rim by simply making the rim more positively charged, or by changing the structure of Hfq to make the rim more accessible.
Tighter RNA binding to the rim appears to shift sRNA recognition from the inner proximal pore, which is highly selective for uridine, to the rim, which is less sequence-specific. Although the uridines at the 3′ end of DsrA are important for the stability of the complex with the WT protein, removing the U-tail had little effect on the stability of the Hfq:D9A-DsrA complex (Table 1). In addition, Hfq:D9A perturbed interactions with DsrA stem-loops II and III (Fig. 3). The rim of Hfq has been long reported to bind double-stranded RNA 35; 36, and is likely important for folding structured RNAs around Hfq in a manner that promotes sRNA-mRNA annealing and gene regulation 17; 20.
This shift toward stronger RNA interactions at the rim may explain why acidic residue mutations allow Hfq to bypass AAN motifs, which are normally required for Hfq to act on rpoS mRNA 29; 31 and other targets. In our annealing assays with unstructured oligonucleotides, Hfq:D9A was as active on non-specific and U-tailed RNAs as on A-tailed RNAs, while WT Hfq is far more active on RNAs containing an A-tail (Fig. 4). Similarly, increased non-specific binding of RNAs at the rim enables Hfq:D9A to form an alternative complex with rpoS which is modestly less active than the WT Hfq·rpoS complex but does not depend on interactions with the (AAN)4 and A6 motifs both in vitro and in vivo. Additionally, the D9A mutation overcame the deleterious effects of mutations on the rim (R16A) and distal face (Y25D and K31A) on rpoS up-regulation in the absence of the AAN motifs in vivo (Fig. S5), further indicating that it bypasses the need for specific recruitment of Hfq to AAN motifs. Together our results suggest that loss of a negatively charged side chain allows Hfq to act on RNAs that lack specific U or A-rich binding sites.
Although replacement of an acidic residue makes Hfq a less discriminating chaperone, this is apparently detrimental to the function of certain sRNA-mRNA pairs. The Hfq D9A allele was first isolated for its failure to support negative regulation of sdhC by RyhB sRNA and negative regulation of cirA by OmrB 21, and in subsequent studies D9A was more deleterious for negative than positive regulation 22. One possibility is that the mutant Hfq proteins fail to properly restructure both positive and negative mRNA targets for optimal base pairing with incoming sRNAs. These mutations could also weaken Hfq binding to mRNA sequence motifs needed for translational repression. Alternatively, the shift from recognition of the sRNA 3′ end to the sRNA body may allow Hfq to prematurely dissociate from sRNA-mRNA complexes, also weakening repression. Finally, the higher expression of rpoS::lacZ in the absence of sRNA overexpression suggests that these Hfq mutations may upset the competition between native sRNAs, elevating rpoS translation by basal levels of ArcZ, DsrA or RprA sRNAs. Regardless of the mechanism, we conclude that conserved negative charges surrounding the rim RNA binding site unexpectedly contribute to selective recognition of sRNAs and their targets by Hfq.
MATERIALS AND METHODS
Hfq purification
Wild type Hfq was overexpressed and purified from E. coli BL21 (DE3) cells using a Hi-Trap Co2+ column (GE Healthcare) and UNO S6 ion-exchange column (Bio-Rad) as previously described 37. Alignment of 145 non-redundant (<90% identity) Hfq sequences showed that D9 and D40 are almost invariant while E18 and E37 are moderately conserved. Hfq mutants (D9A, E18A, E37A, D40A, WT-S38W, D9A-S38W, and E18A-S38W) were made by QuikChange (Stratagene) and purified using the same protocol.
Fluorescence binding assays
Binding constants for Hfq and D16-FAM RNA were measured by fluorescence anisotropy as described before 23. RNA binding to the rim was measured by changes in the fluorescence emission of a single tryptophan Hfq-S38W as previously described 18. Contributions from the inner filter effect were determined by repeating the RNA titrations in buffer containing 1.25 M NaCl, which inhibits RNA binding. The percent quenching of the tryptophan fluorescence emission at 340 nm was calculated as 100% × {(I0 − I)/I0 − (I0′ − I′)/I0′}, in which I0 is the initial tryptophan emission in the absence of RNA, I is the emission in the presence of RNA, and I0′ and I′ are the initial tryptophan emission and the emission in the presence of RNA in 1.25 M NaCl respectively.
RNase If footprinting
5′-32P-DsrA (0.1 μM) and DsrA–Hfq complexes (8 μl) containing 1 μM Hfq monomer were prepared as described 8 and incubated 30 min at 30°C. Each sample was treated with 2 μl 1.25 U/μl of RNase If for 1 min at 37°C. Reactions were stopped by the addition of 10 μl buffered phenol. After phenol-chloroform extraction and ethanol precipitation, RNA pellets were redissolved in 6 μl formamide loading dye (90% (v/v) formamide, TBE, 0.1% (w/v) bromophenol blue, 0.1% (w/v) xylene cyanol) and subsequently loaded on an 8% polyacrylamide sequencing gel. Sequence ladders were obtained by nuclease digestion under denaturing conditions as described 38. Band intensities were integrated with SAFA 39 and normalized to bands with constant intensity (nt 5–7) in different experiments.
Molecular beacon-target annealing kinetics
Reverse-phase HPLC purified RNA molecular beacon, 5′ FAM-GGUCCCCCACUCGA CUCACCACCGGACC-DABCYL (Trilink Biotechnologies), was used without further purification. Target RNAs (Dharmacon) were purified as described previously 28. The target RNA sequences were oligo C, 5′GUGGUCAGUCGAGUGG3′; CU, oligo C plus U6; CA, oligo C plus A18. The annealing kinetics between the molecular beacon (50 nM) and target RNA (100 nM) in TNK buffer (10 mM Tris-HCl pH 7.5, 50 mM NaCl, 50 mM KCl) at 30°C was measured by an Applied Photophysics SX 18MV stopped-flow spectrometer as described previously 28; 40. Reactions were performed with 0–1.5 μM Hfq monomer as shown in the figures.
Native gel mobility shift assays for rpoS RNA and sRNA
DsrA, ArcZ56 and rpoS301 RNA were transcribed in vitro 8. Binding affinities of Hfq variants with 32P labeled DsrA at 30°C were measured by a native gel shift assay as previously described 8. The disappearance of labeled DsrA was plotted against Hfq monomer concentrations and subsequently fit to a Hill equation (θ = A [Hfq]n/([Hfq]n + Kdn), in which θ is the fraction of DsrA bound to Hfq, A is the amplitude, Kd is the binding constant and n is the Hill co-efficient (Fig. S1). The association kinetics of 32P-labeled rpoS mRNA 5′-UTR (rpoS301) with 200 nM sRNA in the presence of WT or mutant Hfq protein was measured by native gel mobility shift as described before 8; 29. The fraction of counts in each lane corresponding to rpoS•sRNA duplex or rpoS•Hfq•sRNA ternary complex versus time was fit to a biphasic rate equation.
β-Galactosidase activity assay
A derivative of wild-type E.coli MG1655 in which the 5′-UTR of rpoS was fused to lacZ, downstream of a pBAD promoter, 29 was further modified by P1 phage transduction 41 to introduce chromosomal Hfq mutations. The mutants were constructed as previously described 22 and confirmed by sequencing (see Supplementary Table S1). These strains were transformed with pLac, pDsrA and pArcZ plasmids 30. Overnight cultures were diluted into fresh LB containing arabinose (0.2%), ampicillin (100 μg/mL), and IPTG (100 μM) for 6 hours. OD600 and β-galactosidase activity were measured as described previously 29. The specific activities were calculated by Vmax/OD600 and were the average of three independent measurements. For plate assays, E. coli strains listed in Table S1 were grown overnight at 37°C on MacConkey agar containing 0.0001% arabinose, if not stated otherwise.
Supplementary Material
Highlights.
Hfq chaperones bacterial small non-coding RNAs during stress and pathogenesis
E. coli Hfq normally selects cellular RNAs containing UUU or AAN sequence motifs
Mutating acidic residues near the Hfq active site makes it less selective
Asp 9 to Ala mutation bypasses recruitment of Hfq to AAN sites in mRNAs
Acidic residues help Hfq discriminate between correct and non-specific targets
Acknowledgments
FUNDING
This work was supported by a grant from the National Institute of General Medicine [R01 GM46686 to S.W.] and in part by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research (to S.G.)
The authors thank N. Majdalani and Y. Peng for technical help, and N. De Lay, G. Storz, T. Updegrove, and A. Zhang for discussion and comments..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris JF, Micheva-Viteva S, Li N, Hong-Geller E. Small RNA-mediated regulation of host-pathogen interactions. Virulence. 2013;4:785–795. doi: 10.4161/viru.26119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaberdin VR, Blasi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weichenrieder O. RNA binding by Hfq and ring-forming (L)Sm proteins: A tradeoff between optimal sequence readout and RNA backbone conformation. RNA Biol. 2014;11:537–549. doi: 10.4161/rna.29144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq Protein Increases OxyS RNA Interaction with Target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 10.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 11.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Updegrove TB, Wartell RM. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA*mRNA duplex formation in rpoS riboregulation. Biochim Biophys Acta. 2011;1809:532–540. doi: 10.1016/j.bbagrm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Dimastrogiovanni D, Frohlich KS, Bandyra KJ, Bruce HA, Hohensee S, Vogel J, Luisi BF. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife. 2014:3. doi: 10.7554/eLife.05375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng Y, Curtis JE, Fang X, Woodson SA. Structural model of an mRNA in complex with the bacterial chaperone Hfq. Proc Natl Acad Sci USA. 2014;111:17134–17139. doi: 10.1073/pnas.1410114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–1189. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in Interaction Surfaces Differentially Impact E. coli Hfq Association with Small RNAs and Their mRNA Targets. J Mol Biol. 2013;425:3678–3697. doi: 10.1016/j.jmb.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 2011;39:5193–5202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panja S, Woodson SA. Hexamer to Monomer Equilibrium of E. coli Hfq in Solution and Its Impact on RNA Annealing. J Mol Biol. 2012;417:406–412. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome research. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinhara A, Matsui M, Hiraoka K, Nomura W, Hirano R, Nakahigashi K, Tomita M, Mori H, Kanai A. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC genomics. 2011;12:428. doi: 10.1186/1471-2164-12-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Updegrove TB, Correia JJ, Galletto R, Bujalowski W, Wartell RM. E. coli DNA associated with isolated Hfq interacts with Hfq’s distal surface and C-terminal domain. Biochim Biophys Acta. 2010;1799:588–596. doi: 10.1016/j.bbagrm.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol. 2014;426:275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panja S, Woodson SA. Fluorescence reporters for Hfq oligomerization and RNA annealing. Methods Mol Biol. 2015;1259:369–383. doi: 10.1007/978-1-4939-2214-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silhavy TJ. In: Experiments with gene fusions. Berman ML, Enquist LW, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1984. [Google Scholar]
- 42.Robinson KE, Orans J, Kovach AR, Link TM, Brennan RG. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014;42:2736–2749. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.