Abstract
Introduction
The “Experimental Data and Geometric Analysis Repository”, or EDGAR is an Internet-based archive of curated data that are freely distributed to the international research community for the application and validation of electrocardiographic imaging (ECGI) techniques. The EDGAR project is a collaborative effort by the Consortium for ECG Imaging (CEI, ecg-imaging.org), and focused on two specific aims. One aim is to host an online repository that provides access to a wide spectrum of data, and the second aim is to provide a standard information format for the exchange of these diverse datasets.
Methods
The EDGAR system is comprised of two interrelated components: 1) a metadata model, which includes a set of descriptive parameters and information, time signals from both the cardiac source and body-surface, and extensive geometric information, including images, geometric models, and measure locations used during the data acquisition/generation; and 2) a web interface. This web interface provides efficient, search, browsing, and retrieval of data from the repository.
Results
An aggregation of experimental, clinical and simulation data from various centers is being made available through the EDGAR project including experimental data from animal studies provided by the University of Utah (USA), clinical data from multiple human subjects provided by the Charles University Hospital (Czech Republic), and computer simulation data provided by the Karlsruhe Institute of Technology (Germany).
Conclusions
It is our hope that EDGAR will serve as a communal forum for sharing and distribution of cardiac electrophysiology data and geometric models for use in ECGI research.
Introduction
Need for EDGAR
Electrocardiography has been the primary mode of diagnosing cardiac arrhythmias and dysfunctions for the better part of a century. The main limitation of the ECG is that it is an integrated measure of cardiac electrical activity, sensitive in a very macroscopic sense to the sum of all electrical sources in the heart. Because of this integration and effects of the electrically conductive thorax, the ECG is a blurred, attenuated image of the underlying cardiac activity. As a result, it has diagnostic limitations especially in localizing sources of irregular electrical behavior. A further limitation comes from the sparse spatial sampling of the standard ECG, which directly covers only a small portion of the torso surface.
Body Surface Potential Mapping (BSPM) provides additional spatial coverage and resolution compared to standard electrocardiography and has been used as a research tool for almost 60 years [1]. Closely linked to BSPM are reconstruction approaches that seek to quantitatively estimate cardiac electrical activity using physics or statistically-based approaches. The resulting family of electrocardiographic inverse problem also has a long history that includes many different formulations and solution approaches [2]. Being able to accurately identify and localize electrical activity within the heart from the ECG (or the BSPM) is extremely useful in cardiovascular diagnostics because it is non-invasive, painless, inexpensive, and capable of continuous sampling. Unfortunately, the inverse problem is ill-posed and may also be non-unique, leading to poorly conditioned systems of equations to solve numerically. Despite the challenges, BSPM combined with a commercial ECGI system (ECVUE™, CardioInsight, Inc., Cleveland) has recently begun to show remarkable progress in clinical applications [3, 4]. A second commercial ECGI system also exists but with less documented clinical use, the AmyCard system (AMYCARD LLC, Moscow, Russia) [5].
This emergence of a viable commercial ECGI system has motivated a concerted and cooperative multidisciplinary effort from researchers to advance the state of the art in this field. One result is the recent creation of the Consortium for ECG Imaging (CEI, ecg-imaging.org), which recognizes that different centers have different areas of specific expertise relevant to ECGI; no single center can possibly develop all the necessary resources, skills, and experience. In some centers, for example, the strengths lie in mathematics while in others they lie in the domain of signal analysis and processing, numerical methods, or efficient software implementation. Other centers conduct animal experiments with disease models or acquire relevant signals and images from patients, from which they create image-based models of the heart and thorax.
Data sharing, a common need
It is self-evident that rapid and robust technical progress in ECGI will be enhanced through shared access to a range of data that is suitable for evaluation and comparison. By being able to compare different mathematical, numerical, and computational approaches on data with gold standard validation, trustworthy solutions can emerge. It is also necessary that such data come from different sources, using different experimental conditions, and with different clinical applications in mind. It is naive to assume that a single approach will succeed under all possible clinical and pathophysiological conditions. There are many parameters necessary to carry out ECGI, starting with lead selection and ending with analysis and visualization of results. Each of these parameters need to be tuned to the target application or adjusted on a case by case basis. A further challenge in the field is to define suitable quantitive metrics of success or error between gold standard and proposed ECGI results. The open availability of a range of standard data sets will enable the exploration and comparison of different metrics.
Experiments, with either animal or human subjects, are challenging to perform, to document, and to conduct with suitable care and accuracy. One aspect of this process highly relevant to sharing and evaluation of associated modeling is the acquisition and organization of the actual signals, images, and associated information. A complete dataset for ECGI must include 1) a source description of the heart, 2) a model of the geometry of the heart and volume conductor (with known electrical conductivities), and 3) the resulting body-surface potentials sampled at a set of identified leads. Acquiring this data requires a diverse skill set and can represent substantial resource cost. Often such datasets are used once by their collectors and then filed away indefinitely to remain inaccessible to the research community at large [6].
EDGAR is an Internet-based archive of curated data that are freely distributed to the international research community for the application and validation of electrocardiographic imaging (ECGI) techniques. The EDGAR project is a collaborative effort by the Consortium for ECG Imaging (CEI, ecg-imaging.org), and focused on two specific aims. One aim is to host an online repository that provides access to a wide spectrum of data. We propose to include a range of high quality samples of cardiac source and body-surface potential signals in order to support evaluation studies including performance of algorithms and computer programs. The second aim is to provide a standard information format for the exchange of these diverse datasets. We propose a framework that includes a metadata model describing the different components that make up a dataset and also the format in which data (e.g., time signals, meshes etc.) is made available, in order to facilitate aggregation and comparison of multidisciplinary datasets.
The members of CEI are committed to open sharing of data and algorithms, thus enabling comparisons of results from common datasets. By capitalizing on the strengths of the partners of this project, we hope to achieve synergy via collaboration and through minimization of redundancies. The aggregation of experimental, clinical, and simulation data from various centers will allow modelers to benefit from a large and diverse data pool, which will add robustness and accuracy to the ECGI approaches they develop.
Methods
EDGAR Framework
The EDGAR system is comprised of two interrelated components: 1) the metadata model, which describes the parameters and descriptions of the time signals and measurement geometry together with the time signals themselves and the geometric models that describe the heart, torso, and measurement leads; and 2) the web interface, a front end for efficient access and retrieval of data from the repository.
Metadata Model
The metadata model describes the different components that make up a dataset and also the format in which data (e.g., time signals, meshes etc.) is made available, in order to facilitate aggregation and comparison of multidisciplinary datasets. Each EDGAR dataset is comprised of five modules: Time Signals, Geometric Models, Forward and Inverse Transforms, Registration Information, and Medical Images. In addition, there is an Experiment Metadata as well as Experiment Documentation associated with each dataset as shown in Figure 1.
Figure 1.
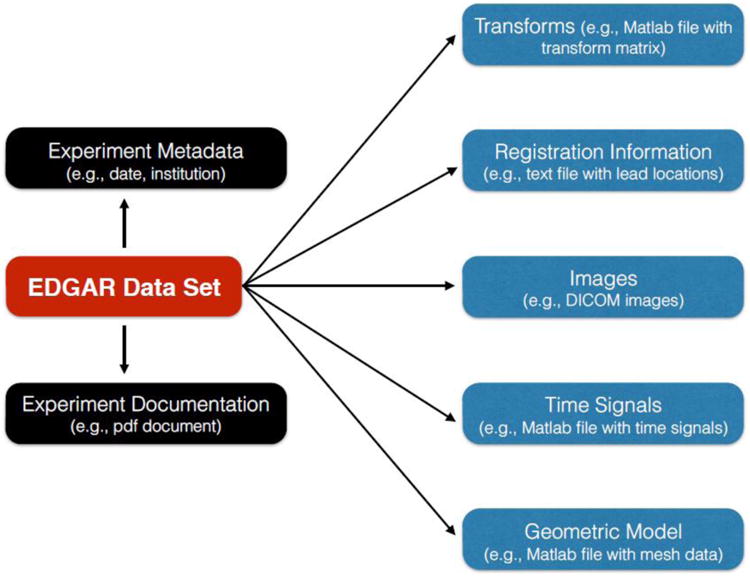
Time Signals
Time Signals (e.g., electrograms and ECG's) are a set of time varying signals sampled at a constant frequency (at least within one experiment), each of which is associated with a known location in the heart or thorax. We organize time signals according to a hierarchy shown in Figure 2, the top level of which is an experiment. An experiment describes a single study with the same subject (patient or animal), sometimes with a single study goal, e.g., to evaluate the effects of pacing intervention on cardiac electric behavior. An experiment consists of one or more interventions, a set of recordings that share a common or at least related set of conditions, typically recorded in close temporal proximity. Examples of an intervention include a sequence of recordings from a single episode of ischemia, a sequence of measurements from a heart undergoing rapid pacing at a set of rates, or a series of recordings that each correspond to a different level of a drug. Each intervention encapsulates a series of one or more “runs” that are continuous recordings of time signals (e.g., 5 s of ECG recording from the body-surface). Each run comprises a set of signals from a single acquisition but from multiple locations (e.g., a set of body-surface ECGs together with simultaneously captured epicardial electrograms). An intervention may contain a single run, e.g., the case of a single captured spontaneous beat (extrasystole). Data need not always come from experiments or clinical studies, but also from simulations, in which case each run is still a set of time signals generated under constant conditions or parameter settings.
Figure 2.
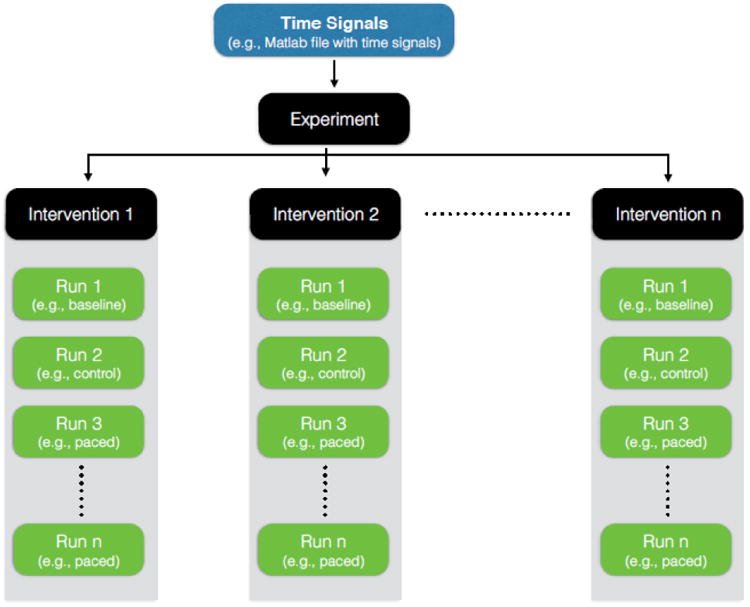
File Formats
Time signals are stored in MATLAB (http://www.mathworks.com) file format organized into structures with following contents:
.potvals, .data, or .field time signals as M × N array, where M is the number of data channels and N is the number of time instants.
numleads the number of channels of data.
numframes the number of time instants of data.
samplefrequency the sampling frequency used for acquisition.
.unit the type of units for the data, “um” for microvolts, “mv” for millivolts and “V” for volts.
.label the name of the time series. This is optional, but is useful in identifying the time series, particularly from a multi-time-series file.
gain a scaling factor that was applied as part of the amplification process during acquisition.
Note that only the “potvals” field is required. A MATLAB array may be one instance of these fields, a cell or struct array of them, or simply an M × N array of data.
Geometric Models
Geometry Models include elements of the subject's anatomy (e.g., torso, heart, and intervening organs) expressed as node locations in the three dimensional (3D) space connected by a network of edges to form polygons (e.g., triangles in 2D or tetrahedra in 3D). These meshes are the basis for numerical solutions and also serve to visualize the associated measured or simulated potentials and currents. Meshes can be generic or subject specific to an experiment, intervention or even an individual run.
File Formats
Geometric models are available in either an ASCII text file or MATLAB file formats. The ASCII format has the following structure:
Points (.pts) file The points (nodes) or “.pts” files contain the locations in 3D space of each point in the model, defined as follows:
ASCII file, no special characters permitted;
Each line contains one triplet, ordered as x, y, and z values; one or more spaces between values, which are assumed to be real, floating point numbers;
Each line may also optionally contain a group number as a fourth element;
The order of points in the file is the implicit order of the nodes in the geometry and of the time signals in those files; links between channels of time signal data and node locations are based on this ordering.
Triangle (.fac) files The characteristics of a triangle facet connectivity or “.fac” file are as follows:
ASCII file, no special characters permitted;
Each line contains a triplet of integer values pointing to the nodes of the geometry. Node numbers begin at 0 not 1!;
The order of triangle vertices (nodes) is not strictly controlled, however, it is recommended that the order reflect a common convention in graphics - a counterclockwise sequence of vertices viewed from the outside of the triangle;
Each line may contain an optional fourth value, which is the group number for the triangle;
Order of triangles in the file is not meaningful except for internal bookkeeping.
Tetrahedra (.tet) files The characteristics of a volumetric tetrahedra connectivity or “.tet” file are as follows:
ASCII file, no special characters permitted;
Each line contains a sequence of integer values pointing to the nodes of the geometry. Node numbers begin at 0 not 1!;
The order of tetrahedral vertices (nodes) is not controlled;
Each line may contain an optional fifth value which is the group number for the tetrahedron, typically associated with a tissue type and eventually an electrical conductivity value in simulations;
Order of tetrahedra in the file is not meaningful except for internal bookkeeping.
MATLAB geometry files The geometric models or meshes in a MATLAB file format are also organized into structures with the following fields:
.pts or .node elements of the structure contain the node locations, usually in a 3 × N array (although most flexible readers will check and accept either 3 × N or its transpose, N × 3), where N is the number of nodes.
.fac or .face elements contain the triangle connectivities, usually in a 3 × M array (again, well crafted reader programs will accept the transpose) where M is the number of triangles.
.seg or .edge elements contain the line segment connectivities, usually in a 2 × E array, where E is the number edges or line segments.
.tet or .tetra elements contain tetrahedral connectivities, and may contain an associated tissue assignment.
Forward and Inverse Transforms
Forward and Inverse Transforms contain the transformation matrices that project signals in either the forward (e.g., heart to body-surface) or inverse (e.g., body-surface to heart) direction. Such transforms are not a required module in EDGAR but are encouraged as they provide for the most complete comparison of algorithms and solutions.
File Formats
Transformation matrices are stored in a MATLAB file format as simple 2D matrices in a A × B array, where A is the number of nodes on the observation geometry (rows) and B is the number of nodes on the source geometry (columns).
Registration Information
Registration is the process of transforming different sets of data into one coordinate system. For example, electrode locations (e.g., epicardial electrical sock) digitally acquired during an experiment can be mapped to a mesh using registration techniques such as translation, scaling and rotation.
File Formats
Registration information can be varied and thus, its organization necessitates supplemental description in the documentation section of the experiment. The file formats are similarly varied and may include both ASCII text and MATLAB files.
Medical Images
Medical Images (e.g., MRI, CT scans etc.) typically form the basis of geometric models (meshes). To progress from images to a model entails identifying regions of interest (e.g., left ventricle) and labeling each of them, a process known as “segmentation.” This segmentation of the image volume into regions is then used to generate a subject specific mesh.
File Formats
EDGAR assumes the medical images are stored in the industry standard DICOM file format (http://www.dicom.nema.org), whereas segmentations, if available, are stored in near regular raster data (NRRD) format (http://teem.sourceforge.net).
Experiment Metadata
Experiment Metadata includes attributes such as the date of the experiment, species, institution, study category, and key words (e.g., ischemia, in situ, etc.). Moreover, the EDGAR system leverages the experiment metadata to allow searching for specific datasets. Preferred file format is ASCII with free text descriptions.
Experiment Documentation
Experiment Documentation is a collection of free form text documents that describe the experiment and the all relevant background information. Preferred file formats are ASCII and PDF. We will encourage data providers to not only describe the origins of the data as precisely as possible, but also to identify and quantify or estimate sources of error in their data.
Web Interface
EDGAR has a web interface (www.ecg-imaging.org/edgar) available through the CEI website (www.ecg-imaging.org) that enables easy, convenient, and efficient access to data. The EDGAR metadata storage scheme is implemented in REDCap [7] a secure database web application specialized for scientific research, and hosted on University of Utah web servers.
Typical interaction with EDGAR web front end would include searching for datasets based on specific metadata criteria or alternately browsing through all the available datasets. Moreover, the user can choose to download an entire dataset or individual components as needed. Each user must establish an identity on the server (i.e., in REDCap) but these are not regular login accounts and do not require passwords. Demographics and logistical information about the user is gathered upon first applying to access data on EDGAR. Moreover, data collected in this manner is secured and used only to document usage of the repository and any associated grants or funded research for which an EDGAR dataset is used.
Each dataset can also include a desired form of acknowledgement for any subsequent use of the data. Such acknowledgement is often in the form of a sentence to be included in the appropriate sections of publications or proposals.
Data Curation and patient privacy
Datasets hosted on EDGAR are carefully reviewed and validated before they become publicly available. All patient data contributed to the repository will undergo stripping of possible identifiers and will be anonymous. Investigators who wish to conduct biomedical studies for which additional clinical information is essential should contact the original authors of the data in order to gain access to this information under IRB approval to ensure patient protection. The management of new dataset contributions will occur through the EDGAR administration team and we do not envision direct upload capabilities. Data are freely available for all uses, academic and commercial, with the sole expectation that the source of the data be properly cited and acknowledged.
To access data from EDGAR will require each use to accept the following conditions:
“I acknowledge my responsibility to inform myself about and adhere to all local laws and regulations pertaining to the use of these data. I further pledge to cite the owner of the data and acknowledge the Consortium for Electrocardiographic Imaging (CEI) in all publications that make use of them.”
All donors of data must agree to the following terms:
“I am the owner of the data I wish to donate. I attest that these data were collected in accordance with all applicable rules and permissions in my home country (open IRB or IACUC or equivalent). I attest that I have the right to give the data in perpetuity to the Consortium of Electrocardiographic Imaging (CEI) for posting and distribution.”
Results
An aggregation of experimental, clinical and simulation data from various centers is being made available through the EDGAR project including experimental data from animal studies provided by the University of Utah (USA), clinical data from multiple human subjects provided by the Charles University Hospital (Czech Republic), and computer simulation data provided by the Karlsruhe Institute of Technology (Germany).
Experimental Data
Experimental data provided by the University of Utah includes epicardial, transmural and torso tank surface potential signals recorded from canine model ischemia studies. In addition, subject specific geometry models, anatomical MRI scans of the heart and forward transformation matrices are also included in the datasets.
Clinical Data
Clinical data provided by the Charles University Hospital includes body-surface potentials associated with endocardially-paced beats, which were recorded from three human subjects. Patient specific meshes for each of the human subjects as well as precomputed forward transformation matrices are also included in the dataset.
Simulation Data
Simulation data provided by Karlsruhe Institute of Technology (KIT) includes simulations (using a cellular automaton) of 15 paced ventricular beats that were applied to patient specific torso and heart geometries. In addition to the geometric models, lead field regularization transforms for the various mesh geometries are also provided as part of the dataset.
Discussion
It is important to put the EDGAR repository for electrocardiographic data archiving in the context of other similar projects. An early example that has had significant impact on ECG feature detection schemes is the MIT-BIH database [8], which is now part of a successful and diverse Physionet project [6]. There is a European standard for storage and exchange of ECG signals known as the “Open European Data Interchange and Processing for Computerized Electrocardiography” [9] that has also been successful. A more recent and also specialized example is the “Telemetric and Holter ECG Warehouse” initiative or THEW [10]. All these examples contain only ECG time signals and fail to support the image data or the geometric models that are essential for ECGI. We know of no existing repository that supports images, geometric models, and signals within a common framework.
Deciding on the appropriate file formats is not a simple question and is constrained by multiple factors, primary of which are ease of access and efficiency of storage and the time required to read and write the files. We chose a binary format simply for space/storage efficiency and to accelerate writing and reading of the time signals. ASCII formats do represent a lowest common denominator but a very inefficient one, especially for large data sets like time signals from hundreds of locations, large meshes, or large transfer matrices. The reason for selecting a format that is readable in MATLAB, is the ubiquity of MATLAB software, i.e., not only the original program from Mathwork's but the many freely available MATLAB file readers. There is even an open source replacement for MATLAB that would support reading and then rewriting the data in any desired internal format. In cases with small data sizes, e.g., a triangulated surface mesh that contains only a few hundred nodes, we have also provided ASCII equivalent formats.
By creating an open platform for high quality ECGI data acquired from clinical and experimental studies, we anticipate many benefits to the research community. The first is to provide researchers with access to hitherto unavailable, fully integrated and documented datasets. By gathering all possible components of complete datasets, e.g., from images to forward transform matrices, researchers can focus on components that are directly related to their own expertise. Each element of modeling pipeline provides technical challenges and also sources of error. EDGAR datasets will include, for example, the original medical images as well as their segmentations and the resulting surface and volume meshes. A researcher with expertise in image analysis can explore novel segmentation or meshing tools and compare the results to those from the repository. Another group can accept geometric models in the repository and focus instead on the numerical implementations of the source models or the regularization of the inverse formulation. All groups will benefit from research into suitable metrics to evaluate the accuracy of ECGI but only some will have the statistical expertise to fully evaluate, compare, and improve on existing metrics. When these groups collaborate, it will be possible to evaluate the impact of all the associated technical elements and approaches on the resulting inverse solutions.
A second benefit of the EDGAR project will be access to a diverse pool of data, i.e., experiments based entirely on simulations of cardiac activity and body-surface potentials with schematic geometries as well as those based entirely on medical imaging, multisite cardiac mapping studies and BSPM from patients. Such diversity will provide opportunities to test algorithms and implementations under a range of conditions. Moreover, techniques that are effective under a range of conditions are also likely to be useful for clinical applications.
A third benefit is that by applying algorithms and numerical methods to the same datasets, researchers can compare results across common data, a possibility never before broadly available. As a result, there have been almost now truly unbiased comparisons of methods or implementations. In addition, by providing a wide range of data examples under different experiment and clinical conditions, any comparison or evaluation will capture dependencies on the data or specific target question; a solution that is optimized for one type of application may not be robust across applications. For example, the optimal regularization parameters for locating ischemic regions in the ventricles will be unlikely to perform as well when reconstructing reentrant rotors in the left atrium.
Conclusions
In summary, the CEI and the EDGAR project seek to establish establish a communal forum for the sharing and distribution of cardiac electrophysiology data and geometric models for use in ECGI research. It is our hope that EDGAR will mark the beginning of a new generation of ECGI approaches. Moreover, we hope it will set a new tone of collaboration and cooperation in scientific research that will enable the creation of increasingly accurate and robust models that will produce novel insights into the physiology and clinical care of the heart.
Acknowledgments
Support for this research comes from the NIH NIGMS Center for Integrative Biomedical Computing (www.sci.utah.edu/cibc), NIH NIGMS Grant No. P41GM103545, the Nora Eccles Treadwell Foundation, and the Nora Eccles Treadwell Foundation for Cardiovascular Research. The authors also thank Bernie Lasalle, Scott Claerhout, James Clements and Mei Xue for their technical support and input to the system. The first Workshop on ECG Imaging in 2015 was financially supported by the German Research Foundation under the grant DO 637/13-1. Support for author LW comes from NIH/NHLBI R21HL125998, NSF/CAREER award ACI-1350374, and ADVANCE-RIT/NSF HRD-1209115.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kedar Aras, Bioengineering Department, Scientific Computing and Imaging Institute (SCI), Cardiovascular Research and Training Institute (CVRTI), University of Utah, Salt Lake City, UT, USA.
Wilson Good, Bioengineering Department, Scientific Computing and Imaging Institute (SCI), Cardiovascular Research and Training Institute (CVRTI), University of Utah, Salt Lake City, UT, USA.
Jess Tate, Bioengineering Department, Scientific Computing and Imaging Institute (SCI), Cardiovascular Research and Training Institute (CVRTI), University of Utah, Salt Lake City, UT, USA.
Brett Burton, Bioengineering Department, Scientific Computing and Imaging Institute (SCI), Cardiovascular Research and Training Institute (CVRTI), University of Utah, Salt Lake City, UT, USA.
Dana Brooks, Department of Electrical and Computer Engineering, Northeastern University, Boston, MA, USA.
Jaume Coll-Font, Department of Electrical and Computer Engineering, Northeastern University, Boston, MA, USA.
Olaf Doessel, Institute of Biomedical Engineering, Karlsruhe Institute of Technology, Karlsruhe, Germany.
Walther Schulze, Institute of Biomedical Engineering, Karlsruhe Institute of Technology, Karlsruhe, Germany.
Danila Potyagaylo, Institute of Biomedical Engineering, Karlsruhe Institute of Technology, Karlsruhe, Germany.
Linwei Wang, Program of Computing and Information Sciences, Rochester Institute of Technology, Rochester, NY, USA.
Peter van Dam, Radboud University, Nijmegen, The Netherlands David Geffen School of Medicine at UCLA (Los Angeles), CA, USA.
Rob MacLeod, Bioengineering Department, Scientific Computing and Imaging Institute (SCI), Cardiovascular Research and Training Institute (CVRTI), University of Utah, Salt Lake City, UT, USA.
References
- 1.de Ambroggi L. Body Surface Potential Mapping. In: Macfarlane PW, van Oosterom A, Pahlm O, Kligfield P, Janse M, Camm J, editors. Comprehensive Electrocardiology. London: Springer-Verlag Ltd; 2010. pp. 1375–1413. [Google Scholar]
- 2.Pullan AJ, Cheng LK, Nash MP, Brooks DH, Ghodrati A, Macleod RS. The inverse problem of electrocardiology. In: Macfarlane PW, van Oosterom A, Pahlm O, Kligfield P, Janse M, Camm J, editors. Comprehensive Electrocardiology. London: Springer-Verlag Ltd; 2010. pp. 299–344. [Google Scholar]
- 3.Rudy Y, Lindsay BD. Electrocardiographic imaging of heart rhythm disorders: from bench to bedside. Cardiac Electrophysiology Clinics. 2015;7(1):17–35. doi: 10.1016/j.ccep.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bear L, Cuculich PS, Bernus O, Efimov I, Dubois R. Introduction to noninvasive cardiac mapping. Cardiac Electrophysiology Clinics. 2015;7(1):1–16. doi: 10.1016/j.ccep.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Bokeriia LA, Revishvili A, Kalinin AV, Kalinin VV, Liadzhina OA, Fetisova EA. Hardware-software system for noninvasive electrocardiographic examination of heart based on inverse problem of electrocardiology. Biomedical Engineering. 2008;42(6):273–279. [PubMed] [Google Scholar]
- 6.Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PC, Mark RG, et al. Physiobank, PhysioToolkit, and PhysioNet components of a new research resource for complex physiologic signals. Circulation. 2000;101:e215–e220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 7.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moody GB, Mark RG. The MIT-BIH arrhythmia database on CD-ROM and software for use with it. Computers in Cardiology. 1993:185–188. [Google Scholar]
- 9.Willems JL, Rubel P, Zywietz C. Standard interchange for computerized electrocardiography. Stud Health Technol Inform. 1993;6:185–194. [PubMed] [Google Scholar]
- 10.Coudrec JP. A unique digital electrocardiographic repository for the development of quantitative electrocardiography and cardiac safety: the telemetric and Holter ECG warehouse (THEW) Journal of Electrocardiology. 2010;43(6):595–600. doi: 10.1016/j.jelectrocard.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]


