Abstract
The Par polarity complex creates mutually exclusive cortical domains in diverse animal cells. Activity of the atypical protein kinase C (aPKC) is a key output of the Par complex as phosphorylation removes substrates from the Par domain. Here we investigate how diverse, apparently unrelated Par substrates, couple phosphorylation to cortical displacement. Each protein contains a basic and hydrophobic (BH) motif that interacts directly with phospholipids and also overlaps with aPKC phosphorylation sites. Phosphorylation alters the electrostatic character of the sequence, inhibiting interaction with phospholipids and the cell cortex. We searched for overlapping BH and aPKC phosphorylation site motifs (i.e. putative PhosphoRegulated BH motifs) in several animal proteomes. Candidate proteins with strong PRBH signals associated with the cell cortex but were displaced into the cytoplasm by aPKC. These findings demonstrate a potentially general mechanism for exclusion of proteins from the Par cortical domain in polarized cells.
Graphical Abstract

Animal cells are polarized in remarkably diverse ways, such as the specialized apical cortex of a simple epithelium and the leading edge of motile cells (Knoblich, 2010; Overeem et al., 2015; Tepass, 2012). Although these cells are dramatically different, they are polarized by the same molecular machinery known as the Par complex (Goehring, 2014; Goldstein and Macara, 2007). The capacity of the Par complex to direct diverse cell polarities derives in part from its ability to act on a multitude of downstream proteins. For example, in asymmetrically dividing neural stem cells, the Par complex polarizes fate determinants (Betschinger et al., 2003; Knoblich, 2010), whereas in epithelia it organizes junctional components (Suzuki et al., 2001; Tepass, 2012). The characteristic adaptability of the Par complex to regulate distinct classes of proteins suggests that there may be a common mechanism by which it acts on downstream factors, yet little is known about how Par activity is coupled to substrate polarity. Knowledge of this mechanism is important not only for our basic understanding of Par-mediated polarity, but it might also allow for the identification of novel Par-regulated proteins and provide insight into the evolutionary pathways underlying the wide range of polarities found among metazoa.
The Par complex polarizes cells by creating and maintaining mutually exclusive cortical domains. Upstream factors that can be cell type specific define the Par domain (Cuenca et al., 2003; Harris and Peifer, 2005; Martin-Belmonte et al., 2007; Rolls et al., 2003; Schober et al., 1999; Wodarz et al., 1999), and downstream proteins are excluded from this cortical area to create the substrate domain. While the Par complex can function through other mechanisms (Cline and Nelson, 2007; von Stein et al., 2005; Zhang and Macara, 2006), a key output is the activity of atypical Protein Kinase C (aPKC). Proteins that are directly downstream of the Par complex are often aPKC substrates, and phosphorylation is both necessary and sufficient for cortical displacement and concomitant removal from the Par domain (Atwood and Prehoda, 2009; Betschinger et al., 2003; Hao et al., 2006; Hurov et al., 2004; Smith et al., 2007; Suzuki et al., 2004; Wirtz-Peitz et al., 2008; Zhang et al., 2001). For proteins polarized by this mechanism, their phosphorylation must be coupled to release from the cortex. Although phosphorylation-coupled cortical release is critical to Par complex-mediated polarity, very little is known about the mechanisms by which Par substrates associate with the cortex and how phosphorylation is linked to this interaction. The lack of sequence homology or shared globular domain structure among Par substrates has made it difficult to identify a “polarity code”.
The Par complex functions at the cell cortex, a complex organelle that includes the phospholipid bilayer and a meshwork of membrane-associated proteins and cytoskeletal elements beneath it (Engelman, 2005; Groves and Kuriyan, 2010; Morone et al., 2006). It has been unclear how these components might contribute to Par-mediated polarity, although actin polymerization is known to be required and protein-protein interactions have been implicated for certain substrates (Betschinger et al., 2005; Dho et al., 1999; Knoblich et al., 1997; Strand et al., 1994). Direct lipid binding has only been demonstrated for Numb PTB domain, however this is not sufficient for cortical localization (Dho et al., 1999). Given the complexity of the cell cortex and the potential for diverse protein-phospholipid and protein-protein interactions, we set out to identify the interactions that retain Par-polarized proteins within their cortical domain and how aPKC phosphorylation regulates these cortical interactions to prevent entry into the Par domain.
Results
Short, charged, hydrophobic motifs target Lgl, Mira, and Numb to the cell cortex
To determine if there might be a general mechanism of Par complex polarization, we selected three substrates with no apparent sequence homology or shared domain structure: Lgl, Mira, and Numb (Figure 1A–A″; (Betschinger et al., 2005; Dho et al., 2006; Fuerstenberg et al., 1998; Matsuzaki et al., 1998). Lgl and Numb each have protein interaction domains: β-propellers, a tomosyn homology region (THR) and a phosphotyrosine-binding (PTB) domain, but they are not sufficient for cortical association in Drosophila (Betschinger et al., 2005; Knoblich et al., 1997). Cortical localization of Mira is specified by its NH2-terminal 1-290 amino acids (Fuerstenberg et al., 1998; Matsuzaki et al., 1998), which does not contain any recognizable globular domains. These three diverse Par complex substrates lack clear globular domains that would mediate interactions with the cortex or membrane (Lemmon, 2008). Short stretches of basic and hydrophobic (BH) amino acids are known to interact with the membrane, so we analyzed the basic and hydrophobic character of the sequences using the BH scoring algorithm (Brzeska et al., 2010). Sequence windows with BH scores exceeding 0.6 are candidate phospholipid binding motifs and we found that Lgl, Mira, and Numb each contain a single region above this threshold (Figure 1A–A″). Interestingly, each of these sequences overlaps with a known or predicted aPKC phosphorylation site (Atwood and Prehoda, 2009; Betschinger et al., 2003; Smith et al., 2007). We identified several aPKC sites with BH scores below the threshold, indicating that not all aPKC sites are BH motifs (see below for a more extensive comparison of BH motifs and the aPKC recognition sequence).
Figure 1. Lgl, Mira, and Numb localize to the cell cortex through BH motifs.
A–A″. Basic-Hydrophobic motif signal in Lgl, Mira, and Numb. Previous work identified domains with multiple potential cortical targeting domains in Lgl, Mira, and Numb. The BH-search algorithm was used to analyze the sequences of Drosophila Lgl, Mira, and Numb. The BH signal is shown across each protein’s domain architecture. BH motifs are defined by peaks with a BH signal greater than 0.6 (denoted by blue line) and the peak area indicates the strength of the peak’s BH character. Each protein has a single BH motif that contains at least one aPKC phosphorylation site. Abbreviations are as follow: THR, tomosyn homology region; CLD, cortical localization domain; Cargo BD, cargo binding domain; PTB, phosphotyrosine binding domain; DPF, α-adaptin-binding motif; NPF, Eps15-binding motif.
B. BH motifs are necessary and sufficient for cortical targeting of Lgl, Mira, and Numb. Deletion constructs were used to determine if the BH motif is necessary and sufficient for localization to the S2 cell cortex. Representative images of the localization are presented for each protein when transiently transfected in S2 cells and characterized by either immunofluorescence for the HA epitope tag or by EGFP fluorescence. Localization to the cell cortex was quantified as an intensity ratio for the cortex versus the cytoplasm. At least 17 cells were quantified and the mean and the standard error of the mean (SEM) are shown. Asterisks indicate p < 0.0001 as assessed by a t-test to compare each ΔBH construct to its respective full-length control or each EGFP-fusion to its EGFP control. Scale bar, 10 μm.
Given their short sequences, the BH scoring algorithm could have a high false positive rate, so we investigated whether the Lgl, Mira, and Numb BH motifs mediate cortical localization. Each of these proteins localizes to the cortex of cultured Drosophila S2 cells consistent with their ability to interact with the cortex opposite Par domains. Deletion of each protein’s candidate BH motif caused a loss of cortical enrichment and strong cytoplasmic signal (Figure 1B–B″). In some cases, BH motifs often require additional multivalent interactions to associate with the membrane (Papayannopoulos et al., 2005; Swierczynski and Blackshear, 1996; Winters et al., 2005), so we tested whether each could function on their own. When EGFP is attached to stabilize these short, unstructured sequences (~30–70 amino acids), each BH motif was enriched at the S2 cortex (Figure 1B–B″), although at reduced levels compared to the full-length proteins. These data indicate that cortical localization of Lgl, Mira, and Numb is mediated by their BH motifs. Although additional interactions are likely needed to reinforce cortical recruitment, this appears to be a common mechanism for BH-mediated membrane association (McLaughlin and Murray, 2005; Rohatgi et al., 1999; Winters et al., 2005).
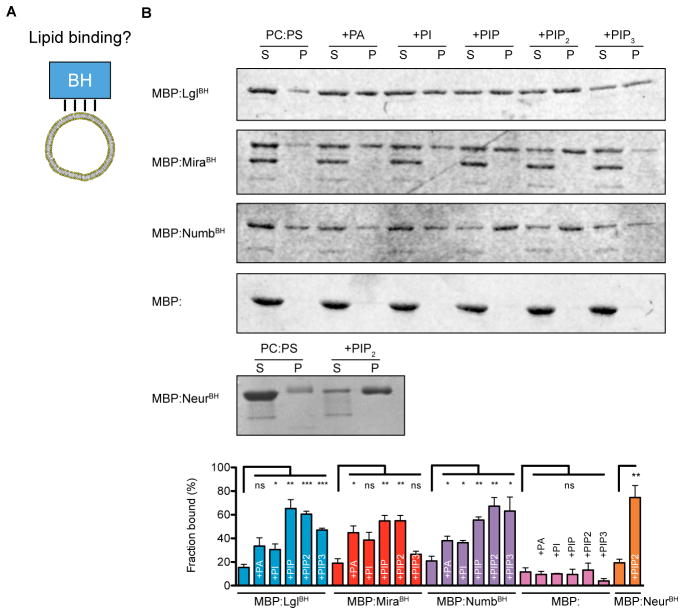
Lgl, Mira, and Numb BH motifs bind directly to phospholipids
The key role of the Lgl, Mira, and Numb BH motifs in cortical localization suggests that protein-phospholipid interactions are, at least in part, responsible for the cortical association of Par substrates. To test for direct protein-phospholipid interactions, we purified the BH motifs as maltose-binding protein (MBP) fusions and tested whether they associate with giant unilamellar vesicles (GUVs) of various lipid compositions via a co-sedimentation assay (Figure 2A). Using the BH motif from Neuralized (Neur) as a positive control (Skwarek et al., 2007), we found that each of the BH motifs from Lgl, Mira, and Numb exhibited very little binding to vesicles containing phosphatidylserine and phosphatidylcholine alone, but interacted with these vesicles when they were doped with various negatively charged phospholipids (Figure 2B, Supplemental Figure 1). Although phosphoinositides with multiple phosphates were slightly preferred over other negatively charged lipids (e.g. phosphatidic acid (PA)), the level of specificity is small enough that we do not expect it to be physiologically relevant.
Figure 2. Phospholipid binding mediates localization to the cell cortex.
A. BH motif phospholipid binding was tested using a cosedimentation phospholipid binding assay.
B. The BH motifs of Lgl, Mira, and Numb bind directly to phospholipid vesicles. A representative Coomassie-stained SDS-PAGE gel from a lipid vesicle-binding cosedimentation assays is presented. Each protein was characterized with an NH2-terminal Maltose Binding Protein (MBP) fusion. The BH motif from Neuralized (Neur residues 68–88) was used as a positive control. The supernatant (S) and pellet (P) contain the unbound and bound fractions, respectively. All vesicles contained a 4-1 mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC) to 1,2-Dioleoyl-sn-Glycero-3-[Phospho-L-Serine] (PS). Lanes marked by +X contained a third lipid that was added to be 10% of the total lipid by mass; abbreviations for these lipids are as follows. PA, L-α-Phosphatidic acid; PI, L-α-phosphatidylinositol; PI4P, L-α-Phosphatidylinositol-4-phosphate; PIP2, Phosphatidlyinositol-4,5-bisphosphate; PIP3, 1,2-dioleoyl-sn-glycero-3-[phosphoinositol-3,4,5-trisphosphate]. The fraction bound expressed as a percentage is shown in the bottom panel. Error bars represent SEM from three independent measurements. Asterisks (*, **, ***) represent significance levels of p < 0.05, 0.01, and 0.001, respectively. See also Supplemental Figure 1 for sequence analysis of Neur BH motif.
Par substrate cortical recruitment is mediated by electrostatic interactions
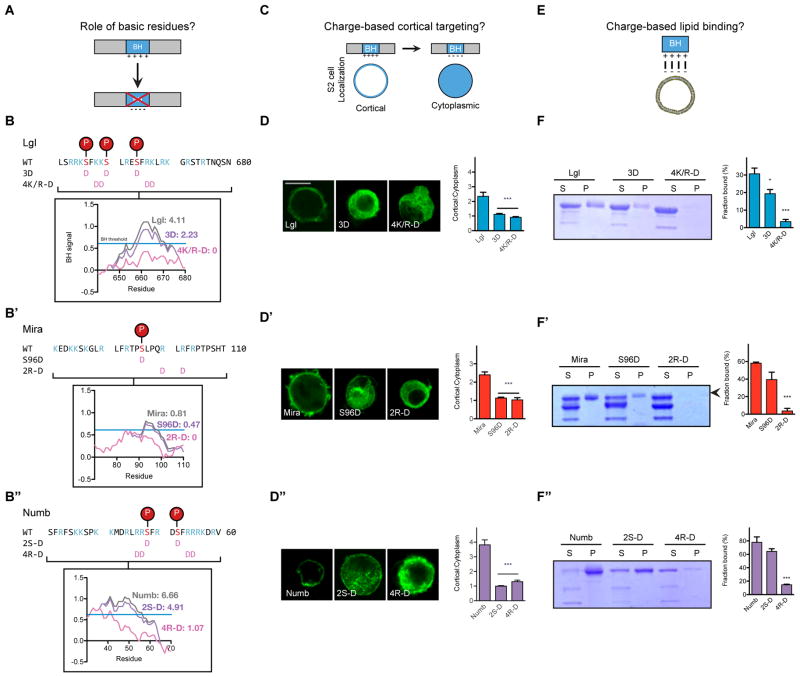
The BH motifs of Lgl, Mira, and Numb have multiple positively charged residues that may confer favorable electrostatics for binding phospholipids. Polybasic regions can bind negatively charged phospholipids, including PS and phosphoinositides. To investigate the role of charge in cortical targeting, we mutated basic residues within the BH motif to the acidic residue aspartic acid; these mutations reduced the calculated BH signal to often eliminate BH motif identification (Figure 3A and 3B–B″). We found that mutations that acidify the BH motif greatly reduce cortical localization of the full-length protein in S2 cells (Figure 3C and 3D–D″). Further, this effect was not site specific because mutations at multiple sites along the BH motif caused similar localization phenotypes (Supplemental Figure 2A).
Figure 3. Electrostatic interactions mediate Lgl, Mira, and Numb interactions with phospholipids and the cell cortex.
A. BH motif point mutants test the role for charged residues in cortical localization.
B–B″. Charged residues contribute to BH motif of Lgl, Mira, and Numb. The sequence of the BH motifs from Lgl, Mira, and Numb are displayed, highlighting basic residues (blue) and aPKC phosphosites (red). Terminology and location of the point mutants are displayed with pink aspartic acids. The inset shows the effect of each mutation on the BH signal and the area of the BH peak, as computed by BH-search. The BH threshold is displayed by a blue line with a BH signal of 0.6.
C. S2 cell localization assay tested if BH motif charge mediates localization to the cell cortex.
D–D″. Mutations to acidic residues reduce localization to the cell cortex in transiently transfected S2 cells. The localization of each full length protein with point mutations was characterized by immunostaining for the HA epitope tag (located on each protein’s NH2-terminus), and the localization was quantified as a cortical to cytoplasmic signal intensity ratio for at least 16 cells (mean ± SEM). Scale bar, 10 μm. See also Supplemental Figure 3A for additional point mutants.
E. Lipid binding assays tested if BH motif charge disrupts phospholipid binding.
F–F″. Acidic mutations reduce phospholipid binding. A Coomassie-stained gel from lipid vesicle binding sedimentation assays with vesicles of 4:1 PC:PS plus 10% PIP2 is shown. S indicates supernatant and P the pellet fraction. Arrowheads mark the full MBP-BH protein while other bands are truncation products (e.g. MBP alone). The fraction of protein bound, quantified as the amount of protein in the pellet over total protein, was quantified in triplicate for each vesicle composition. The mean and SEM of the fraction bound are shown. Asterisks indicate p < 0.05 and 0.0001 for one and three asterisks, respectively. Significance was evaluated using a non-parametric t-test relative to the unmutated BH motif.
See also Supplemental Figure 2.
Consistent with their effect on cortical localization, charge swap mutations reduced BH affinity for liposomes (Figure 3E and 3F–F″). To further test the role of electrostatics, we measured liposome interactions in the presence of 500 mM KCl, which reduces the entropic cost of displacing ions from the bilayer surface (McLaughlin, 1989). High ionic strength reduced the interaction between phospholipids and each BH motif (Supplemental Figure 2B), supporting the conclusion that direct, electrostatic interactions with phospholipids mediate BH motif cortical enrichment.
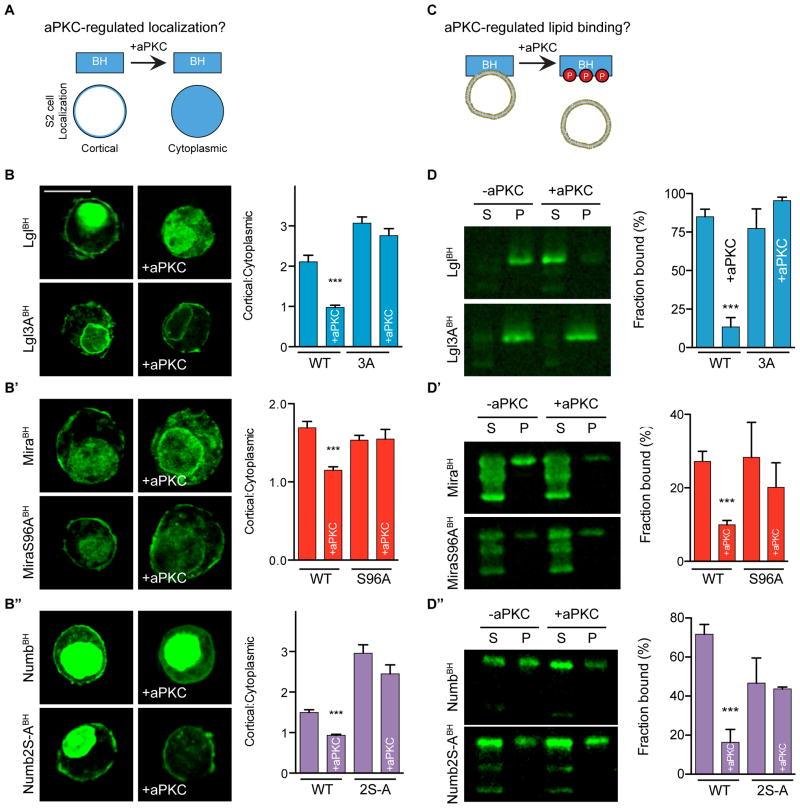
aPKC phosphorylation regulates BH cortical targeting and phospholipid binding
The Lgl, Mira, and Numb BH motifs contain verified and/or predicted aPKC phosphorylation sites. As charge swap mutations significantly reduce BH motif phospholipid binding and cortical association, we predicted that Par-induced polarization results when BH and aPKC phosphorylation site motifs are in close enough proximity that the phosphorylation(s) can sufficiently influence BH electrostatics to reduce the affinity for the membrane. This would allow Par substrates to associate with the cortex via their BH motif (and any accessory interactions) in regions lacking the Par complex. However, upon entering the Par cortical domain, the BH motif would be phosphorylated by aPKC, altering electrostatic character and reducing membrane affinity. Consistent with this model, we observed that phosphomimetic Lgl BH had reduced PIP2 binding, although no statistically significant difference was observed for Mira or Numb BH motifs (Figure 3F–F″). However, expression of aPKC significantly reduced cortical enrichment of the motifs from all three proteins (Figure 4A and 4B–B″), recapitulating the behavior of the full-length proteins. Non-phosphorylatable variants of each BH motif remain localized to the cell cortex when aPKC was expressed (Figure 4B-B″ and Supplemental Figure 3A), indicating that phosphorylation is required for displacement. Furthermore, we found that aPKC phosphorylation inhibits BH interaction with PIP2-containing vesicles, suggesting that disruption of this direct interaction is responsible for cortical displacement (Figure 4C and 4D-D″). Addition of aPKC had no effect on non-phosphorylatable BH motif variants Lgl3A, MiraS96A, or NumbS48AS52A, in the absence of ATP, or with aPKC harboring a kinase dead mutation (K293W) (Supplemental Figure 3B–D). The difference between the phosphomimetic and aPKC-phosphorylated proteins in binding PIP2-containing vesicles likely arises from the higher negative charge density of the phosphorylated proteins. We conclude that aPKC displaces Lgl, Mira, and Numb from the cortex by phosphorylation inhibiting phospholipid binding.
Figure 4. Phosphorylation directly inhibits Par substrate BH motif interactions with phospholipids and the cell cortex.
A. Schematic of aPKC regulation of BH motif cortical localization assay.
B–B″. Phosphorylation by aPKC disrupts Lgl, Mira, and Numb BH motif cortical enrichment. The localization of EGFP fused BH motifs both in the absence and presence (+aPKC) of aPKC is shown, along with that for BH motifs with phosphorylation sites mutated to alanine (3A, S96A, S48AS52A). Expression of aPKC was verified by immunostaining. Representative images from each transfection are shown. Expression of aPKC reduces cortical enrichment, and this reduction is dependent on the presence of the phosphorylation site. The mean and SEM are shown for measurements from at least 16 cells. Asterisks indicate p < 0.0001 as assessed by a non-parametric t-test to compare each aPKC-expressing cell to its respective control without aPKC expression. Scale bar, 10 μm. See also Supplemental Figure 2A.
C. The effect of aPKC phosphorylation on phospholipid binding was tested with lipid cosedimentation assays.
D–D″. Effect of aPKC on Par substrate BH motif binding to PC/PS/PIP2 vesicles. Lipid-binding cosedimentation assays were performed in the presence of aPKC and ATP. Samples were analyzed by immunoblotting for the MBP tag. S marks the supernatant and P marks the pellet. The “S-to-A” mutations are: Lgl3A, MiraS96A and NumbS48AS52A. All vesicles were composed of 4:1 PC:PS plus 10% PIP2. The fraction of protein bound, quantified as the amount of protein in the pellet over total protein, was quantified in triplicate for each vesicle composition. The mean and SEM of the fraction bound are shown. Asterisks indicate p < 0.0001. Significance was evaluated using a non-parametric t-test relative to binding in the absence of aPKC. See also Supplemental Figure 3B–D.
A bioinformatics approach to identifying candidate PRBH motifs
Our analysis of Lgl, Mira, and Numb suggests that Phospho-Regulated BH (PRBH) motifs mediate Par substrate polarization by coupling phosphorylation to membrane affinity. This coupling derives from the overlap of BH and aPKC phosphorylation motifs, and we used this criterion to search for putative PRBH motifs. We implemented an algorithm in the Python programming language (van Rossum) using the maximum BH score within a motif (Brzeska et al., 2010) along with a scoring system based on the aPKC phosphorylation site motif consensus sequence (Wang et al., 2012), modified to include Miranda (Figure 5A). We used the maximum BH score rather than the sum of BH scores within a motif to select for short, highly charged sequences like those found in Lgl, Miranda, and Numb. Additionally, to account for the observation that these sequences often contain multiple phosphorylations sites (Graybill and Prehoda, 2014), the PRBH score is increased by 0.2 times the number of phosphorylation sites. The weighting factor was set such that the BH and aPKC site score contributions to the overall PRBH score remained balanced. We used this algorithm to identify candidate PRBH motifs in several animal proteomes and numerically describe PRBH motifs by a maximum BH score (a metric of the basic character), aPKC site scores, and the number of aPKC sites identified (Figures 5B–C, Supplemental Figure 4A–B, and Supplemental Data Tables 1–3). The Lgl PRBH was identified as one of the top scoring motifs in the proteomes of human, fly, worm, and sponge, demonstrating that its charge and phosphosites are conserved in metazoan (Figures 5B–C, Supplemental Figure 4A, and Supplemental Data Tables). The algorithm also identified Numb and Mira, although with lower PRBH scores. It is important to note that for two reasons Par-polarized proteins likely represent a subset of PRBH-containing proteins. First, polarity is only one cellular process for which phosphorylation regulated membrane association is important. Second, as many kinases have overlapping specificity it is unlikely that all the identified PRBH motifs are solely aPKC substrates (especially those with low aPKC site scores). For example, two bona fide PRBH proteins regulated by the conventional PKC were identified: MARCKS and diacylglycerol kinase (DGK) (Luo et al., 2003; Topham et al., 1998; Überall et al., 1997). Finally, aPKC functions in other processes besides polarity (Farese et al., 2014; Standaert et al., 2001). While these effects increase the false positive rate for identifying polarity proteins, they also suggest that the algorithm will have utility outside of polarity.
Figure 5. PRBHscreen identifies candidate aPKC-regulated PRBH motifs.
A. Schematic representation of bioinformatics identification of PRBH motifs from proteomic files. The BH motif scoring algorithm identifies sequences with strong BH character. Vertical position on a scale from −2 to 2 shows each residues’ BH score. A representative BH plot has been included to depict the maximum BH score. aPKC consensus sites were identified and scored by scanning for S/T residues with preferred residues in the specific NH2- and COOH-terminal sites. The scale shows the aPKC site score (ranging from −0.1 to 0.35) for each residue in the specified site. Unlisted residues have no effect on the overall phosphosite score. An overall PRBH score was assigned to sequences identified in both the BH and aPKC site identifying algorithm.
B–C. PRBH score distribution from in the Drosophila and human proteome. Select PRBH motifs from the fly and human proteome are highlighted in B and C, respectively. Gray points display each identified putative PRBH motif with its respective PRBH max BH score and max aPKC site score. Blue, purple and red denote the number of identified aPKC sites. Blue lines mark the PRBH threshold values, sequences with scores less than these values are not candidate PRBH motifs. See also Supplemental Figure 4 and the Supplemental Data Tables for analysis of the C. elegans and sponge proteomes.
D. aPKC phosphorylates candidate PRBH motifs. An autoradiograph for P-32 emission shows that Amer1BH1-2, CKIγ-2BH, MPP7PRBH-GK, and PIP82PRBH were phosphorylated by aPKC. A Coomassie-stained loading control gel is displayed.
E–H. aPKC inhibits localization to the cell cortex for several candidate PRBH motifs. Representative images are shown with a quantification of the cortical to cytoplasmic signal intensity ratio as the mean ± SEM. Cartoons show the constructs characterized with candidate aPKC phosphosites listed. Each protein was characterized as an EGFP-fusion protein. Statistical significance testing was performed using the t-test of aPKC cotransfected cells to singly transfected cells. Asterisks indicate p < 0.001 and 0.0001 for two and three asterisks, respectively. Significance was evaluated using a non-parametric t-test relative of the singly transfected cells to aPKC cotransfected cells. ns, not significant.
See also Supplemental Figure 4.
To validate the algorithm, we selected several candidate PRBH motifs to determine if they are bona fide regulated membrane association elements. We selected candidates from the fly and human proteomes with a range of PRBH scores, aPKC site scores, and maximum BH scores (Figure 5B–C) to examine how well PRBH score correlates with activity. For each candidate, we tested if they can be phosphorylated by aPKC in vitro, whether they associate with the cortex in S2 cells, and whether aPKC can displace them into the cytoplasm. We found that each protein was indeed phosphorylated by aPKC in an in vitro kinase assay (Figure 5D). Membrane-associated guanylate kinase (MAGUK) p55 subfamily member 7 (MPP7) regulates tight junction formation (Stucke et al., 2007) and has a single, highly scored putative PRBH motif that includes two predicted aPKC phosphosites (Figure 5C and 5E). The candidate MPP7 PRBH was highly enriched at the cell cortex and aPKC expression inhibited cortical localization (Figure 5E). PIP82, a Drosophila protein involved in signal transduction downstream of photoreceptors (Suri et al., 1998), similarly contains two PRBH motifs, the stronger of which has high BH character and two predicted aPKC sites (Figure 5B and 5F). We observed that the PIP82 candidate PRBH sequence also localized to the S2 cell cortex and this localization was antagonized by aPKC (Figure 5F).
We also examined several lower scoring PRBH motifs to determine if they function as aPKC regulated cortical localization motifs. The putative PRBH motif on human casein kinase I γ (CKIγ-2) contains a strongly predicted aPKC site rated similar to MPP7, but has weak basic character as demonstrated by a low maximum BH height (Figure 5C and 5G). To our knowledge, its localization remain uncharacterized in polarized cells, however the Casein Kinase-associated protein Lrp6 localizes basolaterally in the Xenopus neuroectoderm (Davidson et al., 2005; Huang and Niehrs, 2014). CKIγ-2 PRBH localizes weakly to the cell cortex and has punctate localization suggesting it localizes to endomembrane organelles, as seen previously for Casein kinase (Tomishige et al., 2009). Expression of aPKC reduced the cortical localization of the CKIγ-2 candidate PRBH and did not alter the distribution of CKIγ-2 between the cortex and cytoplasm (Figure 5G). The human tumor suppressor Adenomatous polyposis coli membrane recruitment protein (Amer1, also known as WTX) represses WNT/β-catenin signaling (Grohmann et al., 2007; Rivera et al., 2007; Tanneberger et al., 2011); it contains a strong BH signal but a weak aPKC phosphosite score (Figure 5C and 5H). Correspondingly, we found that the Amer1 PRBH is targeted to the cell cortex, but aPKC expression does not reduce its cortical localization (Figure 5H). However, the number of potential phospho-accepting residues within Amer1’s PRBH makes it a prime candidate for regulation by another kinase. We conclude that the PRBH algorithm is a good predictor of aPKC-regulated cortical association, with higher scoring motifs more likely to exhibit this behavior. Furthermore, a more precise assessment of PRBH character can be made be directly comparing BH motif area, the number of aPKC phosphosites, etc.
Discussion
The organization of the animal cell cortex by the Par complex involves two key steps (Goehring, 2014; Goldstein and Macara, 2007; Knoblich, 2010; Prehoda, 2009). The first involves the specification of the Par domain by upstream components such as the Rho GTPase Cdc42. In the second, diverse proteins must “plug into” Par polarity by occupying cortical areas that lack the Par complex. We have examined how the key output of the Par complex, aPKC activity, leads to cortical exclusion of the diverse array of Par-polarized proteins. Our approach was to examine the cortical interactions of three Par substrates that had no described sequence or domain similarities and to determine how aPKC phosphorylation modulates their cortical binding. Although these Par substrates do not have clear sequence homology, they each contain a “Phospho-regulated BH” motif that couples aPKC phosphorylation to membrane affinity. This coupling is a direct consequence of the overlap of BH and aPKC phosphorylation sites in the PRBH motif as this allows phosphorylation to have a significant effect on the electrostatic character of the sequence. Using this defining feature, we developed a computational approach for identifying candidate PRBH motifs, which we validated that aPKC regulates cortical association of several hits from the human and fly proteomes. We expect this technique will be useful for identifying candidate polarity proteins, and proteins involved in other cellular processes where regulated membrane association is important.
Multivalent interactions mediate Par substrate cortical localization
We have found that Par substrate BH motifs localize to the cortex, but it is important to note that robust targeting requires elements outside of the BH sequence. While our data suggests that PRBH motifs are key regulatory elements for polarity, the requirement of additional interactions for high affinity cortical interactions means that they are unlikely to lead to substrate polarity on their own. Similar “multivalent” interaction mechanisms has been observed with other BH motif-containing proteins that are regulated by phosphorylation and utilize accessory interactions to enhance their cortical targeting, including the yeast pheromone signaling protein Ste5 (Pryciak and Huntress, 1998; Winters et al., 2005) and the actin filament crosslinker MARCKS (Hartwig et al., 1992; McLaughlin and Murray, 2005; Thelen et al., 1991). In Ste5, protein-protein interactions provide additional cortex affinity, whereas MARCKS contains a myristoyl modification that works with its BH motif (George and Blackshear, 1992). The multivalent nature of protein-phospholipid interactions mediated by BH motifs may explain why protein-protein interactions are important to polarize some Par substrates. For example, Lgl interacts with non-muscle myosin II (Betschinger et al., 2005), and this interaction may cooperate with BH-phospholipid binding to yield the increased cortical interaction of full-length Lgl compared to its BH alone. Phospholipid binding by the BH motif and the PTB domain may robustly target Numb to the cell cortex (Dho et al., 1999). However, in order for cortical localization to be regulated, the accessory interactions must not be stable enough to mediate cortical targeting on their own, or alternatively they must themselves be regulated by phosphorylation (McLaughlin and Murray, 2005; Strickfaden et al., 2007). Future studies of candidate PRBH-containing proteins will determine which regulated interactions are used for Par-mediate polarity and how factors such as cortical binding affinities, multiple PRBH motifs, and cortical mobility cooperate to mediate substrate polarization.
PRBH: a mechanism for convergent evolution of Par-mediated polarity?
Phosphorylation can regulate protein activity by several means, including allostery and conformational changes (Cohen, 2000; Johnson and Barford, 1993; Serber and Ferrell, 2007). In this type of regulation, phosphate attachment to a specific side-chain alters protein structural features and/or dynamics that are important for catalytic or binding activity (Barford et al., 1991). This coupling requires a connection between the phosphorylation site and the native state energy landscape, such that the phosphorylation site sequence is typically highly conserved (Holt et al., 2009). Allosteric effects or conformational changes may induce the polarization of substrates that lack PRBH motifs (e.g. Baz/Par-3 and Par-1) (Hurov et al., 2004; Morais-de-Sá et al., 2010; Sotillos et al., 2004; Suzuki et al., 2004). In contrast to complex mechanisms like these, phosphorylation of many Par substrates alters activity by changing the bulk electrostatics of the phospholipid binding sequence (Serber and Ferrell, 2007). Here, phosphorylation must occur close to the binding sequence but does not have to be coupled to structural or dynamic changes in the protein (Holt et al., 2009). Because many sequences satisfy the requirements of 1) an electrostatic character sufficient for binding negatively charged phospholipids and 2) one or more aPKC phosphorylation sites, we propose that the sequence path for convergent evolution of PRBH motifs could be fairly simple. An existing motif, either BH or aPKC phosphosite is likely to require a small number of mutations to retain existing function while causing a gain of the missing function. For example, a protein that is advantageously targeted to the membrane via a BH motif could require only a small number of mutations to become polarized with the introduction of one or more aPKC phosphosites.
Functions of candidate PRBH-containing proteins
We identified several proteins whose cortical localization is regulated by aPKC: MPP7 and PIP82, and CKIγ-2. In epithelial cells, MPP7 localizes to the lateral cortex and mediates tight junction formation (Stucke et al., 2007). While previous work has not implicated aPKC in its regulation, its localization and aPKC’s inhibition of cortical localization make it a strong candidate Par-polarized substrate. The localization of several Casein kinase paralogs that lack predicted PRBH motifs (Extended Data Table) have been described (Davidson et al., 2005; Gross et al., 1997), although the localization of the PRBH-containing CKIγ-2 in a polarized in vivo context is uncharacterized to our knowledge. However, recent work demonstrated that the CKIγ-associated protein Lrp6 localizes to the basolateral cortex in the neuroepithelium (Davidson et al., 2005; Huang and Niehrs, 2014), suggesting that CKIγ-2 may also exhibit this pattern of localization. Future work will need to address if aPKC indeed regulates CKIγ-2 localization in vivo. The localization and molecular function of PIP82 in Drosophila photoreceptor cells has not been described, but it is intriguing that light exposure causes it to be dephosphorylated (Suri et al., 1998). If the PIP82 PRBH is dephosphorylated during this process, it would be a mechanism for coupling cortical localization to light exposure. While we have identified many putative aPKC-regulated PRBH motifs, future work will need to address how this regulatory sequence is used cellular processes besides polarity. One particularly enticing PRBH protein is the retromer component Vps26 as it has been implicated in Par- and Scribble-mediated endosomal trafficking (de Vreede et al., 2014). Characterization of Vps26 and other PRBH proteins will likely emphasize the diverse functions of aPKC phosphorylation.
A model for protein polarization directed by the Par complex
From these studies, we propose a model for Par complex function (Figure 6). In this model, Par-polarized substrates localize to the cortex via direct interactions between their BH motifs and membrane phospholipids. When a substrate enters the Par domain, either from the cytoplasm or by diffusion along the membrane, it becomes phosphorylated by aPKC. Addition of phosphates alters the electrostatics of the PRBH motif to reduce its affinity for phospholipids causing it to be displaced into the cytoplasm. Future work will be required to complete the “life cycle” of these substrates to understand the fate of Par substrates once displaced into the cytoplasm. Phosphorylation may be coupled to inactivation, degradation, or phosphatase-mediated re-association to the cortex.
Figure 6. Model for Par polarization by phosphoregulated BH motifs.

PRBH motifs mediate localization to regions of the cell cortex opposite the Par complex. The box detail shows the molecular interactions occurring at the interface between the Par and substrate domains. Par substrates will localize to the cortex by electrostatic interactions with phospholipids until they encounter aPKC and become phosphorylated. Phosphorylation reduces their membrane affinity. Weak accessory interactions mediate localization to the cell cortex but they must allow the substrate to dissociate from the cell cortex, when the BH motif is phosphorylated.
Experimental procedures
Sequence analysis and computational work
BH motifs were initially identified using BH-search: a computational algorithm described in Brzeska et al. and is available online (http://helixweb.nih.gov/bhsearch/) (Brzeska et al., 2010). Domain analyses were performed using SMART (Letunic et al., 2015; Schultz et al., 1998). Figures were assembled using the Adobe Creative Suite. The PRBH algorithm was implemented using the Python programming language with standard Python libraries including numpy and matplotlib. The Drosophila and human non-redundant proteome were analyzed from the EMBL-EBI reference proteome files. Please see the Supplemental Information for the full script and a detailed description of the PRBH identification program. Briefly, BH motifs were identified using a previously described BH scoring metric (Brzeska et al., 2010). Residues surrounding S/T residues were scored based on the aPKC consensus sequence (Wang et al., 2012) with Miranda’s aPKC recognition sequence included. Composite scores were assigned to sequences identified as BH motifs and aPKC consensus sequences. PRBH scores above a threshold value were analyzed.
Molecular cloning and cell culture
All molecular cloning was performed as previously described (Graybill et al., 2012). Please see the Extended Experimental Procedures in the Supplement for details. Cell culture was performed as previously described (Lu and Prehoda, 2013). Briefly, cells were grown according to manufacture’s protocol, transiently transfected with Effectene (QIAGEN). Transfected cells were fixed and immunostained as previously described (Lu and Prehoda, 2013), and imaged with the following confocal microscopes: Olympus Fluoview FV1000 BX61 with a PlanApo N 60×/1.42 oil and a Leica SP2 confocal microscope with a 63×/1.40–0.60 oil CS objective. All proteins were tagged at their NH2-terminus. Image analysis was performed with ImageJ.
Protein expression and purification
Protein expression was performed as previously described (Graybill et al., 2012). Briefly, MBP-tagged proteins were purified from BL21 (DE3) cells by amylose affinity purification. aPKC was purified from HEK293F cells transiently transfected with 293fectin transfection reagent (Life Technology). All His6-tagged aPKC constructs were purified with Ni2+-nitrilotriacetic acid resin purification from HEK293F cell lysates. Please see the Extended Experimental Procedures for a detailed description of this protein purification.
Biochemical assays
All lipids were purchased from Avanti Polar Lipids. Giant unilamellar vesicles were prepared as previously described (Winters et al., 2005). Please see the Extended Experimental Procedures for details of on vesicle preparation, and lipid cosedimentation assays. Pelleting assays were performed as previously described (Prehoda et al., 2000). Giant unilamellar vesicles and associated bound proteins were pelleted by ultracentrifugation and analyzed by SDS-PAGE. Kinase assays were performed as previously described (Atwood and Prehoda, 2009; Graybill et al., 2012). Briefly, aPKC and substrates were incubated in assay buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2) at 30°C for 5 min before addition of 1 mM ATP doped with [γ-32P]ATP (~1.0 X 105/nmol of ATP). The kinase reaction proceeded for 30 minutes before quenching with SDS loading dye. Samples were run on a 12.5% acrylamide SDS-PAGE gel, then analyzed with the Storm 860 Molecular Imager and a phosphor screen (Molecular Dynamics).
Supplementary Material
Acknowledgments
Plasmids were generously provided by Roland Le Borgne (Numb), Gabrielle Boulianne (Neuralized), Kentaro Hanada (CKIγ-2), and Jürgen Behrens (Amer1). Thanks to Robert Lyle McPherson for his assistance with cell culture and protein purification. We would like to thank Michael Drummond, Chiharu Graybill, Ryan Holly, Kimberly Jones, Robert Lyle McPherson, and Brett Wee for useful discussions, and J. Andy Berglund and Raghuveer Parthasarathy for equipment use. Thanks to Tom Stevens, Brad Nolen, and Chris Doe for comments on the manuscript and to Peter Pryciak for useful discussions about this work. This work was supported by the National Institutes of Health Predoctoral Training Grant GM007759 (M.J.B.) and the National Institutes of Health Grant GM068032 (K.E.P.).
Footnotes
Author contributions
All experiments were designed and conceived by M.J.B and K.E.P. M.J.B. performed all experiments. Programming was performed by M.J.B and K.E.P. The manuscript was written by M.J.B and K.E.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atwood SX, Prehoda KE. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991;218:233–260. doi: 10.1016/0022-2836(91)90887-c. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Eisenhaber F, Knoblich JA. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr Biol. 2005;15:276–282. doi: 10.1016/j.cub.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Brzeska H, Guag J, Remmert K, Chacko S, Korn ED. An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J Biol Chem. 2010;285:5738–5747. doi: 10.1074/jbc.M109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline EG, Nelson WJ. Characterization of mammalian Par 6 as a dual-location protein. Mol Cell Biol. 2007;27:4431–4443. doi: 10.1128/MCB.02235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation – a 25 year update. Trends in Biochemical Sciences. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- de Vreede G, Schoenfeld JD, Windler SL, Morrison H, Lu H, Bilder D. The Scribble module regulates retromer-dependent endocytic trafficking during epithelial polarization. Development. 2014;141:2796–2802. doi: 10.1242/dev.105403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho SE, French MB, Woods SA, McGlade CJ. Characterization of Four Mammalian Numb Protein Isoforms IDENTIFICATION OF CYTOPLASMIC AND MEMBRANE-ASSOCIATED VARIANTS OF THE PHOSPHOTYROSINE BINDING DOMAIN. J Biol Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- Dho SE, Trejo J, Siderovski DP, McGlade CJ. Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: Structural determinants of numb association with the cortical membrane. Mol Biol Cell. 2006;17:4142–4155. doi: 10.1091/mbc.E06-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- Farese RV, Lee MC, Sajan MP. Atypical PKC: a target for treating insulin-resistant disorders of obesity, the metabolic syndrome and type 2 diabetes mellitus. Expert Opin Ther Targets. 2014;18:1163–1175. doi: 10.1517/14728222.2014.944897. [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol Cell Neurosci. 1998;12:325–339. doi: 10.1006/mcne.1998.0724. [DOI] [PubMed] [Google Scholar]
- George DJ, Blackshear PJ. Membrane association of the myristoylated alanine-rich C kinase substrate (MARCKS) protein appears to involve myristate-dependent binding in the absence of a myristoyl protein receptor. J Biol Chem. 1992;267:24879–24885. [PubMed] [Google Scholar]
- Goehring NW. PAR polarity: from complexity to design principles. Exp Cell Res. 2014;328:258–266. doi: 10.1016/j.yexcr.2014.08.009. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill C, Prehoda KE. Ordered multisite phosphorylation of lethal giant larvae by atypical protein kinase C. Biochemistry. 2014;53:4931–4937. doi: 10.1021/bi500748w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill C, Wee B, Atwood SX, Prehoda KE. Partitioning-defective Protein 6 (Par-6) Activates Atypical Protein Kinase C (aPKC) by Pseudosubstrate Displacement. J Biol Chem. 2012;287:21003–21011. doi: 10.1074/jbc.M112.360495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann A, Tanneberger K, Alzner A, Schneikert J, Behrens J. AMER1 regulates the distribution of the tumor suppressor APC between microtubules and the plasma membrane. J Cell Sci. 2007;120:3738–3747. doi: 10.1242/jcs.011320. [DOI] [PubMed] [Google Scholar]
- Gross SD, Simerly C, Schatten G, Anderson RA. A casein kinase I isoform is required for proper cell cycle progression in the fertilized mouse oocyte. J Cell Sci. 1997;110(Pt 24):3083–3090. doi: 10.1242/jcs.110.24.3083. [DOI] [PubMed] [Google Scholar]
- Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Boyd L, Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJC, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH, Thelen M, Rosen A, Janmey PA, Nairn AC, Aderem A. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Niehrs C. Polarized Wnt signaling regulates ectodermal cell fate in Xenopus. Dev Cell. 2014;29:250–257. doi: 10.1016/j.devcel.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Barford D. The effects of phosphorylation on the structure and function of proteins. Annu Rev Biophys Biomol Struct. 1993;22:199–232. doi: 10.1146/annurev.bb.22.060193.001215. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc Natl Acad Sci USA. 1997;94:13005–13010. doi: 10.1073/pnas.94.24.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucl Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MS, Prehoda KE. A NudE/14-3-3 pathway coordinates dynein and the kinesin Khc73 to position the mitotic spindle. Dev Cell. 2013;26:369–380. doi: 10.1016/j.devcel.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Prescott SM, Topham MK. Protein kinase C alpha phosphorylates and negatively regulates diacylglycerol kinase zeta. J Biol Chem. 2003;278:39542–39547. doi: 10.1074/jbc.M307153200. [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128:383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. The electrostatic properties of membranes. Annu Rev Biophys Biophys Chem. 1989;18:113–136. doi: 10.1146/annurev.bb.18.060189.000553. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Morais-de-Sá E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morone N, Fujiwara T, Murase K, Kasai RS, Ike H, Yuasa S, Usukura J, Kusumi A. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J Cell Biol. 2006;174:851–862. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overeem AW, Bryant DM, van IJzendoorn SCD. Mechanisms of apical-basal axis orientation and epithelial lumen positioning. Trends Cell Biol. 2015;25:476–485. doi: 10.1016/j.tcb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA. A Polybasic Motif Allows N-WASP to Act as a Sensor of PIP2 Density. Molecular Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Prehoda KE. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb Perspect Biol. 2009;1:a001388. doi: 10.1101/cshperspect.a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MN, Kim WJ, Wells J, Driscoll DR, Brannigan BW, Han M, Kim JC, Feinberg AP, Gerald WL, Vargas SO, et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science. 2007;315:642–645. doi: 10.1126/science.1137509. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih H-P, Lee C-Y, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum G. The Python Programming Language [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. PNAS. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber Z, Ferrell JE. Tuning bulk electrostatics to regulate protein function. Cell. 2007;128:441–444. doi: 10.1016/j.cell.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Skwarek LC, Garroni MK, Commisso C, Boulianne GL. Neuralized contains a phosphoinositide-binding motif required downstream of ubiquitination for delta endocytosis and notch signaling. Dev Cell. 2007;13:783–795. doi: 10.1016/j.devcel.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillos S, Díaz-Meco MT, Caminero E, Moscat J, Campuzano S. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J Cell Biol. 2004;166:549–557. doi: 10.1083/jcb.200311031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert ML, Bandyopadhyay G, Kanoh Y, Sajan MP, Farese RV. Insulin and PIP3 activate PKC-zeta by mechanisms that are both dependent and independent of phosphorylation of activation loop (T410) and autophosphorylation (T560) sites. 2001;40:249–255. doi: 10.1021/bi0018234. [DOI] [PubMed] [Google Scholar]
- von Stein W, Ramrath A, Grimm A, Müller-Borg M, Wodarz A. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 2005;132:1675–1686. doi: 10.1242/dev.01720. [DOI] [PubMed] [Google Scholar]
- Strand D, Jakobs R, Merdes G, Neumann B, Kalmes A, Heid HW, Husmann I, Mechler BM. The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain. J Cell Biol. 1994;127:1361–1373. doi: 10.1083/jcb.127.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM, Timmerman E, Vandekerckhove J, Gevaert K, Hall A. The MAGUK protein MPP7 binds to the polarity protein hDlg1 and facilitates epithelial tight junction formation. Mol Biol Cell. 2007;18:1744–1755. doi: 10.1091/mbc.E06-11-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V, Qian Z, Hall JC, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, et al. aPKC Acts Upstream of PAR-1b in Both the Establishment and Maintenance of Mammalian Epithelial Polarity. Current Biology. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Swierczynski SL, Blackshear PJ. Myristoylation-dependent and electrostatic interactions exert independent effects on the membrane association of the myristoylated alanine-rich protein kinase C substrate protein in intact cells. J Biol Chem. 1996;271:23424–23430. doi: 10.1074/jbc.271.38.23424. [DOI] [PubMed] [Google Scholar]
- Tanneberger K, Pfister AS, Brauburger K, Schneikert J, Hadjihannas MV, Kriz V, Schulte G, Bryja V, Behrens J. Amer1/WTX couples Wnt-induced formation of PtdIns(4,5)P2 to LRP6 phosphorylation. EMBO J. 2011;30:1433–1443. doi: 10.1038/emboj.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U. The Apical Polarity Protein Network in Drosophila Epithelial Cells: Regulation of Polarity, Junctions, Morphogenesis, Cell Growth, and Survival. 2012;28:655–685. doi: 10.1146/annurev-cellbio-092910-154033. [DOI] [PubMed] [Google Scholar]
- Thelen M, Rosen A, Nairn AC, Aderem A. Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature. 1991;351:320–322. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- Tomishige N, Kumagai K, Kusuda J, Nishijima M, Hanada K. Casein kinase I{gamma}2 down-regulates trafficking of ceramide in the synthesis of sphingomyelin. Mol Biol Cell. 2009;20:348–357. doi: 10.1091/mbc.E08-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham MK, Bunting M, Zimmerman GA, McIntyre TM, Blackshear PJ, Prescott SM. Protein kinase C regulates the nuclear localization of diacylglycerol kinase-zeta. Nature. 1998;394:697–700. doi: 10.1038/29337. [DOI] [PubMed] [Google Scholar]
- Überall F, Giselbrecht S, Hellbert K, Fresser F, Bauer B, Gschwendt M, Grunicke HH, Baier G. Conventional PKC-α, Novel PKC-ε and PKC-θ, but Not Atypical PKC-λ Are MARCKS Kinases in Intact NIH 3T3 Fibroblasts. J Biol Chem. 1997;272:4072–4078. doi: 10.1074/jbc.272.7.4072. [DOI] [PubMed] [Google Scholar]
- Wang C, Shang Y, Yu J, Zhang M. Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKCι in complex with a substrate peptide from Par-3. Structure. 2012;20:791–801. doi: 10.1016/j.str.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu SL, Rubin CS. A novel adapter protein employs a phosphotyrosine binding domain and exceptionally basic N-terminal domains to capture and localize an atypical protein kinase C: characterization of Caenorhabditis elegans C kinase adapter 1, a protein that avidly binds protein kinase C3. J Biol Chem. 2001;276:10463–10475. doi: 10.1074/jbc.M008990200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.