Abstract
Ichthyoses are a group of various different types of hereditary disorders affecting skin cornification. They are characterized by hyperkeratoses of different severity levels and are associated with a dry and scaling skin. Genome-wide association analysis of nine affected and 13 unaffected Great Danes revealed a genome-wide significant peak on chromosome 9 at 57–58 Mb in the region of SLC27A4. Sequence analysis of genomic DNA of SLC27A4 revealed the non-synonymous SNV SLC27A4:g.8684G>A in perfect association with ichthyosis-affection in Great Danes. The mutant transcript of SLC27A4 showed an in-frame loss of 54 base pairs in exon 8 probably induced by a new splice acceptor site motif created by the mutated A- allele of the SNV. Genotyping 413 controls from 35 different breeds of dogs and seven wolves revealed that this mutation could not be found in other populations except in Great Danes. Affected dogs revealed high amounts of mutant transcript but only low levels of the wild type transcript. Targeted analyses of SLC27A4 protein from skin tissues of three affected and two unaffected Great Danes indicated a markedly reduced or not detectable wild type and truncated protein levels in affected dogs but a high expression of wild type SLC27A4 protein in unaffected controls. Our data provide evidence of a new splice acceptor site creating SNV that results in a reduction or loss of intact SLC27A4 protein and probably explains the severe skin phenotype in Great Danes. Genetic testing will allow selective breeding to prevent ichthyosis-affected puppies in the future.
Introduction
Hereditary ichthyoses represent a heterogenous group of skin cornification disorders that affect the entire integument and are characterized by hyperkeratosis and blistering or scaling [1, 2]. In human, a wide variety of mutations have been shown to be responsible for different types of ichthyoses [3, 4]. They were classified into syndromic and non-syndromic forms and further subgroups with regard to clinical signs, onset of the disease and genetic background. On the whole 36 genes have been detected which were supposed to be involved in non-syndromic and/or syndromic human ichthyoses [1]. For the investigation of the causative mutations for human ichthyoses domestic animals have been shown to be useful model organisms. Ichthyosiform dermatoses have been described in cattle, mice, rats, dogs and pigs [5–15].
In dogs, several ichthyoses-associated mutations have been detected in specific breeds which revealed in most cases an autosomal recessive mode of inheritance [10–13, 15]. In Golden Retrievers lamellar ichthyosis was shown to be associated with a mutant PNPLA1 (patatin-like phospholipase domain containing 1) which was supposed to harbor potential causative mutations for human ichthyosis in European and North African populations as well [4, 10]. A similar type of lamellar ichthyosis with a generalized severe hyperkeratosis was detected in Jack Russell Terriers. A 1980-bp insertion in transglutaminase 1 (TGM1) could be shown to disrupt splicing of exon 10 and result in a premature stop codon and a significant decrease of mRNA expression [13, 16]. In Great Danes, a primary disorder of cornification has been detected in different litters as well. Patho-histological examinations indicated a congenital non-epidermolytic ichthyosis of a lamellar type [15]. A close relationship in-between affected dogs in pedigree analysis suggested an ichthyosis type resembling the autosomal recessive ichthyosis in humans.
The objective of this study was to perform a genome-wide association analysis and candidate gene study in order to elucidate the genetic background of ichthyosis in Great Danes and to identify the potential causative mutation.
Results
Phenotype
We examined 15 cases of Great Dane puppies affected with ichthyosiform dermatosis from five different litters. All affected dogs were Fédération Cynologique Internationale (FCI) breed standard Great Danes of the color type black/harlequin and were all distantly related (S1 Fig). The familial pattern of the 15 cases supports a recessive simple Mendelian mode of inheritance. Dominant and X-linked modes of inheritance can be excluded because parents were normal and both sexes were affected. All affected puppies shared one male ancestor.
Clinical examination revealed signs of a generalized severe hyperkeratosis in all cases with a formation of a strongly wrinkled, thickened and scaling skin especially in the region of the eyes and nose (Fig 1). These changes led to a dry inelastic and lichenified skin of an untidy appearance in the affected dogs and a markedly swollen periocular skin which impeded the opening of the puppy’s eyes in some cases. In-between the wrinkles the exudative character of the skin promoted secondary infections. Due to the poor prognosis, all affected dogs were euthanized at the age of 7–40 days. Additional computer tomographic and endoscopic examinations after euthanasia in two five week old affected dogs revealed a ventrally displaced auditory canal with an atypically wrinkled shape but no signs of other anomalies.
Fig 1. Clinical picture of ichthyosiform dermatosis in Great Danes.

(A) A five week old puppy with a characteristically wrinkled and squamous skin is shown. (B) In a one week old puppy prominent wrinkles in the face impede the opening of the dog’s eyes.
In addition to clinical examinations all affected dogs were carefully studied in necropsy [15]. The wrinkled skin showed a patho-histological feature of a marked epidermal and follicular hyperkeratosis (S2 Fig).
Genome-wide association analysis
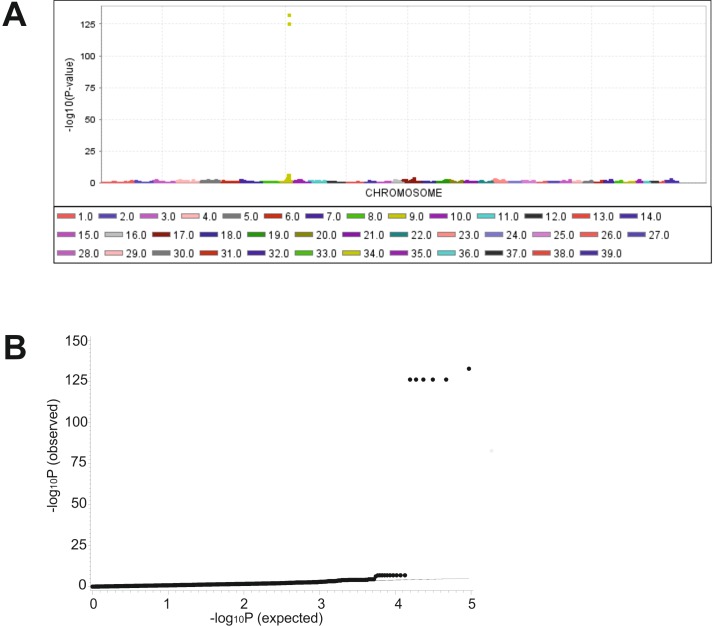
In order to map the locus underlying ichthyosiform dermatosis, a genome-wide association study (GWAS) was performed using 93,882 SNPs in nine affected and 13 unaffected Great Danes. A genome-wide significant association was detected on canine chromosome (CFA) 9 for a region of 740 kb from 54,031,320 to 54,772,131 bp on the dog genome assembly CanFam3.1 (Fig 2). The highest P-values reached six SNPs (P-value = 1e-126.3 to 1e-132.8) located within this region. The quantile-quantile (Q-Q) plots illustrated that inflation due to stratification effects had not increased P-values. After correction for cryptic relatedness, an association for the same genomic region was confirmed showing the highest peaks for these SNPs (P-values from 1e-88.1 to 1e-94.3). Removing of these most significant SNPs did not reveal a further genome-wide significantly associated region. All six associated SNPs within this genomic region showed a perfect co-segregation with the phenotype (S1 Table). The nine affected dogs were homozygous and the unaffected dogs were heterozygous (n = 10) or homozygous for the alternate allele (n = 3). All obligate carriers (parents of affected dogs, n = 3) were heterozygous. The minor allele frequency (MAF) of these highly associated SNPs was 0.38 for all Great Danes, 0.0 for affected and 0.36 for unaffected dogs. We used a candidate gene approach and investigated all 16 genes in the associated region and further 37 genes at a distance of 1 Mb from this associated region for known involvement in ichthyosis affection in mammals. Adjacent to this significantly associated region, SLC27A4 (Solute Carrier Family 27) is annotated at 55,164,817–55,177,741 bp (Ensembl, http://www.ensembl.org), approximately 393 kb downstream of this region.
Fig 2. Genome-wide association analysis (GWAS) for ichthyosis in Great Danes.
(A) Manhattan-plot of the -log10P-values from GWAS. A significant peak is located on canine chromosome (CFA) 9 in the region of 54,031,320–54,772,131 bp (CanFam3.1). (B) Q-Q plot of observed versus expected -log10P-values. Six SNPs show highly significant -log10P-values.
We analyzed the haplotype structure and haplotype association of this associated genomic region. The six highly associated SNPs are located within two different haplotype blocks (S3 Fig). Two SNPs (BICF2S23340470, BICF2G630473300) are located within a haplotype block at 54,010,938–54,144,992 bp of 134 kb, whereas the other four highly associated SNPs (BICF2G630473617, BICF2G630473695, BICF2G630473702, BICF2G630473744) can be found within a haplotype block at 54,426,765–54,772,131 bp of 343 kb length. Haplotypes with the disease-related alleles were significantly associated (P-value <0.0001) with the affection status.
Sequence and mutation analysis
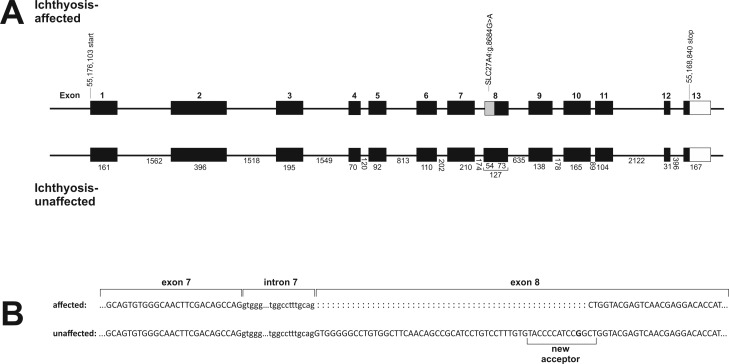
Sequence analysis of exons and exon/intron boundaries of SLC27A4 revealed two variants, the exonic SNV SLC27A4:g.8684G>A (ss1536958154) and the intronic indel SLC27A4:g.9852del (ss1536958155, Table 1). Functional analysis of the indel showed no significant reference to any modifying effect like a variation of acceptor sites that could be induced by this variant. However, the SNP SLC27A4:g.8684G>A in exon 8 was predicted to be a non-synonymous mutation resulting in a substitution of glutamine for arginine (p.Arg417Gln). The incorporation of a polar and neutral amino acid instead of a basic amino acid in this region was predicted to be deleterious with a probability of approximately 50% (subPSEC score = -2.5, substitution position-specific evolutionary conservation). Human splicing finder [17] tool suggested that this mutation within exon 8 created a new splice acceptor site motif ‘taccccatccagCT’, located 52 bp downstream of the authentic 3’ splice acceptor site (Fig 3).
Table 1. Variants detected by sequencing exons and intron boundaries of the genomic and complementary DNA (cDNA) of SLC27A4 in Great Danes.
| SNV name | Type | Base change | Predicted effect | Amino acid change |
|---|---|---|---|---|
| SLC27A4:g.8684G>A | SNV | G>A | non-synonymous | R>Q |
| SLC27A4:g.9852delC | Deletion | C | intron | N/A |
Fig 3. Experimental data derived from Sanger sequencing of SLC27A4 complementary DNA.
(A) Gene model of ichthyosis-affected in comparison to unaffected Great Danes. In the region of the non-synonymous SNV SLC27A4:g.8684G>A affected dogs show aberrantly spliced transcript with an in-frame loss of the first 54 bp of exon 8. (B) A new acceptor site is created by the A-allele of SLC27A4:g.8684G>A that results in a shorter RNA product. Affected dogs show only a small amount of RNA of the wild type size.
Sequencing of the complementary DNA (cDNA) of SLC27A4 revealed a loss of 54 bp from the beginning of exon 8 in affected dogs, and thus an exon 8 of 73 bp in length, whereas unaffected dogs showed an exon 8 of 127 bp in length as expected by the reference sequence. The differences in the size of exon 8 in-between affected and unaffected dogs could be significantly shown by PCR-products spanning the affected region (Fig 4). Obligate carriers revealed two different PCR-products. Nevertheless, some affected dogs also showed a very weak band of the wild type size. Placing an additional cDNA-primer directly into the 54 bp from the 5’ end of exon 8 confirmed that also low amounts of wild type transcript could be amplified in all affected dogs. Sequencing of this wild type PCR-product in affected dogs confirmed that it was identical with the wild type transcript in unaffected dogs. The experimental sequence showed an identity of 98.55% with Ensembl (ENSCAFT00000031925, transcript 2) and 100% with the NCBI (XM_548438.4) reference sequence. The predicted transcript 1 sequence of SLC27A4 provided from Ensembl (ENSCAFT00000049296) could not be amplified in any of the investigated hair root samples of affected and unaffected dogs. Nevertheless, NetStart 1.0 (http://www.cbs.dtu.dk/services/NetStart/) suggested a shorter open reading frame of SLC27A4 as expected by the reference sequence starting at position 169 (NCBI XM_548438.4).
Fig 4. Mutation analysis of SLC27A4 using complementary DNA.
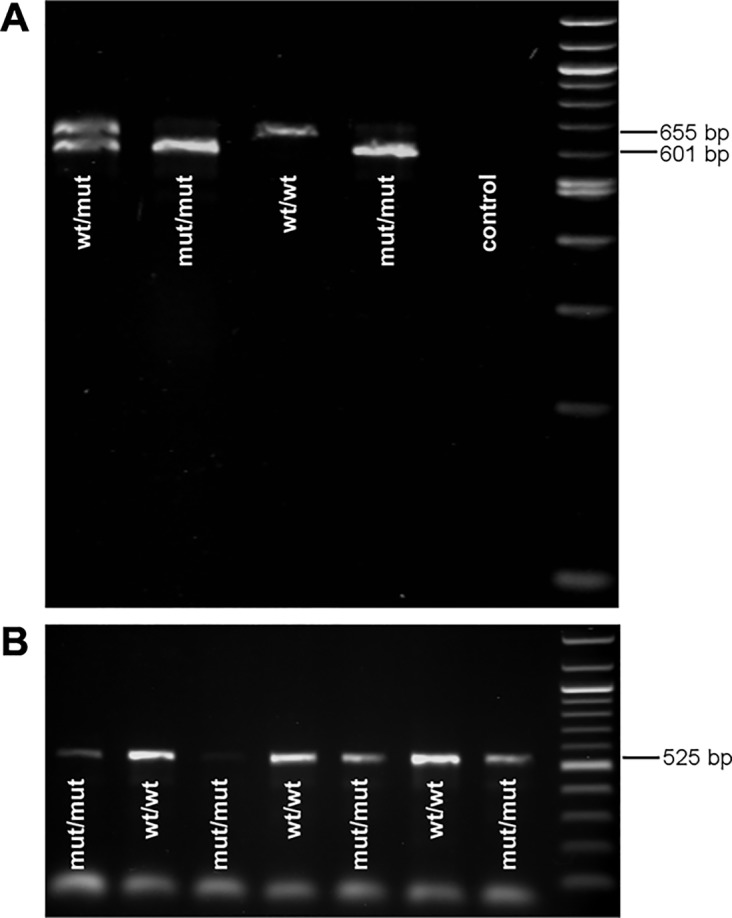
(A) A loss of 54 bp is present in affected Great Danes homozygous for allele A (mut/mut) in the SNV SLC27A4:g.8684G>A. The PCR-product spans 601 bp from exon 7 to 11 in affected dogs and 655 bp in unaffected dogs homozygous for the wild type (wt/wt) whereas heterozygous dogs show both products (wt/mut). A very small amount of wild type PCR-product (655 bp) can also be seen in affected dogs. (B) PCR-product of affected and unaffected Great Danes using a primer placed into the first 54 bp of exon 8. It confirms that affected dogs also have small amount of RNA of wild type size.
Protein expression
Protein analysis was performed to estimate the consequences of the 54 bp deletion and thus a loss of 18 amino acids (p.Val344_Arg361del) on SLC27A4 protein expression. Samples of skin tissue protein from affected dogs revealed either low or no SLC27A4 protein levels of expected wild type size (66.09 kDa) and very low levels of mutant protein (64.11 kDa) in western blot analysis. In unaffected dogs only wild type SLC27A4 protein could be shown to be expressed matching the experimental protein size of 66.09 kDa (Fig 5). Functional analysis of SLC27A4 using the protein sequence and classification tool InterPro suggested that the truncation leads to a size reduction of the AMP-dependent synthetase/ligase domain (IPR000873) [18, 19].
Fig 5. Western blot analysis of SLC27A4 in affected and unaffected Great Danes.

Wild type SLC27A4 was detected at 66.09 kDa and mutant SLC27A4 at 64.11 kDa using a polyclonal anti-SLC27A4 antibody corresponding to a region within amino acids 1 and 202 of the dog SLC27A4. Anti-Calnexin (CANX) polyclonal antibody was used as control. Affected Great Danes (A1-A3) show either low or no protein levels of mutant and wild type SLC27A4. In unaffected controls (C1-C2) only wild type protein is expressed.
Validation
Both mutations detected in SLC27A4 were genotyped in 28 Great Danes including the detection sample (n = 22) and further six Great Danes with patho-histologically confirmed ichthyosis. We found a perfect association of the SNV SLC27A4:g.8684G>A with the affected phenotype but no segregation of the phenotypes for the indel SLC27A4:g.9852del. Additional verification of SLC27A4:g.8684G>A in further 220 Great Danes, 413 controls of 35 different dog breeds and seven wolves confirmed the perfect association (Table 2) and a total absence of the disease-associated allele A in any other tested dog breed or wolves (S2 Table).
Table 2. Distribution of the SLC27A4:g.8684G>A genotypes in 249 Great Danes, 413 controls of other breeds, and seven wolves.
| Animals | G/G | G/A | A/A |
|---|---|---|---|
| Ichthyosis-affected (n = 15) | 0 | 0 | 15 |
| Obligate carriers (n = 5) | 0 | 5 | 0 |
| Unaffected relatives (n = 85) | 47 | 38 | 0 |
| Controls—Great Danes (n = 144) | 144 | 0 | 0 |
| Controls—other breeds (n = 413) | 413 | 0 | 0 |
| Controls—wolves (n = 7) | 7 | 0 | 0 |
Discussion
In this study a genome-wide association analysis for autosomal recessive ichthyosis in Great Danes showed a highly significant peak on chromosome 9 near SLC27A4. Sequencing of the genomic DNA of SLC27A4 revealed the exonic variant SLC27A4:g.8684G>A, 52 bp downstream of the beginning of exon 8, which was proposed to create a new and strong splice acceptor site, and thus an in-frame loss of 54 bp in the SLC27A4 transcript of ichthyosis-affected puppies. We found that all 15 ichthyosis-affected Great Danes were homozygous for SLC27A4:g.8684G>A mutant allele A whereas none of the genotyped 413 dogs of 35 different breeds and seven wolves showed a SNV allele different from the wild type. Affected dogs could be shown to express low or no levels of mutant truncated protein and very low or no levels of wild type SLC27A4 protein as well.
Functional analyses of SLC27A4 revealed this protein to be the major enzyme for activating very long chain fatty acid substrates [20]. SLC27A4 was shown to drive indirectly the uptake of fatty acids at the plasma membrane by catalyzing esterification from its location in the endoplasmatic reticulum [21]. Analyses of skin biopsies in ichthyosis patients suggested the mutant SLC27A4 protein to induce an uneven distribution of lipids and disturb the formation of the skin barrier [22]. The importance of SLC27A4 for skin and hair development was also demonstrated by the mouse model of wrinkle-free mice that revealed a defective skin barrier by a retrotransposon insertion in SLC27A4 [23]. Fatp4-/- mice showed a thick and extremely tight skin similar to the human restrictive dermopathy [23, 24]. In our study, we could show that a splice site modifying mutation and a subsequent lack or marked reduction of SLC27A4 wild type protein could also play a crucial role for skin developmental defects in dogs.
Modifying splice site mutations affecting SLC27A4 protein could also be observed in human. Human SLC27A4, the fatty acid transport protein 4 (FATP4), was shown to play a role in ichthyosis prematury syndrome [22, 25]. Sequence analyses of affected patients from North Africa, Middle East and Scandinavia revealed in total ten mutations in SLC27A4 that could not be found in healthy controls [22, 25]. Two of these mutations were predicted to result in splice site changes which in turn induce the truncation of SLC27A4 protein. Patients homozygous for p.C168X were even supposed to express no SLC27A4 protein showing no detectable band of SLC27A4 in western blot analysis [22]. Nevertheless, none of the mutations found in human were located in the homologous region near SLC27A4:g.8684G>A detected in Great Danes.
Our results of western blot analysis suggest that due to low transcription of wild type RNA in affected dogs, only a small amount or no stable intact SLC27A4 protein was produced. However, a high amount of mutant transcript with an in-frame loss of 54 bp could be verified in skin tissues in affected dogs. We propose that the strong decrease of protein levels was a result of degradation of the major proportion of the resulting shortened protein by the ubiquitin-proteasome pathway. Analyses in human gynecological cancer cells suggested in-frame insertions or deletions to have a strong impact on the stability of proteins [26]. Normal mRNA levels of AT-rich interactive domain 1A resulted in decreased protein levels due to increased proteasomal degradation. Quality-control mechanisms have been shown to be sensitive systems in eukaryotic cells which have been able to ensure quality of intracellular proteins and to degrade mutant proteins that could considerably disturb folding processes [27]. Defects in this sensitive system were proposed to result in various malfunctions in skin regulative processes. We suppose that the impact of the truncation in the AMP-dependent synthetase/ligase domain of SLC27A4 was markedly high and could have negatively influenced protein function. In human Dorfman-Chanarin syndrome, a truncation of CGI-58 protein could be shown to affect the lipase/esterase/thioesterase activity of CGI-58 and result in severe ichthyosis [28]. A lack of protein expression could also be detected in Norfolk Terriers affected with ichthyosis [12]. It was supposed that the mutation in the consensus donor splice site of intron 5 resulted in a nonsense-mediated decay of KRT10 transcripts. In lamellar ichthyosis-affected Jack Russell Terriers only a small amount of transglutaminase 1 (TGM1) protein could be detected on immunoblotting triggered by an insertion that disrupted the expression of TGM1 [13]. It can be assumed that a reduced SLC27A4 protein expression could result in analogous skin damaging effects in Great Danes.
In conclusion, we identified a SLC27A4 missense variant as the most likely causative mutation for ichthyosis in Great Danes. Together with data from Fatp4 transgenic mice our data confirm that a lack of SLC27A4 protein causes epidermal hyperplasia resulting from increased proliferation of suprabasal cells. Genetic testing of this mutation will open the opportunity for selective breeding to avoid mating among genetic carriers and producing ichthyosis-affected puppies.
Materials and Methods
Ethics statement
All animal work has been conducted according to the national and international guidelines for animal welfare. The Lower Saxony state veterinary office at the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany, was the responsible Institutional Animal Care and Use Committee (IACUC) for this specific study. The EDTA-blood sampling of the Great Danes for the present study had been approved by the IACUC of Lower Saxony, the state veterinary office Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany (registration number 33.9-42502-05-14A465). Tissue samples were taken immediately after euthanasia of the affected puppies. EDTA-blood samples of control breeds derived from the bio-bank for diagnostic purposes and were taken from accredited veterinarians.
Animals
For the GWAS, EDTA-blood samples of 22 Great Danes were employed. Validation of the mutations identified was performed in the detection sample (n = 22) and further six Great Danes with histo-pathologically confirmed ichthyosis. In addition, the SNV SLC27A4:g.8684G>A was validated in 221 EDTA-blood samples of unaffected Great Danes and 413 controls of 35 different breeds of dog and seven wolves (S2 Table). Skin tissues were collected from 6 affected Great Dane puppies that had to be euthanized due to the poor prognosis and two Great Dane controls euthanized due to other fatal diseases. All affected dogs were examined by veterinarians at the University of Veterinary Medicine Hannover. Euthanized animals were submitted for necropsy and histopathological examination to the University of Veterinary Medicine Hannover. Skin tissues from affected Great Danes and controls were fixed in 10% buffered formalin (pH 7.4) and embedded in paraffin. Sections of three μm were stained with hematoxylin and eosin (HE) [15].
Genotyping
Genomic DNA was extracted using 500 μl EDTA-blood and a standard ethanol fraction [29]. Samples of nine dogs representing four litters with ichthyosis-affected puppies were available for analysis (S1 Fig). Genotyping of these 9 affected and 13 unaffected Great Danes was done using the canine Illumina high density beadchip (Illumina). Quality criteria for analyses of the 173,662 SNPs genotyped were minor allele frequencies (MAF) >0.05, genotyping rate per SNP >0.90 and tests for Hardy-Weinberg equilibrium (P-value<0.000001). After filtering for quality criteria, 93,882 SNPs have been left for analysis. The mean genotyping rate per individual was 99 percent.
Raw and processed data is available for the 22 Great Danes with genotypes and can be obtained through the GEO website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE73400.
The variant SLC27A4:g.8684G>A identified within SLC27A4 was genotyped in 249 Great Danes and 413 controls of 35 different dog breeds and seven wolves using restriction fragment length polymorphisms according to a standard PCR- and digestion-protocol (S3 Table) [30]. The indel mutation SLC27A4:g.9852del was visualized using gel electrophoresis on 6% polyacrylamide denaturing gels (RotiphoreseGel 40, Carl Roth, Karlsruhe, Germany) and an automated capillary sequencer (LI-COR 4300, LI-COR Biotechnology, Bad Homburg, Germany).
Sequence analysis
The genomic regions of SLC27A4 exons and exon/ intron boundaries as well as the cDNA of SLC27A4 (473 bp 5’UTR to 477 bp 3’UTR) were sequenced in two ichthyosis-affected and two unaffected Great Danes. RNA was isolated from hair roots in stabilization RNALater reagent (Qiagen, Maryland, USA) and transcribed into cDNA according to standard protocols [30]. For complete DNA removal, RNA was additionally treated with DNase (RNase-Free DNase Set, Qiagen). Primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) and PCR was prepared according to a standard mastermix protocol [30] using Q-solution (Qiagen) as enhancer reagent (S4 Table). The PCR-reaction for cDNA and genomic DNA was run on a thermocycler TProfessional 96 (Biometra, Göttingen, Germany) according to the following protocol: 95°C for 5 minutes, 38 cycles of 94°C for 30 seconds, primer specific annealing temperature for 30 seconds and 72°C for 45 seconds. Finally, the reaction remained for 5 minutes at 72°C and cooled down to 4°C. PCR-products were cleaned up using Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase (Thermo Scientific, Darmstadt, Germany) according to the manufacturers’ protocol (http://www.thermoscientificbio.com/dna-and-rna-modifying-enzymes/exonuclease-i). Sequencing and alignments were performed by the automated sequencer Genetic Analyzer 3500 (Applied Biosystems by Life Technologies, Darmstadt, Germany) and the analysis software Sequencher 4.8 (Genes Codes, Ann Arbor, MI, USA).
Western blot analysis
For western blot analysis we isolated protein from skin tissues of three affected and two unaffected Great Danes using Ultra Thurax Homogenizer. Equal protein amounts were loaded and separated by 12% SDS-PAGE and blotted onto a polyvinylidene fluoride membrane (Roti-PVDF, Carl Roth). A polyclonal rabbit anti-FATP4 antibody (Thermo Scientific) at a 1:1000 dilution was used for hybridization of the membrane. Anti-Calnexin antibody (CANX, 1:1000, Sigma-Aldrich Chemie GmbH, Munich, Germany) served as control for a second hybridization of the same membrane. After removing the unbound primary antibody, the membrane was exposed to anti-rabbit antibody at 1:5000 dilution (goat anti-rabbit IgG, Dianova, Hamburg, Germany) and detected by enhanced chemiluminescence (SuperSignal West Femto Chemiluminescent Substrate, Thermo Scientific) on a Gel Doc XR+ system.
Bioinformatic analysis
Further estimation of the functional impact of the detected non-synonymous mutation on the protein was performed using Panther classification system (www.pantherdb.org) [31, 32] and Human Splicing Finder tool (version 2.4.1, http://www.umd.be/HSF) [17]. Sequence comparison data were obtained from Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo) [33]. Estimation of the open reading frame was done using NetStart 1.0 (http://www.cbs.dtu.dk/services/NetStart/). Functional analysis of the protein was performed using InterPro [18, 19] and ExPASy (http://web.expasy.org/compute_pi/).
Statistical analysis
The ALLELE procedure of SAS/Genetics, version 9.4 (SAS Institute, Cary, NC, USA) was used to calculate polymorphism information content, heterozygosity, allelic diversity, allele and genotype frequencies and χ2-tests for Hardy-Weinberg-Equilibrium for the genotyped SNPs. The genome-wide association analyses were performed using a generalized linear model. The analyses were run using TASSEL [34]. The model explained for sex effects and the SNP genotypes. Further models with the first two to five principal components (PC) as covariates had been run to test the robustness of the models. PCs were parameterized to capture cryptic population structure. PCs were derived from the quality filtered set of SNPs. The results for the extended models were consistent with the model omitting PCs. We applied the Bonferroni correction using the MULTIPLE TEST procedure of SAS to determine the threshold for genome-wide significance. Quantile-quantile (Q-Q) plots for observed versus expected —log10P-values to be used for control of population stratification were calculated using SAS.
Distribution of genotypes was calculated using the FREQ procedure of SAS. Genotypic and allelic associations and allelic trends as well as odds ratios (ORs) with their 95% confidence intervals were calculated using the CASECONTROL procedure of SAS/Genetics. We analysed the haplotype structure of the identified genomic regions using Haploview 4.0 [35].
Supporting Information
The affected dogs were derived from five different litters.
(TIF)
Skin sections from an unaffected neonatal puppy (A) and from a 28-days old puppy affected with ichthyosis (B). The skin of the affected puppy is characterized by marked epidermal and follicular hyperkeratosis (Hematoxylin and eosin staining, 100 x).
(TIF)
The LD coefficients (r2) between the markers are shown. Red fields display r² values greater than 0.50, white and blue fields display r² values less than 0.15. Two associated SNPs (BICF2S23340470, BICF2G630473300) can be found in a 134 kb haplotype block at 54 Mb and further four associated SNPs (BICF2G630473617, BICF2G630473695, BICF2G630473702, BICF2G630473744) are located in a haplotype block at 54,4–55 Mb of 343 kb length.
(TIF)
All six SNPs (BICF2S23340470, BICF2G630473300, BICF2G630473617, BICF2G630473695, BICF2G630473702 and BICF2G630473744) show a perfect co-segregation with the phenotype.
(DOCX)
The number of dogs and wolves genotyped for the SNV SLC27A4:g.8684G>A of the SLC27A4 gene and the frequency for the mutant A allele are shown. None of these control samples showed a genotype different from wild type.
(DOCX)
SNV SLC27A4:g.8684G>A was validated by the use of restriction fragment length polymorphism (RFLP) whereas the deletion SLC27A4:g.9852del could be investigated by gel-electrophoresis using the LI-COR automated sequencing system. Primer pairs, amplicon size (AS) in base pairs (bp), annealing (AT), restriction enzyme and incubation temperature (IT) are given.
(DOCX)
Target regions, product sizes in base pairs (bp) and annealing temperatures (AT) are shown.
(DOCX)
Acknowledgments
We are grateful to all dog breeders donating us samples from their dogs and sharing pedigree data of their dogs. We thank especially Dr. M. Peters, Hela Jark, Marion Rawert and Dr. Horst Hollensteiner for their support in collecting samples. We acknowledge expert technical support by Jörn Wrede, Heike Klippert-Hasberg, Stefan Neander, Mogens Drabert and Stephanie Geveke.
Data Availability
Raw and processed data is available for the 22 Great Danes with genotypes and can be obtained through the GEO website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE73400.
Funding Statement
The authors have no support or funding to report.
References
- 1. Oji V, Tadini G, Akiyama M, Blanchet Bardon C, Bodemer C, Bourrat E, et al. Revised nomenclature and classification of inherited ichthyoses: results of the first ichthyosis consensus conference in Soreze 2009. J Am Acad Dermatol. 2010;63(4):607–41. 10.1016/j.jaad.2009.11.020 . [DOI] [PubMed] [Google Scholar]
- 2. Mecklenburg L, Hetzel U, Ueberschar S. Epidermolytic ichthyosis in a dog: clinical, histopathological, immunohistochemical and ultrastructural findings. J Comp Pathol. 2000;122(4):307–11. 10.1053/jcpa.1999.0371 . [DOI] [PubMed] [Google Scholar]
- 3. Dahlqvist J, Klar J, Hausser I, Anton-Lamprecht I, Pigg MH, Gedde-Dahl T Jr., et al. Congenital ichthyosis: mutations in ichthyin are associated with specific structural abnormalities in the granular layer of epidermis. J Med Genet. 2007;44(10):615–20. 10.1136/jmg.2007.050542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fachal L, Rodríguez Pazos L, Ginarte M, Carracedo A, Toribio J, Vega A. Identification of a novel PNPLA1 mutation in a Spanish family with autosomal recessive congenital ichthyosis. Br J Dermatol. 2014;170(4):980–2. 10.1111/bjd.12757 [DOI] [PubMed] [Google Scholar]
- 5. Dardano S, Gandolfi B, Parma P, Polli M, Bighignoli B, Strillacci MG, et al. Characterization of bovine TGM1 and exclusion as candidate gene for ichthyosis in Chianina. J Hered. 2008;99(1):81–3. 10.1093/jhered/esm101 [DOI] [PubMed] [Google Scholar]
- 6. Molteni L, Dardano S, Parma P, Polli M, De Giovanni A, Sironi G, et al. Ichthyosis in Chianina cattle. Vet Rec. 2006;158(12):412 [DOI] [PubMed] [Google Scholar]
- 7. Carter T, Phillips RS. Ichthyosis, a new recessive mutant in the house mouse. J Hered. 1950;41:297–300. [DOI] [PubMed] [Google Scholar]
- 8. Knox WE, Lister-Rosenoer LM. Infantile ichthyosis in rats: a new model of hyperkeratotic skin disease. J Hered. 1978;69(6):391–4. [DOI] [PubMed] [Google Scholar]
- 9. O’Goshi K-I, Tabata N, Sato Y, Tagami H. Comparative study of the efficacy of various moisturizers on the skin of the ASR miniature swine. Skin Pharmacol Physiol. 2000;13(2):120–7. [DOI] [PubMed] [Google Scholar]
- 10. Grall A, Guaguere E, Planchais S, Grond S, Bourrat E, Hausser I, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44(2):140–7. 10.1038/ng.1056 . [DOI] [PubMed] [Google Scholar]
- 11. Mauldin EA, Wang P, Evans E, Cantner CA, Ferracone JD, Credille KM, et al. Autosomal Recessive Congenital Ichthyosis in American Bulldogs Is Associated With NIPAL4 (ICHTHYIN) Deficiency. Vet Pathol. 2014. 10.1177/0300985814551425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Credille KM, Barnhart KF, Minor JS, Dunstan RW. Mild recessive epidermolytic hyperkeratosis associated with a novel keratin 10 donor splice-site mutation in a family of Norfolk terrier dogs. Br J Dermatol. 2005;153(1):51–8. 10.1111/j.1365-2133.2005.06735.x . [DOI] [PubMed] [Google Scholar]
- 13. Credille K, Minor J, Barnhart K, Lee E, Cox M, Tucker K, et al. Transglutaminase 1-deficient recessive lamellar ichthyosis associated with a LINE-1 insertion in Jack Russell terrier dogs. Br J Dermatol. 2009;161(2):265–72. 10.1111/j.1365-2133.2009.09161.x [DOI] [PubMed] [Google Scholar]
- 14. Forman OP, Penderis J, Hartley C, Hayward LJ, Ricketts SL, Mellersh CS. Parallel mapping and simultaneous sequencing reveals deletions in BCAN and FAM83H associated with discrete inherited disorders in a domestic dog breed. PLoS Genet. 2012;8(1):e1002462 10.1371/journal.pgen.1002462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffmann A, Metzger J, Wohlke A, Peters M, Junginger J, Mischke R, et al. Congenital Ichthyosis in 14 Great Dane Puppies With a New Presentation. Vet Pathol. 2015. 10.1177/0300985815595516 . [DOI] [PubMed] [Google Scholar]
- 16. Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, et al. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet. 1995;9(3):279–83. [DOI] [PubMed] [Google Scholar]
- 17. Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67 Epub 2009/04/03. gkp215 [pii] 10.1093/nar/gkp215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40(Database issue):D306–12. 10.1093/nar/gkr948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jia Z, Moulson CL, Pei Z, Miner JH, Watkins PA. Fatty acid transport protein 4 is the principal very long chain fatty acyl-CoA synthetase in skin fibroblasts. J Biol Chem. 2007;282(28):20573–83. 10.1074/jbc.M700568200 . [DOI] [PubMed] [Google Scholar]
- 21. Milger K, Herrmann T, Becker C, Gotthardt D, Zickwolf J, Ehehalt R, et al. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J Cell Sci. 2006;119(22):4678–88. [DOI] [PubMed] [Google Scholar]
- 22. Klar J, Schweiger M, Zimmerman R, Zechner R, Li H, Torma H, et al. Mutations in the fatty acid transport protein 4 gene cause the ichthyosis prematurity syndrome. Am J Hum Genet. 2009;85(2):248–53. 10.1016/j.ajhg.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moulson CL, Martin DR, Lugus JJ, Schaffer JE, Lind AC, Miner JH. Cloning of wrinkle-free, a previously uncharacterized mouse mutation, reveals crucial roles for fatty acid transport protein 4 in skin and hair development. Proc Natl Acad Sci U S A. 2003;100(9):5274–9. 10.1073/pnas.0431186100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moulson CL, Lin MH, White JM, Newberry EP, Davidson NO, Miner JH. Keratinocyte-specific expression of fatty acid transport protein 4 rescues the wrinkle-free phenotype in Slc27a4/Fatp4 mutant mice. J Biol Chem. 2007;282(21):15912–20. 10.1074/jbc.M701779200 . [DOI] [PubMed] [Google Scholar]
- 25. Sobol M, Dahl N, Klar J. FATP4 missense and nonsense mutations cause similar features in Ichthyosis Prematurity Syndrome. BMC Res Notes. 2011;4:90 10.1186/1756-0500-4-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guan B, Gao M, Wu C-H, Wang T-L, Shih I-M. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia. 2012;14(10):986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–9. 10.1038/nature02263 . [DOI] [PubMed] [Google Scholar]
- 28. Akiyama M, Sawamura D, Nomura Y, Sugawara M, Shimizu H. Truncation of CGI-58 protein causes malformation of lamellar granules resulting in ichthyosis in Dorfman-Chanarin syndrome. J Invest Dermatol. 2003;121(5):1029–34. [DOI] [PubMed] [Google Scholar]
- 29. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215 Epub 1988/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Metzger J, Schrimpf R, Philipp U, Distl O. Expression Levels of LCORL Are Associated with Body Size in Horses. PLoS ONE. 2013;8(2): e56497 10.1371/journal.pone.0056497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci U S A. 2004;101(43):15398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007; 23:2633–5. [DOI] [PubMed] [Google Scholar]
- 35. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The affected dogs were derived from five different litters.
(TIF)
Skin sections from an unaffected neonatal puppy (A) and from a 28-days old puppy affected with ichthyosis (B). The skin of the affected puppy is characterized by marked epidermal and follicular hyperkeratosis (Hematoxylin and eosin staining, 100 x).
(TIF)
The LD coefficients (r2) between the markers are shown. Red fields display r² values greater than 0.50, white and blue fields display r² values less than 0.15. Two associated SNPs (BICF2S23340470, BICF2G630473300) can be found in a 134 kb haplotype block at 54 Mb and further four associated SNPs (BICF2G630473617, BICF2G630473695, BICF2G630473702, BICF2G630473744) are located in a haplotype block at 54,4–55 Mb of 343 kb length.
(TIF)
All six SNPs (BICF2S23340470, BICF2G630473300, BICF2G630473617, BICF2G630473695, BICF2G630473702 and BICF2G630473744) show a perfect co-segregation with the phenotype.
(DOCX)
The number of dogs and wolves genotyped for the SNV SLC27A4:g.8684G>A of the SLC27A4 gene and the frequency for the mutant A allele are shown. None of these control samples showed a genotype different from wild type.
(DOCX)
SNV SLC27A4:g.8684G>A was validated by the use of restriction fragment length polymorphism (RFLP) whereas the deletion SLC27A4:g.9852del could be investigated by gel-electrophoresis using the LI-COR automated sequencing system. Primer pairs, amplicon size (AS) in base pairs (bp), annealing (AT), restriction enzyme and incubation temperature (IT) are given.
(DOCX)
Target regions, product sizes in base pairs (bp) and annealing temperatures (AT) are shown.
(DOCX)
Data Availability Statement
Raw and processed data is available for the 22 Great Danes with genotypes and can be obtained through the GEO website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE73400.