Dear Editor,
The ubiquitin specific protease 7 (USP7), also known as herpes virus associated ubiquitin specific protease (HAUSP), is a well-characterized deubiquitinase (Reyes-Turcu et al., 2009). USP7 plays important roles in various biological processes, including cell survival, proliferation, apoptosis, tumorigenesis, viral infection, and epigenetic regulation through regulating the protein stability of tumor suppressors (p53, PTEN, FOXO, claspin), E3 ligases (MDM2, Mule, viral proteins ICP0), epigenetic regulators (DNMT1, Tip60, UHRF1) (Pfoh et al., 2015). USP7 is comprised of three recognizable domains: the N-terminal TRAF domain, the catalytic domain, and the C-terminal Tandem UBL domain (designated TUDUSP7) (Fig. 1A). Previous study shows that the TRAF domain binds to P/AxxS motif (from p53 and MDM2) and is responsible for substrate recognition (Sheng et al., 2006). However, how USP7 recognizes other substrates remains largely unknown.
Figure 1.
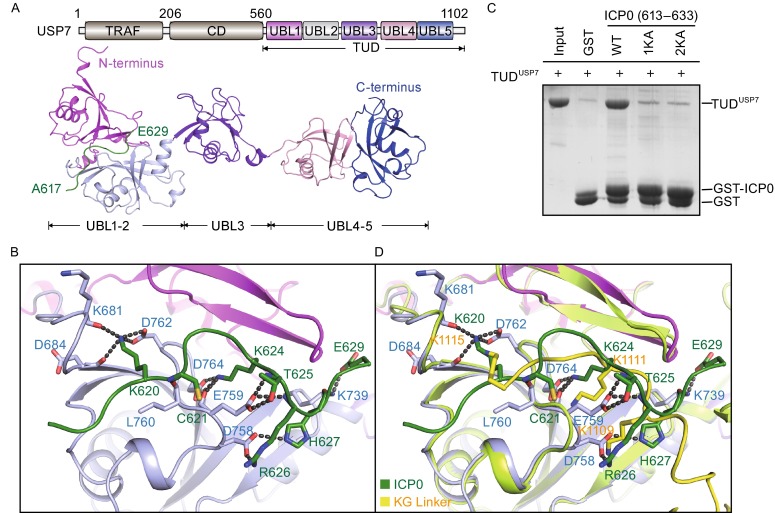
Crystal structure of the USP7-ICP0 complex. (A) Color-coded domain architecture of human USP7 (top) and ribbon representations of the overall structure of the TUDUSP7-ICP0 complex (bottom). The color scheme is used in all structural figures. The ICP0 peptide is colored in forest. The N- and C-termini of USP7 and ICP0 are indicated. (B) Closed-up view of the intermolecular interaction. USP7 and ICP0 are shown in ribbon representations. Critical residues for the interaction are shown in stick representation. Hydrogen bonds are shown as dashed lines. (C) GST pull-down assay for the interaction between TUDUSP7 and ICP0. Wild type and mutants of GST-tagged ICP0 (613–633) were incubated with TUDUSP7 and immobilized on glutathione resin. The bound proteins were subjected to SDS-PAGE and visualized by Coomassie blue staining. The free GST is resulted from protein purification and would not affect the results. (D) Comparison of TUDUSP7-ICP0 and USP7-DNMT1 structures. In USP7-DNMT1 structure, USP7 is colored in lemon and the KG-Linker in yellow. Critical residues for the interactions are shown in stick representation. Hydrogen bonds are shown as dashed lines
Previous study shows that TUDUSP7 is important for the regulation of USP7 enzymatic activity (Faesen et al., 2011). We have previously showed that USP7 stabilizes UHRF1 through the interaction between TUDUSP7 and a Spacer region between the SRA and RING domain of UHRF1 (Ma et al., 2012). Our recent study indicates that USP7 binds to and stabilizes DNMT1 through the interaction between an acidic pocket of TUDUSP7 and the KG repeat region (KG-Linker) of DNMT1. We therefore speculate that TUDUSP7 may function as a binding site for some other USP7-binding proteins or substrates. To test this hypothesis, we first investigated the interaction between USP7 and the Herpes simplex virus type 1 (HSV1) regulatory protein ICP0 (also known as Vmw110), which is the first identified USP7-interacting protein and regulates the lytic and latent viral infection through inducing proteasomal degradation of various cellular proteins (Boutell and Everett, 2013).
It has been shown that ICP0 peptide (residues 594–633) interacts with the TUD of USP7 (Everett et al., 1999). To investigate how ICP0 binds to TUDUSP7, we determined the crystal structure of TUDUSP7 in complex with ICP0 (613–633) at 2.70 Å resolution (Fig. 1A, Table S1). In the TUDUSP7-ICP0 complex structure, the TUDUSP7 adopts an extended conformation, and shares similar fold with the TUDUSP7 alone (2YLM.PDB) (Faesen et al., 2011) (Fig. S1). As observed in previous studies (Faesen et al., 2011; Cheng et al., 2015), UBL1–2, UBL3, and UBL4–5 of TUDUSP7 form three separate modules and are connected by flexible regions flanking the UBL3. The ICP0 packs against an acidic groove on the surface of UBL1–2. Most of the residues involved in intermolecular interaction are well covered by the electron density, indicating the model was correctly built (Fig. S2).
As indicated in the TUDUSP7-ICP0 structure, the interaction is mediated by a network of hydrogen bonds and salt bridge contacts. Residue K620ICP0 forms hydrogen bonds with the carbonyl oxygen atoms of residues K681USP7, D684USP7 and side chain of D762USP7. Residue K624ICP0 snugly inserts into an acidic cavity, with the side chain stabilized by residues E759USP7 and D764USP7. Hydroxyl group of T625ICP0 forms hydrogen bond with E759USP7. Residues R626ICP0 and H627ICP0 form hydrogen bonds with side chain of residue D758USP7. The carboxyl group of residue E629 interacts with the side chain of K739USP7. The main chains of ICP0 (residues C621, T625, and R626) form hydrogen bonds with residue K739USP7, L760USP7, and E759USP7 (Fig. 1B). Consistent with the observation from the structural analyses, mutations K624AICP0 (1KA) and K620A/K624AICP0 (2KA) of ICP0, largely decreased the binding affinities to TUDUSP7, compared with the wild-type ICP0 (Fig. 1C), supporting the key role of these residues in mediating the intermolecular interaction.
Our previous study indicates that the KG-Linker of DNMT1 binds to TUDUSP7 and plays an important role in mediating DNMT1-USP7 interaction and USP7-mediated stabilization of DNMT1. Structural comparison of USP7-DNMT1 and USP7-ICP0 indicates that ICP0 and KG-LinkerDNMT1 pack against the same acidic pocket of TUDUSP7 in an opposite orientation (Cheng et al., 2015). As shown in Fig. 1D, residues K624ICP0 and K1111DNMT1 both bind to the acidic cavity formed by residues E736USP7, D758USP7, E759USP7, and D764USP7, and residues K620ICP0 and K1115DNMT1 form hydrogen bonds with D684USP7 and D762USP7. In support of above observation, mutation D758A/E759A/E764A of TUDUSP7 (designated M1USP7-TUD) largely reduced the binding affinity to ICP0 and DNMT1 peptides in GST pull down experiment (Fig. 2A). The above analyses suggest that TUDUSP7 may recognize consensus primary sequence containing KxxxK and therefore the acidic pocket of TUDUSP7 serves as another binding site (besides the TRAF) for recognition of USP7-interacting proteins.
Figure 2.
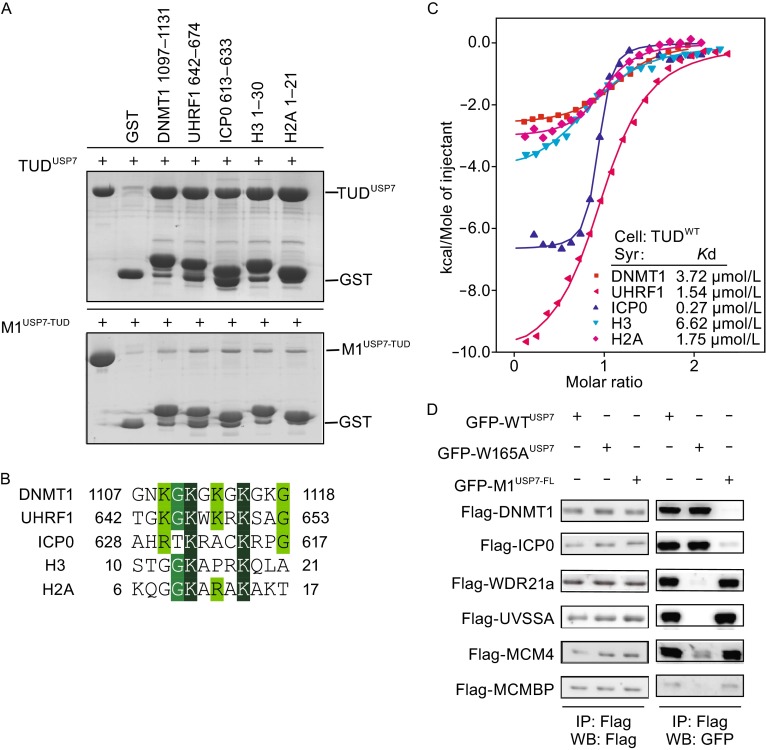
TUD is a new substrate-binding domain. (A) DNMT1, UHRF1, ICP0, histone H3 and H2A bind to the same acidic pocket of TUDUSP7. The GST pull down assay were performed as in Fig. 1C. (B) The alignment of the corresponding sequences of DNMT1, UHRF1, ICP0, H3, and H2A. Identical and highly conserved residues are highlighted in dark green and conserved residues in light green. This indicated that all these protein have the conserved KxxxK motif. (C) ITC measurement of the interactions between TUDUSP7 and various peptides. (D) The interactions between full-length USP7 and its binding proteins. HEK293T cells were transiently co-transfected with FLAG-tagged proteins and GFP-tagged USP7 (wild-type and mutants) followed by immunoprecipitation. The proteins were detected by immunoblotting using the indicated antibodies
To test the hypothesis, we searched human proteins database and found a number of candidates (including UHRF1, histone H3, histone H2A) contain such consensus motif and potentially bind to TUDUSP7 (Fig. 2B). We therefore used TUDUSP7 and M1USP7-TUD (defective binding to KxxxK motif) to investigate that whether these proteins bind to USP7 on the same site. As shown in the GST pull down assays, wild type TUDUSP7 binds to the KG-Linker of DNMT1, ICP0 (613–633), UHRF1 (642–674), histone H3 (1–30), and histone H2A (1–21) (Fig. 2A). We also confirmed the interaction using the ITC measurement (Fig. 2C). In contrast, mutation of the residues within the acidic pocket (M1USP7-TUD) largely decreased the binding affinity to all the five peptides (Fig. 2A). In support of this observation, USP7 has been shown to bind to and stabilize UHRF1 and this interaction is regulated by phosphorylation of UHRF1 during cell cycle (Ma et al., 2012). However, whether histones H3 and H2A are regulated by USP7 in vivo needs further investigation.
USP7 is one of the best-characterized deubiquitinases (Nicholson and Suresh Kumar, 2011). Previous studies have identified a number of USP7-binding proteins, including several substrates for deubiquitination (Sowa et al., 2009). Mutation W165AUSP7 on the TRAF domain has been shown to impair its interaction with p53 and MDM2 (Sheng et al., 2006). We next verified the interaction between USP7 and its binding proteins in cells using co-immunoprecipitation to investigate which site (TRAF or the acidic pocket on TUDUSP7) on USP7 is responsible for the interaction with distinct binding proteins. Consistent with above analyses, M1USP7-FL abolished its interaction with DNMT1 and ICP0 (Fig. 2D). In contrast, WDR21a, UVSSA, MCMBP, and MCM4 showed comparable USP7-binding affinity to M1USP7-FL, but impaired the interaction with W165AUSP7 (Fig. 2D), indicating that these proteins mainly bind to USP7 on the TRAF domain. Taken together, USP7 binds to distinct proteins on two separate sites, TRAF and the acidic pocket on TUDUSP7, respectively.
Intriguingly, WDR21a, UVSSA, MCMBP, MCM4, p53, and MDM2 (binds to TRAF domain) are involved in cell cycle, DNA repair or DNA replication, whereas DNMT1 and UHRF1 (binds to TUDUSP7) are involved in the maintenance of DNA methylation. Thus, our study indicates that the existence of two substrate-binding sties allows USP7 to recognize distinct groups of proteins/substrates for the regulations of different biological pathways. The proteins that bind to the same substrate-binding site may compete with each other for regulation if they exist in the same cellular compartment. Our study also provides a structural basis for designing inhibitors of USP7 that specifically regulates the maintenance of DNA methylation.
Electronic supplementary material
Acknowledgments
FOOTNOTES
We thank staff members of beamline BL17U and BL19U at SSRF for assistance in data collection, and staff members of Biomedical Core Facility, Fudan University for their help on biochemical analyses. This work was supported by grants from the National Basic Research Program of China (No. 2011CB965300), the National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” of China (2014ZX09507-002), the National Natural Science Foundation of China (Grant Nos. 31425008, 31270779, 31100526, 31000325), Basic Research Project of Shanghai Science and Technology Commission (12JC1402700), the Program of Shanghai Subject Chief Scientist (14XD1400500), Shanghai Pujiang program (13PJD006) and Shanghai Municipal Commission of Health and Family Planning (20144Y0103).
The atomic coordinate and structure factor have been deposited in the Protein Date Bank with the accession code 5C56.
Jingdong Cheng, Ze li, Rui Gong, Jian Fang, Yi Yang, Chang Sun, Huirong Yang, and Yanhui Xu declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by the any of the authors.
Footnotes
Jingdong Cheng and Ze Li contributed equally to this work.
References
- Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, Li Z, Xu Y. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat Commun. 2015;6:7023. doi: 10.1038/ncomms8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci USA. 2012;109:4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- Pfoh R, Lacdao IK, Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr Relat Cancer. 2015;22:T35–54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.