Abstract
Introduction
Keratoconus (KC) is a non-inflammatory thinning of the cornea and a leading indication for corneal transplantation. Oxidative stress plays a role in the pathogenesis of this disease. The products of the hOGG1 and MUTYH genes play an important role in the repair of oxidatively modified DNA in the base excision repair pathway. We hypothesized that variability in these genes may change susceptibility to oxidative stress and predispose individuals to the development of KC. We investigated the possible association between the c.977C>G polymorphism of the hOGG1 gene (rs1052133) and the c.972G>C polymorphism of the MUTYH gene (rs3219489) and KC occurrence as well as the modulation of this association by some KC risk factors.
Material and methods
A total of 205 patients with KC and 220 controls were included in this study. The polymorphisms were genotyped with polymerase chain reaction (PCR) restriction fragment length polymorphism and PCR-confronting two-pair primer techniques. Differences in genotype and allele frequency distributions were evaluated using the χ2 test, and KC risk was estimated with an unconditional multiple logistic regression with and without adjustment for co-occurrence of visual impairment, allergies, sex and family history for KC.
Results
We did not find any association between the genotypes and combined genotypes of the c.977C>G polymorphism of the hOGG1 gene and the c.972G>C polymorphism of the MUTYH gene and the occurrence of KC.
Conclusions
Our findings suggest that the c.977C>G-hOGG1 polymorphism and the c.972G>C-MUTYH polymorphism may not be linked with KC occurrence in this Polish subpopulation.
Keywords: oxidative stress, genetic variability, DNA glycosylases, oxidative DNA damage, DNA repair
Introduction
Keratoconus (KC; MIM #148300) is a progressive, non-inflammatory disease of the cornea that usually develops in adolescence and can lead to severe visual impairment or blindness. In about 20% of cases, it progresses to the point of legal blindness and then can be treated only by corneal transplantation [1–3]. To date, KC is a major cause of cornea transplantation in Western countries [4, 5]. Keratoconus usually presents in the second decade of life and progresses into the third and fourth decades, with decreasing visual function [3–6]. The estimated prevalence of this disease is ~50–230 per 100 000, depending on the clinical detection methods and diagnostic criteria [3]. Keratoconus is observed in all ethnic groups, with no female or male predominance [7]. It was found that the incidence of KC in Asians was 25 per 100 000 (1 per 4000) per year, compared with 3.3 per 100 000 (1 in 30 000) per year in Caucasians [8].
Although the aetiology of KC remains unclear, a growing body of evidence suggests that both genetic and environmental factors and interaction between them are responsible for the disease induction and progression. It was shown that environmental factors include contact lens wear, chronic eye rubbing and atopy of the eye [9, 10]. Genetic factors include familial inheritance, discordance between dizygotic twins and KC association with other genetic disorders. The prevalence of KC in first degree relatives is 3.34%, which is 15–67 times higher than that of general population [11]. Keratoconus is found in 0.5–15% of patients with Down syndrome, and was also reported in patients with Leber's congenital amaurosis, Ehlers-Danlos syndrome and osteogenesis imperfecta [12–14]. In addition, KC occurs with a higher concordance rate of the trait in monozygotic twins than in dizygotic ones [15]. A number of gene loci/chromosomal regions for familial KC have been described [16–19]. Taken together, these data suggest that KC is a multifactorial disease.
Results of several studies indicate that oxidative stress may play a role in the development of KC [20–26]. The cornea is exposed to a wide spectrum of light, including ultraviolet (UV) radiation. UV exposure is a well-characterized environmental stress factor that generates reactive oxygen species (ROS), including free radicals [27]. As the cornea absorbs most of the UV reaching the eye, it would be especially susceptible to ROS-induced damage. In the healthy cornea, several defence mechanisms are present to minimize such damage [23]. In general, the cornea has antioxidant enzymes, including superoxide dismutase, glutathione reductase and glutathione peroxidase, that neutralize ROS before they damage cells [28]. It was demonstrated that KC corneas displayed altered activity of antioxidant enzymes [20, 21], accumulation of cytotoxic ROS [23] and mitochondrial DNA damage [29]. Oxidation may involve various biomolecules – membrane phospholipids, proteins and DNA [30] – and has been implicated in a wide range of pathological conditions, including premature aging and systemic diseases, such as cancer, neurodegenerative and ocular diseases [31–36]. Reactive oxygen species can cause a variety of DNA damage, including single and double strand breaks (SSBs and DSBs, respectively) and DNA base modifications; therefore efficient repair of ROS-induced DNA damage is important for preventing mutations and maintaining the stability of the genome [37]. One of the most commonly produced DNA base lesions, frequently applied as a hallmark of oxidative DNA damage, is 8-oxoguanine (8-OH-G) [38]. About one thousand 8-OH-Gs are estimated to be generated in normal human cells per day [39]. 8-OH-G can pair with adenine, instead of cytosine, during DNA replication. If this mispairing is not repaired, it leads to a G:C to T:A transversion mutation [40]. In mammalian cells two base excision repair (BER) DNA glycosylases, hOGG1 and MUTYH, initiate the repair of such lesions – hOGG1 removes 8-OH-G and MUTYH removes adenine paired with 8-OH-G, leaving an apurinic site in the DNA.
In recent years, the role of genetic factors in KC has been extensively investigated. Several case-control studies have confirmed the association between single nucleotide polymorphisms (SNPs)/mutations and this disease [41–46]. In the present study we assessed the association between polymorphism of two BER genes, MUTYH (c.972G>C; rs3219489) and hOGG1 (c.977C>G; rs1052133), and KC occurrence as well as modulation of this association by some demographic and risk factors for KC. Both polymorphisms are located in the coding regions of their genes, resulting in amino acid substitutions. The c.977G>C polymorphism causes change of serine to cysteine at position 326 (Ser326Cys) of the hOGG1 protein, whereas the c.972C>G polymorphism is associated with substitution of glutamine to histidine at position 324 (Gln324His) of the MUTYH protein. These SNPs may exert a functional effect according to results of association and/or in vitro studies [47–55].
Material and methods
Ethics
The study design was approved by the Bioethics Committee of the Medical University of Warsaw and each patient and control individual gave a written informed consent and approval form for genetic analysis.
Patients and controls
A total of 205 patients with KC and 220 individuals with normal corneas (controls) were enrolled in this study. All patients and controls were examined in the Department of Ophthalmology, Medical University of Warsaw (Warsaw, Poland). They underwent ophthalmic examination, including best-corrected visual acuity, intraocular pressure, slit lamp examination, fundus examination, corneal topography (TMS4, Tomey, Nagoya, Japan), and Orbscan corneal topographical and pachymetric maps (Orbscan IIz, Bausch & Lomb, USA). The diagnosis of KC was based on clinical signs and topographical and pachymetric parameters on Topographic Modeling System (TMS) topography and Orbscan examinations [3, 56, 57]. The map patterns were carefully interpreted manually in all cases. The patients with an unmistakable diagnosis of KC were included in the study group, and the patients with normal corneal topography and pachymetry were included in the control group. Medical history was obtained from all subjects, and no one reported any genetic disease.
After informed consent, 5 ml of venous blood from participants was collected into EDTA tubes, coded and stored at –20°C until further use. In addition, all subjects were interviewed using a structural questionnaire to determine demographic and potential risk factors for KC. Study subjects and controls provided information on their age, lifestyle habits, including smoking, body mass index (BMI), allergy, co-occurrence of visual impairment (hyperopia, astigmatism, myopia) and family history among 1st degree relatives for KC. Smoking was categorized as current, former or never smokers. The validity and reliability of the questionnaires were checked whenever possible. Characteristics of patients and controls are presented in Table I. All individuals employed in our research were unrelated and were of Polish nationality, belonging to the Polish ethnic group.
Table I.
Characteristics of KC patients and controls
| Feature | Controls (n = 220) | KC (n = 205) | Value of p | ||
|---|---|---|---|---|---|
| Number | Frequency | Number | Frequency | ||
| Gender: | |||||
| Females | 129 | 0.59 | 64 | 0.31 | < 0.001 |
| Males | 91 | 0.41 | 141 | 0.69 | |
| Age: | |||||
| Mean ± SD | 60.71 ±20.25 | 36.39 ±11.87 | < 0.001* | ||
| Range | 19–91 | 14–63 | |||
| Smoking: | |||||
| Current/former | 68 | 0.31 | 67 | 0.33 | 0.7732 |
| Never | 152 | 0.69 | 138 | 0.67 | |
| KC in family: | |||||
| Yes | 9 | 0.04 | 22 | 0.11 | 0.0145 |
| No | 21 | 0.96 | 183 | 0.89 | |
| BMI [kg/m2]: | |||||
| ≤ 25 | 94 | 0.44 | 92 | 0.45 | 0.9227 |
| 25–30 | 69 | 0.32 | 66 | 0.32 | |
| ≥ 30 | 53 | 0.25 | 47 | 0.23 | |
| Visual impairment: | |||||
| Yes | 106 | 0.48 | 150 | 0.26 | < 0.001 |
| No | 114 | 0.52 | 55 | 0.74 | |
| Allergies: | |||||
| Yes | 32 | 0.15 | 53 | 0.26 | 0.0053 |
| No | 188 | 0.85 | 152 | 0.74 | |
Values of p for a two-sided χ2 test;
p values for t-test; p values < 0.05 are in bold.
Selection of SNPs and primer design
We searched the public domain of the National Center for Biotechnology Information Single Nucleotide Polymorphisms database (NCBI dbSNP) at http://www.ncbi.nlm.nih.gov/snp to identify potentially functional polymorphisms in the DNA repair genes. We chose to genotype the c.972G>C and the c.977C>G polymorphisms with minor allele frequency (MAF) of 0.284 and 0.224 in the European population, respectively (submitter population ID: HapMap-CEU for both; http://www.ncbi.nlm.nih.gov/snp).
Primers were designed according to the published nucleotide sequence in the ENSEMBL database (gene ID for MUTYH ENSG00000132781 and ENSG00000114026 for hOGG1) and using Primer3 software for hOGG1 SNP (http://frodo.wi.mit.edu/) and Web-based allele-specific primer software for MUTYH SNP (http://bioinfo.biotec.or.th/WASP).
DNA isolation
Genomic DNA was extracted from venous blood using the commercially available AxyPrep Blood Genomic DNA Miniprep Kit (Axygen Biosciences, Union City, CA, USA), according to the manufacturer's instructions. DNA was directly isolated from the white blood cells. DNA purity and concentration were determined by comparing the absorbance at 260 and 280 nm. The purified genomic DNA was stored in TE buffer (5 mM Tris-HCl, 0.1 mM EDTA, pH 8.5), at –20°C until further analysis.
SNPs genotyping
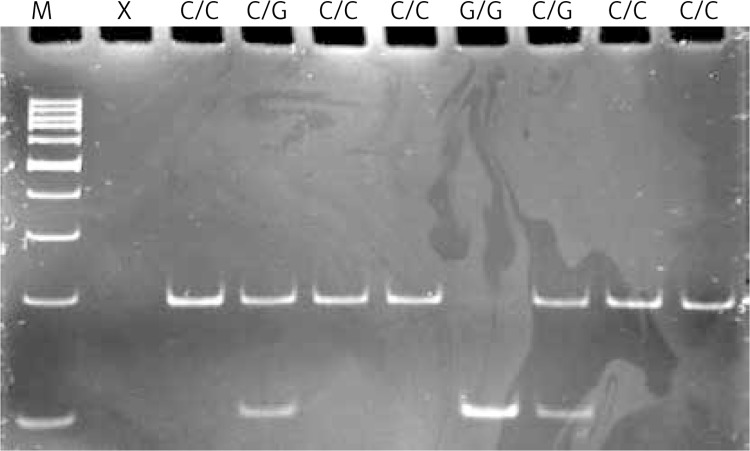
Genotyping of the c.972G>C-MUTYH polymorphism was carried out by polymerase chain reaction (PCR) with confronting two-pair primers (PCR-CTPP). The amplification of allele-specific DNA products of different lengths by adding four (two pairs) primers into one tube containing a PCR mixture enables direct genotyping by gel electrophoresis [58]. Polymerase chain reaction was performed in a 25 μl reaction volume. The reaction mixture contained 50 ng of genomic DNA, 1 U of Biotools DNA polymerase (Biotools, Madrid, Spain), 1 × reaction buffer (750 mM Tris-HCl, pH 9.0, 500 mM KCl, 200 mM (NH4)2SO4), 0.2 mM of each dNTP, 1.5 mM MgCl2, and 0.25 μM of each primer (Metabion, Martinsried, Germany). The primers designed to detect the c.972G>C SNP were: F1: 5’-CCTGTCGGGCAGTCCTGACG-3’ and R2: 5’-GAGGCAGGC ACAGGTGGCAC-3’ for the amplification of the G allele (241-bp band), and F2 5’-CCCAGC TCCCAACACTGGACAC-3’ and R1 5’-CGCTGAAGCTGCTCTGAGGGC-3’ for the amplification of the C allele (362-bp band). F1 and R1 give a common 562-bp PCR product. F2 and R2 confront each other at the 3’ end with the base specific to the allele. The PCR-CTPP conditions were as follows: 5 min of initial denaturation at 95°C, followed by 30 cycles of 30 s denaturation at 95°C, 30 s annealing at 65°C and 1 min extension at 72°C. A final extension step at 72°C for 10 min was included. The PCR products were fractionated by electrophoresis on 3% agarose gel, stained with ethidium bromide and viewed under UV light. The samples were genotyped according to the size of the PCR products: products amplified with the G/G genotype were 562 and 241 bp in length, those with the C/C genotype were 562 and 362 bp, and those with the G/C genotype were 562, 362 and 241 bp. Figure 1 presents a representative gel for analysis of this polymorphism.
Figure 1.
Detection of the MUTYH – c.972G>C (rs3219489) polymorphism. Genotypes are indicated in the upper part of the picture. Lane M shows GeneRuler 100 bp molecular length marker; lane X shows a negative control comprising reaction mixture without target DNA
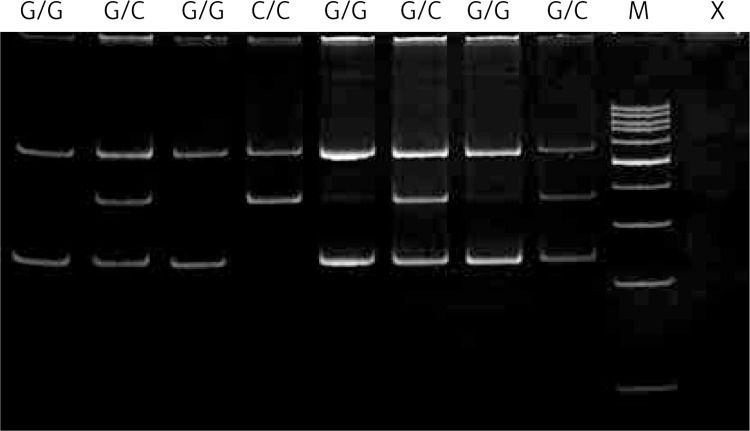
The polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was used to determine the genotypes of the c.977C>G-hOGG1 polymorphism. PCR assay was performed in a total reaction volume of 25 μl containing the same reagents as in the previous analysis except for primers. 200-bp length fragments containing the polymorphic site were amplified using the following primers: sense 5’-ACTGTCACTAGTCTCACCAG-3’ and antisense 5’-GGAAGGTGCTTGGGGAAT-3’. The PCR profile contained an initial denaturation step for 5 min at 95°C, 30 cycles at 95°C for 30 s, 30 s at 58°C annealing temperature and 60 s at 72°C and the final extension step for 5 min at 72°C. After amplification, 200-bp PCR products were analyzed on 3% agarose gel and digested with 2U of SatI (Fnu4HI) restriction endonuclease (Fermentas, Hanover, MD, USA) in a final volume of 15 μl for 16 h at 37°C. PCR products with a G at the polymorphic site were digested into two 100-bp fragments, while those with a C were not, because of the lack of a SatI restriction site. The G/G genotype produced one fragment (100 bp), whereas the G/C genotype yielded two fragments (200, 100 bp) and the homozygote C/C resulted in one 200 bp fragment. Digested PCR products were separated by electrophoresis on 8% polyacrylamide gel and visualized by ethidium bromide staining using a GeneRuler 100 bp ladder (Fermentas, Hanover, MD, USA) as a length marker. A representative gel for this polymorphism is presented in Figure 2.
Figure 2.
Detection of the hOGG1 – c.977C>G (rs1052133) polymorphism. Genotypes are indicated in the upper part of the picture. Lane M shows GeneRuler 100 bp molecular length marker; lane X shows a negative control comprising reaction mixture without target DNA
All PCR amplifications were conducted in a C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). Positive and negative (no template) controls were included in all sets. For quality control, 10% of samples were randomly genotyped again and the results were 100% concordant.
Statistical analysis
The statistical analyses were performed using Statistica 9.0 software (StatSoft, Tulsa, OK, USA) and SigmaPlot v11.0 software (Systat Software, Inc., San Jose, CA, USA). To compare the distributions of demographic variables and potential risk factors between patients and controls the χ2 test was used. Hardy-Weinberg equilibrium was checked using the χ2 test to compare the observed genotype frequencies with the expected frequencies among the case and control subjects. The χ2 analysis was also used to test the significance of the differences between distributions of genotypes and alleles in KC patients and controls. The association between case-control status and each polymorphism, measured by the odds ratio (OR) and its corresponding 95% confidence interval (CI), was estimated using an unconditional multiple logistic regression model, both with and without adjustment for co-occurrence of visual impairment, allergies, sex and family status of KC.
Results
The PCR-RFLP and PCR-CTPP analyses were successful for all 425 DNA samples (205 KC patients and 220 KC-free controls).
Characteristics of study subjects
Table I presents demographic and risk factors for KC of the study patients and controls. The mean ± SD age for KC patients were 36.39 ±11.87 (range: 14–63) and 60.71 ±20.25 (range: 19–91) for controls. Moreover, there were significantly more subjects with a positive family history for KC (1st degree relatives) among the patients in comparison to controls (11% vs. 4%, p = 0.0145). We observed a significant difference between distribution of family history for KC (positive vs. negative family history), gender (females vs. males), allergies (yes vs. no) and co-occurrence of visual impairment (yes vs. no) among patients and controls. These parameters were further adjusted in the multivariate logistic regression model for possible confounding factors of the main effect of the SNPs.
The c.972G>C polymorphism of the MUTYH gene and KC occurrence
The genotype and allele distributions of the c.972G>C polymorphism of the MUTYH gene in KC patients and controls are presented in Table II. The observed genotype frequencies were all in agreement with the Hardy-Weinberg equilibrium calculated for the controls and patients (p>0.05, data not shown). The difference in the frequency distributions of genotypes of the polymorphism between the cases and controls was not statistically significant (p>0.05). We did not find any correlation between genotypes/alleles of this polymorphism and KC occurrence.
Table II.
Distribution of genotypes and alleles of the c.972G>C polymorphism of the MUTYH gene and odds ratio (OR) with 95% confidence interval (95% CI) in patients with KC and controls
| Genotype/allele | Controls (n = 220) | KC (n = 205) | Crude OR (95% CI) | Value of p | Adjusted ORa (95% CI) | Value of p | ||
|---|---|---|---|---|---|---|---|---|
| Number | Frequency | Number | Frequency | |||||
| G/G | 142 | 0.65 | 136 | 0.66 | 1.08 (0.73–1.62) | 0.697 | 1.28 (0.82–2.01) | 0.286 |
| G/C | 73 | 0.33 | 62 | 0.30 | 0.87 (0.58–1.32) | 0.516 | 0.73 (0.46–1.16) | 0.184 |
| C/C | 5 | 0.02 | 7 | 0.04 | 1.52 (0.48–4.87) | 0.481 | 1.72 (0.41–7.26) | 0.464 |
| χ2 = 0.83; p = 0.6601 | ||||||||
| G | 357 | 0.81 | 334 | 0.81 | 1.02 (0.72–1.46) | 0.901 | 1.17 (0.78–1.74) | 0.456 |
| C | 83 | 0.19 | 76 | 0.19 | 0.98 (0.69–1.39) | 0.901 | 0.86 (0.57–1.28) | 0.456 |
OR adjusted for co-occurrence of visual impairment, allergies, sex and family history for KC.
The c.977C>G polymorphism of the hOGG1 gene and KC occurrence
Details of genotype and allele frequencies of the c.977C>G polymorphisms of the hOGG1 gene and summary statistics are shown in Table III. There was no difference in the frequency distributions of genotypes of this SNP between patients and controls (p>0.05), and the observed genotype frequencies did not differ significantly from Hardy-Weinberg equilibrium (p>0.05; data not shown). We did not find any correlation between genotypes/alleles of this polymorphism and KC occurrence.
Table III.
Distribution of genotypes and alleles of the c.977C>G polymorphism of the hOGG1 gene and odds ratio (OR) with 95% confidence interval (95% CI) in patients with KC and controls
| Genotype/allele | Controls (n = 220) | KC (n = 205) | Crude OR (95% CI) | Value of p | Adjusted ORa (95% CI) | Value of p | ||
|---|---|---|---|---|---|---|---|---|
| Number | Frequency | Number | Frequency | |||||
| C/C | 144 | 0.65 | 125 | 0.61 | 0.83 (0.55–1.22) | 0.415 | 0.83 (0.54–1.29) | 0.415 |
| C/G | 69 | 0.31 | 70 | 0.34 | 1.14 (0.76–1.70) | 0.541 | 1.18 (0.75–1.85) | 0.483 |
| G/G | 7 | 0.03 | 10 | 0.05 | 1.56 (0.58–4.18) | 0.376 | 1.96 (0.41–3.53) | 0.746 |
| χ2 = 1.35; p = 0.5089 | ||||||||
| C | 357 | 0.81 | 320 | 0.78 | 0.83 (0.59–1.16) | 0.263 | 0.86 (0.59–1.25) | 0.422 |
| G | 83 | 0.19 | 90 | 0.22 | 1.21 (0.87–1.70) | 0.263 | 1.17 (0.80–1.70) | 0.422 |
OR adjusted for co-occurrence of visual impairment, allergies, sex and family history for KC.
Gene-gene interaction and KC occurrence
We also investigated the association between the occurrence of KC and combined genotypes of the c.977C>G-hOGG1 and c.972G>C-MUTYH polymorphisms. The distribution of such genotypes is shown in Table IV. We did not find any correlation between combined genotypes of both polymorphisms and the occurrence of KC.
Table IV.
Distribution of combined genotypes of the c.977C>G polymorphism of hOGG1 and c.972G>C polymorphism of MUTYH and odds ratio (OR) with 95% confidence interval (95% CI) in patients with KC and controls
| Genotype/allele | Controls (n = 220) | KC (n = 205) | Crude OR (95% CI) | Value of p | Adjusted ORa (95% CI) | Value of p | ||
|---|---|---|---|---|---|---|---|---|
| Number | Frequency | Number | Frequency | |||||
| G/G–C/C | 91 | 0.41 | 80 | 0.39 | 0.91 (0.62–1.34) | 0.623 | 0.99 (0.64–1.52) | 0.952 |
| G/G–C/G | 46 | 0.21 | 51 | 0.25 | 1.25 (0.80–1.97) | 0.330 | 1.45 (0.87–2.41) | 0.156 |
| G/G–G/G | 5 | 0.02 | 5 | 0.02 | 1.08 (0.31–3.77) | 0.910 | 0.75 (0.19–3.05) | 0.688 |
| G/C–C/C | 48 | 0.22 | 43 | 0.21 | 0.95 (0.60–1.51) | 0.832 | 0.81 (0.49–1.35) | 0.423 |
| G/C–C/G | 23 | 0.10 | 14 | 0.07 | 0.63 (0.31–1.26) | 0.188 | 0.55 (0.25–1.22) | 0.141 |
| G/C–G/G | 2 | 0.01 | 5 | 0.02 | 2.73 (0.52–14.20) | 0.234 | 2.27 (0.40–12.94) | 0.355 |
| C/C–C/C | 5 | 0.02 | 2 | 0.01 | 0.42 (0.08–2.21) | 0.308 | 0.76 (0.11–5.41) | 0.789 |
| C/C–C/G | 0 | – | 5 | 0.02 | – | – | – | – |
| C/C–G/G | 0 | – | 0 | – | – | – | – | – |
OR adjusted for co-occurrence of visual impairment, allergies, sex and family history for KC.
Discussion
As mentioned in the Introduction, both environmental and genetic factors and the interaction between them are involved in the pathogenesis of KC. The susceptibility of KC cornea to cellular damage due to chronic oxidative stress was supported by detection of oxidant-antioxidant imbalance and oxidant-induced DNA damage, mainly mitochondrial DNA (mtDNA) in KC corneas. Several studies have found a decreased antioxidant capacity in KC corneas [23, 29]. Mitochondrial DNA is particularly prone to oxidative damage compared with its nuclear counterpart because of its close proximity to oxidative phosphorylation (OXPHOS) [59]. It does not contain introns and has a high transcription rate, resulting in a high probability of oxidative modification of the coding region of genes [29]. When mitochondria are damaged, the level of OXPHOS is decreased, but ROS production is usually increased [60]. It has been shown that mitochondrial dysfunction led to altered gene expression, apoptosis and loss of cell viability [61–63]. It was shown that human KC corneas displayed increased levels of mtDNA deletions and point mutations compared with age-matched normal corneas [29]. Furthermore, it was found that sequence variations in the mitochondrial complex I were the main cause of elevated ROS production [64]. Taking into account these facts, antioxidant-directed therapy may be considered in KC patients to minimize ROS-related damage.
Although KC is recognized as a multifactorial disease, genetic factors may play an essential role in its development [40, 65–68], which was confirmed by segregation analyses, genetic epidemiological data and gene mapping studies. Several gene loci/chromosomal regions for familial KC have been mapped by genome-wide linkage analysis, including 16q22.3-q23.1 [69], 3p14-q13 [17], 2p24 [70], 1p36.23-36.21 [71], 5q14.3-q21.1 [19], 5q21.2, 5q32-q33 [72], 8q13.1-q21.11 [71], 9q34 [18], 14q11.2 [72], 14q24.3 [73], 15q2.32 [72], 15q22.33-q24.2 [16], 17p13 [74] and 20q12 [75]. The first gene to be causally linked with KC was the visual system homeobox 1 (VSX1) gene [41]. It is located on 20p11-q11 [76, 77] and plays a role in craniofacial and ocular development. VSX1 is a member of the paired-like homeodomain transcription factors (TFs), which regulate expression of the cone opsin genes during embryonic development [78, 79]. Although VSX1 plays a role in the development of retinal bipolar interneurons, several studies did not confirm its expression in the mouse and human corneas [41, 80]. In addition, a mouse model with the loss of VSX1 function did not support its role in the cornea [79, 81]. It was shown that two missense mutations in VSX1, R166W and L159M, were associated with KC [41]. However, a similar number of studies did not confirm the presence of these mutations in KC patients [82–85], suggesting that VSX1 might not play a major role and other genetic factors exert a more powerful effect on the development of KC. These conflicting results may be partly explained by low frequency of changes, ethnic variation, and increasing evidence that KC is likely a multifactorial and polygenic disease [86, 87].
Besides VSX1, several other candidate genes for KC have been suggested, including superoxide dismutase 1 (SOD1) [42], lipoxygenase (LOX) [88], transforming growth factor β1 (TGFB1) [43], human leukocyte antigen (HLA) [89], mitochondrial complex 1 genes [64], interleukin 1β (IL1B) [90], collagen type IV, α3 and collagen type IV (COL4A3), α4 (COL4A4) [46], and calpastatin (CAST) [91]. Gajecka et al. [65] identified a novel locus for familial KC at 13q32, by using GeneChip Mapping to genotype 10 affected and 11 unaffected individuals from a large Ecuadorian KC (KTCN-014) family. In a subsequent study [44], they revealed that the c.2262A>C (p.Gln754His) mutation in the DOCK9 gene, located at 13q32, may contribute to the KC phenotype in that family. In a recent study, an association of KC with the hepatocyte growth factor (HGF) gene [92] and microRNA (MIR184) [93] gene was recognized. Other studies have indicated that the rs4954218 SNP near the RAB3GAP1 gene (located at 2q21.3), coding for Rab3 GTPase-activating protein subunit 1, suggests a new susceptibility locus for KC [94].
In the present study, we investigated whether polymorphisms of the MUTYH gene (c.972G>C) and the hOGG1 gene (c.977C>G) are associated with KC. Both SNPs are located in the coding regions of these genes and change amino acids in their products. These changes can directly affect protein function, and therefore they can have serious phenotypic consequences [95, 96]. The c.977G>C polymorphism causes change of serine to cysteine at the 326 codon, whereas c.972C>G polymorphism is associated with substitution of glutamine instead histidine at the 324 codon (Gln324His) of the MUTYH protein. Genetic polymorphisms in DNA repair genes can cause inter-individual difference in preventing mutagenesis induced by DNA damage [47]. The polymorphisms under study may result in changes in the levels and the activities of the proteins they affect, which can lead to reduced protection against oxidative stress [50, 97, 98].
The c.972G>C polymorphism of the MUTYH gene has been implicated in functional changes in the DNA damage response pathway – it was found that the activity of the SNP c.972G>C MUTYH enzyme was decreased by 36% compared with the wild type protein [50]. Recent studies have demonstrated that the G/G genotype increased the risk of end-stage renal disease [99], while the C/C genotype increased the risk of rectal cancer [100]. In this study, we did not find any correlation between genotypes/alleles of this polymorphism and the occurrence of KC.
The c.977C>G (p.Ser326Cys) polymorphism is the most commonly studied hOGG1 gene variation. The 326Cys allele was less effective in preventing G:C to T:A transversion in E. coli mutMmutY mutants than the 326Ser allele, indicating that 326Cys had a lower ability to remove 8-OH-G in DNA [97, 98]. In addition, the allele frequency of 326Cys was markedly different among different ethnic groups [97, 101, 102]. In the Japanese population, about 25% of individuals are homozygous at codon 326 (Cys/Cys), but the frequency of this allele seems to be lower in the Caucasian population [48]. It was demonstrated that the 326Cys allele was positively correlated with an increased risk of lung, oesophagus, prostate and a subset of stomach cancers [47, 52, 103, 104]. In our previous study we found that the C/G genotype and the G allele significantly increased the risk of another eye disease – age-related macular degeneration (AMD) [105]. On the other hand, the C/C genotype and the C allele were positively correlated with a decreased risk of this disease. In the present study, we found that this polymorphism was not associated with occurrence of KC, but it should be stressed that our results are limited to a small Polish subpopulation and can be extrapolated at most to the European populace.
In conclusion, the c.972G>C polymorphism of the MUTYH gene and the c.977C>G polymorphism of the hOGG1 gene may not be associated with keratoconus in the Polish population. Further replication studies performed on other ethnic populations are needed to verify this conclusion.
Acknowledgments
This study was supported by grant number N N402 591940 of the Polish Ministry of Science and Higher Education.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Lass JH, Lembach RG, Park SB, et al. Clinical management of keratoconus. A multicenter analysis. Ophthalmology. 1990;97:433–445. doi: 10.1016/s0161-6420(90)32569-1. [DOI] [PubMed] [Google Scholar]
- 2.Crews MJ, Driebe WT, Jr, Stern GA. The clinical management of keratoconus: a 6 year retrospective study. CLAO J. 1994;20:194–7. doi: 10.1097/00140068-199407000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 4.Kang PC, Klintworth GK, Kim T, et al. Trends in the indications for penetrating keratoplasty, 1980-2001. Cornea. 2005;24:801–3. doi: 10.1097/01.ico.0000157407.43699.22. [DOI] [PubMed] [Google Scholar]
- 5.Washington: Eye Bank Association of America; 2008. Eye Banking Statistical Report. [Google Scholar]
- 6.Olivares Jiménez JL, Guerrero Jurado JC, Bermudez Rodriguez FJ, Serrano Laborda D. Keratoconus: age of onset and natural history. Optom Vis Sci. 1997;74:147–51. doi: 10.1097/00006324-199703000-00025. [DOI] [PubMed] [Google Scholar]
- 7.Fink BA, Sinnott LT, Wagner H, Friedman C, Zadnik K, CLEK Study Group The influence of gender and hormone status on the severity and progression of keratoconus. Cornea. 2010;29:65–72. doi: 10.1097/ICO.0b013e3181ac0518. [DOI] [PubMed] [Google Scholar]
- 8.Georgiou T, Funnell CL, Cassels-Brown A, O'Conor R. Influence of ethnic origin on the incidence of keratoconus and associated atopic disease in Asians and white patients. Eye (Lond) 2004;18:379–83. doi: 10.1038/sj.eye.6700652. [DOI] [PubMed] [Google Scholar]
- 9.Barr JT, Wilson BS, Gordon MO, et al. CLEK Study Group Estimation of the incidence and factors predictive of corneal scarring in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study. Cornea. 2006;25:16–25. doi: 10.1097/01.ico.0000164831.87593.08. [DOI] [PubMed] [Google Scholar]
- 10.Jafri B, Lichter H, Stulting RD. Asymmetric keratoconus attributed to eye rubbing. Cornea. 2004;23:560–4. doi: 10.1097/01.ico.0000121711.58571.8d. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000;93:403–9. [PubMed] [Google Scholar]
- 12.Hyams SW, Kar H, Neumann E. Blue sclerae and keratoglobus. Ocular signs of a systemic connective tissue disorder. Br J Ophthalmol. 1969;53:53–8. doi: 10.1136/bjo.53.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh SZ. Keratoconus and blindness in 469 institutionalised subjects with Down syndrome and other causes of mental retardation. J Ment Defic Res. 1981;4:243–51. doi: 10.1111/j.1365-2788.1981.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 14.Flanders M, Lapointe ML, Brownstein S, Little JM. Keratoconus and Leber's congenital amaurosis: a clinicopathological correlation. Can J Ophthalmol. 1984;19:310–4. [PubMed] [Google Scholar]
- 15.Parker J, Ko WW, Pavlopoulos G, Wolfe PJ, Rabinowitz YS, Feldman ST. Videokeratography of keratoconus in monozygotic twins. J Refract Surg. 1996;12:180–3. doi: 10.3928/1081-597X-19960101-31. [DOI] [PubMed] [Google Scholar]
- 16.Hughes AE, Dash DP, Jackson AJ, Frazer DG, Silvestri G. Familial keratoconus with cataract: linkage to the long arm of chromosome 15 and exclusion of candidate genes. Invest Ophthalmol Vis Sci. 2003;44:5063–6. doi: 10.1167/iovs.03-0399. [DOI] [PubMed] [Google Scholar]
- 17.Brancati F, Valente EM, Sarkozy A, et al. A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J Med Genet. 2004;41:188–92. doi: 10.1136/jmg.2003.012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Rabinowitz YS, Tang YG, et al. Two-stage genome-wide linkage scan in keratoconus sib pair families. Invest Ophthalmol Vis Sci. 2006;47:3791–5. doi: 10.1167/iovs.06-0214. [DOI] [PubMed] [Google Scholar]
- 19.Tang YG, Rabinowitz YS, Taylor KD, et al. Genomewide linkage scan in a multigeneration Caucasian pedigree identifies a novel locus for keratoconus on chromosome 5q14.3-q21.1. Genet Med. 2005;7:397–405. doi: 10.1097/01.gim.0000170772.41860.54. [DOI] [PubMed] [Google Scholar]
- 20.Gondhowiardjo TD, van Haeringen NJ. Corneal aldehyde dehydrogenase, glutathione reductase, and glutathione S-transferase in pathologic corneas. Cornea. 1993;12:310–4. doi: 10.1097/00003226-199307000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Kenney MC, Chwa M, Atilano SR, et al. Increased levels of catalase and cathepsin V/L2 but decreased TIMP-1 in keratoconus corneas: evidence that oxidative stress plays a role in this disorder. Invest Ophthalmol Vis Sci. 2005;46:823–32. doi: 10.1167/iovs.04-0549. [DOI] [PubMed] [Google Scholar]
- 22.Behndig A, Karlsson K, Johansson BO, Brännström T, Marklund SL. Superoxide dismutase isoenzymes in the normal and diseased human cornea. Invest Ophthalmol Vis Sci. 2001;42:2293–6. [PubMed] [Google Scholar]
- 23.Buddi R, Lin B, Atilano SR, Zorapapel NC, Kenney MC, Brown DJ. Evidence of oxidative stress in human corneal diseases. J Histochem Cytochem. 2002;50:341–51. doi: 10.1177/002215540205000306. [DOI] [PubMed] [Google Scholar]
- 24.Chwa M, Atilano SR, Reddy V, Jordan N, Kim DW, Kenney MC. Increased stress-induced generation of reactive oxygen species and apoptosis in human keratoconus fibroblasts. Invest Ophthalmol Vis Sci. 2006;47:1902–10. doi: 10.1167/iovs.05-0828. [DOI] [PubMed] [Google Scholar]
- 25.Chwa M, Atilano SR, Hertzog D, et al. Hypersensitive response to oxidative stress in keratoconus corneal fibroblasts. Invest Ophthalmol Vis Sci. 2008;49:4361–9. doi: 10.1167/iovs.08-1969. [DOI] [PubMed] [Google Scholar]
- 26.Shoham A, Hadziahmetovic M, Dunaief JL, Mydlarski MB, Schipper HM. Oxidative stress in diseases of the human cornea. Free Radic Biol Med. 2008;45:1047–55. doi: 10.1016/j.freeradbiomed.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Wenk J, Brenneisen P, Meewes C, et al. UV-induced oxidative stress and photoaging. Curr Probl Dermatol. 2001;29:83–94. doi: 10.1159/000060656. [DOI] [PubMed] [Google Scholar]
- 28.Squadrito GL, Pryor WA. Oxidative chemistry of nitric oxide: the roles of superoxide, peroxynitrite, and carbon dioxide. Free Radic Biol Med. 1998;25:392–403. doi: 10.1016/s0891-5849(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 29.Atilano SR, Coskun P, Chwa M, et al. Accumulation of mitochondrial DNA damage in keratoconus corneas. Invest Ophthalmol Vis Sci. 2005;46:1256–63. doi: 10.1167/iovs.04-1395. [DOI] [PubMed] [Google Scholar]
- 30.McCord JM. Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med. 1985;312:159–63. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- 31.Ohia SE, Opere CA, Leday AM. Pharmacological consequences of oxidative stress in ocular tissues. Mutat Res. 2005;579:22–36. doi: 10.1016/j.mrfmmm.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 32.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704:152–9. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49:1603–16. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomic S, Brkic S, Maric D, Mikic AN. Lipid and protein oxidation in female patients with chronic fatigue syndrome. Arch Med Sci. 2012;8:886–91. doi: 10.5114/aoms.2012.31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burchardt P, Zurawski J, Zuchowski B, et al. Low-density lipoprotein, its susceptibility to oxidation and the role of lipoprotein-associated phospholipase A2 and carboxyl ester lipase lipases in atherosclerotic plaque formation. Arch Med Sci. 2013;9:151–8. doi: 10.5114/aoms.2013.33176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 39.Kunkel TA. The high cost of living. American Association for Cancer Research Special Conference: endogenous sources of mutations, Fort Myers, Florida, USA, 11-15 November 1998. Trends Genet. 1999;15:93–4. doi: 10.1016/s0168-9525(98)01664-3. [DOI] [PubMed] [Google Scholar]
- 40.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–5. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 41.Héon E, Greenberg A, Kopp KK, et al. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet. 2002;11:1029–36. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- 42.Udar N, Atilano SR, Brown DJ, et al. SOD1: a candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006;47:3345–51. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 43.Burdon KP, Macgregor S, Bykhovskaya Y, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52:8514–9. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czugala M, Karolak JA, Nowak DM, et al. Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus. Eur J Hum Genet. 2012;20:389–97. doi: 10.1038/ejhg.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan T, Liu C, Ma Z, Ding S. The point mutation and polymorphism in keratoconus candidate gene TGFBI in Chinese population. Gene. 2012;503:137–9. doi: 10.1016/j.gene.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 46.Stabuc-Silih M, Ravnik-Glavac M, Glavac D, Hawlina M, Strazisar M. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis. 2009;15:2848–60. [PMC free article] [PubMed] [Google Scholar]
- 47.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–30. [PubMed] [Google Scholar]
- 48.Hamajima N, Takezaki T, Tajima K. Allele frequencies of 25 polymorphisms pertaining to cancer risk for Japanese, Koreans and Chinese. Asian Pac J Cancer Prev. 2002;3:197–206. [PubMed] [Google Scholar]
- 49.Choi JY, Hamajima N, Tajima K, et al. hOGG1 Ser326Cys polymorphism and breast cancer risk among Asian women. Breast Cancer Res Treat. 2003;79:59–62. doi: 10.1023/a:1023305826726. [DOI] [PubMed] [Google Scholar]
- 50.Ali M, Kim H, Cleary S, Cupples C, Gallinger S, Bristow R. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135:499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–42. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 52.Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog. 2005;42:127–41. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 53.Kasahara M, Osawa K, Yoshida K, et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49. doi: 10.1186/1756-9966-27-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao H, Shinmura K, Suzuki M, et al. Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Sci. 2008;99:355–60. doi: 10.1111/j.1349-7006.2007.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyaishi A, Osawa K, Osawa Y, et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pflugfelder SC, Liu Z, Feuer W, Verm A. Corneal thickness indices discriminate between keratoconus and contact lens-induced corneal thinning. Ophthalmology. 2002;109:2336–341. doi: 10.1016/s0161-6420(02)01276-9. [DOI] [PubMed] [Google Scholar]
- 57.Holladay JT. Keratoconus detection using corneal topography. J Refract Surg. 2009;25:S958–62. doi: 10.3928/1081597X-20090915-11. [DOI] [PubMed] [Google Scholar]
- 58.Hamajima N, Saito T, Matsuo K, Tajima K. Competitive amplification and unspecific amplification in polymerase chain reaction with confronting two-pair primers. J Mol Diagn. 2002;4:103–7. doi: 10.1016/S1525-1578(10)60688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace DC, Brown MD, Melov S, Graham B, Lott M. Mitochondrial biology, degenerative diseases and aging. Biofactors. 1998;7:187–90. doi: 10.1002/biof.5520070303. [DOI] [PubMed] [Google Scholar]
- 60.Brown MD, Wallace DC. Molecular basis of mitochondrial DNA disease. J Bioenerg Biomembr. 1994;26:273–89. doi: 10.1007/BF00763099. [DOI] [PubMed] [Google Scholar]
- 61.Petit PX, Susin SA, Zamzami N, Mignotte B, Kroemer G. Mitochondria and programmed cell death: back to the future. FEBS Lett. 1996;396:7–13. doi: 10.1016/0014-5793(96)00988-x. [DOI] [PubMed] [Google Scholar]
- 62.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;28:1309–12. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 63.Richter C. Oxidative damage to mitochondrial DNA and its relationship to ageing. Int J Biochem Cell Biol. 1995;27:647–53. doi: 10.1016/1357-2725(95)00025-k. [DOI] [PubMed] [Google Scholar]
- 64.Pathak D, Nayak B, Singh M, et al. Mitochondrial complex 1 gene analysis in keratoconus. Mol Vis. 2011;17:1514–25. [PMC free article] [PubMed] [Google Scholar]
- 65.Gajecka M, Radhakrishna U, Winters D, et al. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci. 2009;50:1531–9. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paliwal P, Tandon R, Dube D, Kaur P, Sharma A. Familial segregation of a VSX1 mutation adds a new dimension to its role in the causation of keratoconus. Mol Vis. 2011;17:481–5. [PMC free article] [PubMed] [Google Scholar]
- 67.Saee-Rad S, Hashemi H, Miraftab M, et al. Mutation analysis of VSX1 and SOD1 in Iranian patients with keratoconus. Mol Vis. 2011;17:3128–36. [PMC free article] [PubMed] [Google Scholar]
- 68.Nowak DM, Gajecka M. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011;18:2–6. doi: 10.4103/0974-9233.75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyynismaa H, Sistonen P, Tuupanen S, et al. A locus for autosomal dominant keratoconus: linkage to 16q22.3-q23.1 in Finnish families. Invest Ophthalmol Vis Sci. 2002;43:3160–4. [PubMed] [Google Scholar]
- 70.Hutchings H, Ginisty H, Le Gallo M, et al. Identification of a new locus for isolated familial keratoconus at 2p24. J Med Genet. 2005;42:88–94. doi: 10.1136/jmg.2004.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burdon KP, Coster DJ, Charlesworth JC, et al. Apparent autosomal dominant keratoconus in a large Australian pedigree accounted for by digenic inheritance of two novel loci. Hum Genet. 2008;124:379–86. doi: 10.1007/s00439-008-0555-z. [DOI] [PubMed] [Google Scholar]
- 72.Bisceglia L, De Bonis P, Pizzicoli C, et al. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive Loci. Invest Ophthalmol Vis Sci. 2009;50:1081–6. doi: 10.1167/iovs.08-2382. [DOI] [PubMed] [Google Scholar]
- 73.Liskova P, Hysi PG, Waseem N, Ebenezer ND, Bhattacharya SS, Tuft SJ. Evidence for keratoconus susceptibility locus on chromosome 14: a genome-wide linkage screen using single-nucleotide polymorphism markers. Arch Ophthalmol. 2010;128:1191–5. doi: 10.1001/archophthalmol.2010.200. [DOI] [PubMed] [Google Scholar]
- 74.Hameed A, Khaliq S, Ismail M, et al. A novel locus for Leber congenital amaurosis (LCA4) with anterior keratoconus mapping to chromosome 17p13. Invest Ophthalmol Vis Sci. 2000;41:629–33. [PubMed] [Google Scholar]
- 75.Fullerton J, Paprocki P, Foote S, Mackey DA, Williamson R, Forrest S. Identity-by-descent approach to gene localisation in eight individuals affected by keratoconus from north-west Tasmania, Australia. Hum Genet. 2002;110:462–70. doi: 10.1007/s00439-002-0705-7. [DOI] [PubMed] [Google Scholar]
- 76.Hayashi T, Huang J, Deeb SS. Expression of rinx/vsx1 during postnatal eye development in cone-bipolar, differentiating ganglion, and lens fiber cells. Jpn J Ophthalmol. 2005;49:93–105. doi: 10.1007/s10384-004-0162-z. [DOI] [PubMed] [Google Scholar]
- 77.Semina EV, Mintz-Hittner HA, Murray JC. Isolation and characterization of a novel human paired-like homeodomain-containing transcription factor gene, VSX1, expressed in ocular tissues. Genomics. 2000;63:289–93. doi: 10.1006/geno.1999.6093. [DOI] [PubMed] [Google Scholar]
- 78.Chow RL, Volgyi B, Szilard RK, et al. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci USA. 2004;101:1754–9. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohtoshi A, Wang SW, Maeda H, et al. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–6. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 80.Watson T, Chow RL. Absence of Vsx1 expression in the normal and damaged mouse cornea. Mol Vis. 2011;17:737–44. [PMC free article] [PubMed] [Google Scholar]
- 81.Verma A, Das M, Srinivasan M, Prajna NV, Sundaresan P. Investigation of VSX1 sequence variants in South Indian patients with sporadic cases of keratoconus. BMC Res Notes. 2013;6:103. doi: 10.1186/1756-0500-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aldave AJ, Yellore VS, Salem AK, et al. No VSX1 gene mutations associated with keratoconus. Invest Ophthalmol Vis Sci. 2006;47:2820–2. doi: 10.1167/iovs.05-1530. [DOI] [PubMed] [Google Scholar]
- 83.Tanwar M, Kumar M, Nayak B, et al. VSX1 gene analysis in keratoconus. Mol Vis. 2010;16:2395–401. [PMC free article] [PubMed] [Google Scholar]
- 84.Abu-Amero KK, Kalantan H, Al-Muammar AM. Analysis of the VSX1 gene in keratoconus patients from Saudi Arabia. Mol Vis. 2011;17:667–72. [PMC free article] [PubMed] [Google Scholar]
- 85.Jeoung JW, Kim MK, Park SS, et al. VSX1 gene and keratoconus: genetic analysis in Korean patients. Cornea. 2012;31:746–50. doi: 10.1097/ICO.0b013e3181e16dd0. [DOI] [PubMed] [Google Scholar]
- 86.McGhee CN. 2008 Sir Norman McAlister Gregg Lecture: 150 years of practical observations on the conical cornea: what have we learned? Clin Experiment Ophthalmol. 2009;37:160–76. doi: 10.1111/j.1442-9071.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 87.Vincent AL, Jordan C, Sheck L, Niederer R, Patel DV, McGhee CN. Screening the visual system homeobox 1 gene in keratoconus and posterior polymorphous dystrophy cohorts identifies a novel variant. Mol Vis. 2013;19:852–60. [PMC free article] [PubMed] [Google Scholar]
- 88.Bykhovskaya Y, Li X, Epifantseva I, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci. 2012;53:4152–7. doi: 10.1167/iovs.11-9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adachi W, Mitsuishi Y, Terai K, et al. The association of HLA with young-onset keratoconus in Japan. Am J Ophthalmol. 2002;133:557–9. doi: 10.1016/s0002-9394(01)01368-x. [DOI] [PubMed] [Google Scholar]
- 90.Mikami T, Meguro A, Teshigawara T, et al. Interleukin 1 beta promoter polymorphism is associated with keratoconus in a Japanese population. Mol Vis. 2013;19:845–851. [PMC free article] [PubMed] [Google Scholar]
- 91.Li X, Bykhovskaya Y, Tang YG, et al. An association between the calpastatin (CAST) gene and keratoconus. Cornea. 2013;32:696–701. doi: 10.1097/ICO.0b013e3182821c1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burdon KP, Macgregor S, Bykhovskaya Y, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52:8514–9. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hughes AE, Bradley DT, Campbell M, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89:628–33. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li X, Bykhovskaya Y, Haritunians T, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012;21:421–9. doi: 10.1093/hmg/ddr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 96.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–4. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kohno T, Shinmura K, Tosaka M, et al. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–25. doi: 10.1038/sj.onc.1201872. [DOI] [PubMed] [Google Scholar]
- 98.Yamane A, Kohno T, Ito K, et al. Differential ability of polymorphic OGG1 proteins to suppress mutagenesis induced by 8-hydroxyguanine in human cell in vivo. Carcinogenesis. 2004;25:1689–94. doi: 10.1093/carcin/bgh166. [DOI] [PubMed] [Google Scholar]
- 99.Cai Z, Chen H, Tao J, et al. Association of base excision repair gene polymorphisms with ESRD risk in a Chinese population. Oxid Med Cell Longev. 2012;2012:928421. doi: 10.1155/2012/928421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Picelli S, Zajac P, Zhou XL, et al. Common variants in human CRC genes as low-risk alleles. Eur J Cancer. 2010;46:1041–8. doi: 10.1016/j.ejca.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 101.Le Marchand L, Donlon T, Lum-Jones A, Seifried A, Wilkens LR. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:409–12. [PubMed] [Google Scholar]
- 102.Yoshimura K, Hanaoka T, Ohnami S, et al. Allele frequencies of single nucleotide polymorphisms (SNPs) in 40 candidate genes for gene-environment studies on cancer: data from population-based Japanese random samples. J Hum Genet. 2003;48:654–8. doi: 10.1007/s10038-003-0096-1. [DOI] [PubMed] [Google Scholar]
- 103.Takezaki T, Gao CM, Wu JZ, et al. hOGG1 Ser(326)Cys polymorphism and modification by environmental factors of stomach cancer risk in Chinese. Int J Cancer. 2002;99:624–7. doi: 10.1002/ijc.10400. [DOI] [PubMed] [Google Scholar]
- 104.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–42. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 105.Synowiec E, Blasiak J, Zaras M, Szaflik J, Szaflik JP. Association between polymorphisms of the DNA base excision repair genes MUTYH and hOGG1 and age-related macular degeneration. Exp Eye Res. 2012;98:58–66. doi: 10.1016/j.exer.2012.02.008. [DOI] [PubMed] [Google Scholar]