Summary
To develop a culture system for large-scale production of mature hepatocytes, liver progenitor cells (LPCs) with a high proliferation potential would be advantageous. We have found that carboxypeptidase M (CPM) is highly expressed in embryonic LPCs, hepatoblasts, while its expression is decreased along with hepatic maturation. Consistently, CPM expression was transiently induced during hepatic specification from human-induced pluripotent stem cells (hiPSCs). CPM+ cells isolated from differentiated hiPSCs at the immature hepatocyte stage proliferated extensively in vitro and expressed a set of genes that were typical of hepatoblasts. Moreover, the CPM+ cells exhibited a mature hepatocyte phenotype after induction of hepatic maturation and also underwent cholangiocytic differentiation in a three-dimensional culture system. These results indicated that hiPSC-derived CPM+ cells share the characteristics of LPCs, with the potential to proliferate and differentiate bi-directionally. Thus, CPM is a useful marker for isolating hiPSC-derived LPCs, which allows development of a large-scale culture system for producing hepatocytes and cholangiocytes.
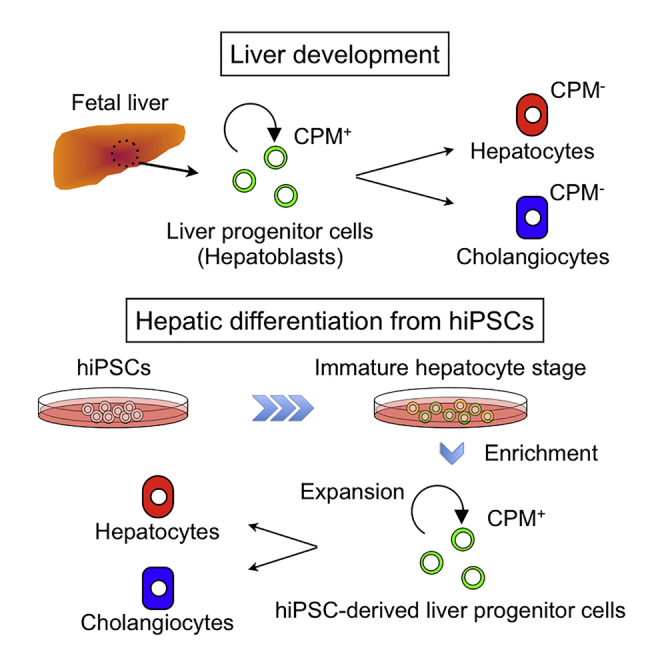
Graphical Abstract
Highlights
-
•
CPM is a novel marker for hepatoblasts during liver development
-
•
Self-renewing hiPSC-derived CPM+ LPCs exhibit bi-potency in vitro
-
•
An efficient and convenient protocol for the generation of hiPSC-derived LPCs
Miyajima, Kido, and colleagues identify CPM as a novel cell surface marker specific for liver progenitor cells (LPCs) in fetal liver. CPM+ LPCs can be isolated from hiPSC-derived immature liver cells, expanded, and differentiated to hepatocytes and cholangiocytes in vitro. CMP+ LPCs are useful for efficient large-scale production of hepatocytes and cholangiocytes.
Introduction
The liver is a central organ for metabolism, and the parenchymal cells, or hepatocytes, play key roles for homeostasis by expressing numerous metabolic and synthetic enzymes. As they express a number of cytochrome P450 oxidases (CYP450s) responsible for the oxidative biotransformation of many endogenous compounds as well as drugs, primary cultures of hepatocytes have been used for drug discovery and toxicology. However, primary hepatocytes exhibit low metabolic activity in vitro, and the supply of human hepatocytes is also limited and variable. To overcome these challenges, human embryonic stem cells (hESCs) and human-induced pluripotent stem cells (hiPSCs) have been considered as an alternative cell source for production of human hepatocytes. To date, there are many studies reporting hepatic differentiation of hiPSCs/hESCs (Ogawa et al., 2013, Si-Tayeb et al., 2010, Takayama et al., 2012). However, in most cases, differentiation of hepatocytes from hiPSCs is accomplished by a time-consuming culture protocol with multiple differentiation steps using expensive cytokines. Also, hepatocytes derived from hiPSCs possess a limited capacity for proliferation and functional maturation. Thus, it is beneficial to develop a simplified culture system for large-scale production of mature hepatocytes from hiPSCs. As liver progenitor cells (LPCs) such as hepatoblasts proliferate extensively in vitro, it would be useful if such cells could be derived from hiPSCs.
The development of the mouse liver begins with early endoderm development. The cells of the ventral foregut endoderm are induced to the hepatoblast stage by fibroblast growth factor (FGF) and bone morphogenetic protein (BMP) signaling from the heart and septum transversum mesenchyme (STM). Following induction, hepatoblasts proliferate and migrate into the STM to form the liver bud with non-parenchymal cells, such as endothelial progenitor cells and hepatic mesenchymal cells (Zaret and Grompe, 2008). Importantly, hepatoblasts isolated from fetal liver can be cultured long-term while maintaining the potential to differentiate into both hepatocytes and cholangiocytes, two types of liver epithelial cell (Suzuki et al., 2000, Tanimizu et al., 2003). LPCs can also be isolated from normal as well as injured adult livers and maintained in culture for long term, although their role in vivo remains elusive (Miyajima et al., 2014).
It has been reported that LPC-like cells were established from hESCs/hiPSCs (Takayama et al., 2013, Yanagida et al., 2013, Zhao et al., 2009), and these cells were shown to proliferate and differentiate into hepatocyte-like cells or cholangiocyte-like cells. These LPCs were either isolated by cell sorting using a combination of specific cell surface markers or generated by adenovirus-mediated gene transfer to promote hepatic lineage differentiation. To develop an efficient culture system for large-scale production of mature functional hepatocytes, our aim was to identify a specific cell surface marker for isolating hiPSC-derived LPCs. In this study, we identified carboxypeptidase M (CPM) as a cell surface marker for hepatoblasts. CPM was also upregulated in hiPSC-derived cells during hepatic differentiation, and the sorted CPM+ cells exhibited features typical of hepatoblasts. Moreover, we developed a highly efficient and reliable culture system for hiPSC-derived LPCs capable of proliferating and differentiating into both hepatocytes and cholangiocytes in vitro.
Results
Identification of CPM as a Hepatoblast Marker
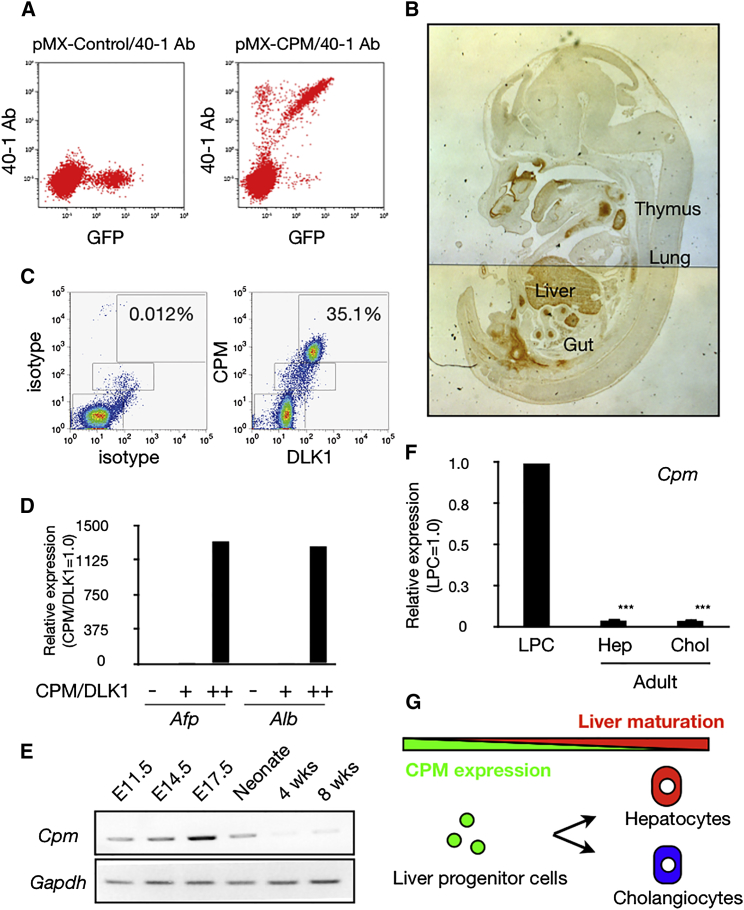
In order to isolate LPCs from hiPSCs effectively, we searched for cell surface molecules expressed in hepatoblasts. Although CXCR4 is known to be expressed in hepatoblasts, it is also detected in endodermal progenitors, thus implying that additional markers would be required to isolate LPCs. DLK1 is an excellent marker for hepatoblasts and has been extensively used to isolate hepatoblasts. However, although immunocytochemical staining using an anti-DLK1 antibody revealed that DLK1-expressing cells were present in hiPSC-derived cells at the immature hepatocyte stage (Figure S1A), flow cytometric (FCM) analysis showed no expression of DLK1 on the cell surface (Figure S1B). We therefore searched for other hepatoblast cell surface molecules. Among 627 monoclonal antibodies established against mouse fetal liver cells (Suzuki et al., 2008), we previously found that 40-1 antibody binds to an unidentified cell surface protein expressed on mouse hepatoblasts (Tanaka et al., 2009). By employing the expression cloning procedure, we identified CPM as the specific antigen for the 40-1 antibody (Figure 1A). Immunohistochemical studies showed that CPM is mainly expressed in endodermal tissues such as liver, thymus, lung, and gut in mouse fetus at E14.5 (Figure 1B), confirming the previous studies that CPM is widely expressed in endodermal tissues during mouse development (Tamplin et al., 2008).
Figure 1.
CPM Expression on Mouse Hepatoblasts
(A) FCM analysis of Ba/F3 cells (control) (left) and Ba/F3 cells expressing CPM (right). GFP-positive CPM expressing cells were identified by using the 40-1 antibody.
(B) Localization of CPM (brown) in E14.5 fetal mouse sagittal section.
(C) FCM analysis of mouse fetal liver cells at E14.5 using antibodies against CPM and DLK1.
(D) Relative Afp and Alb expression in the CPM/DLK1neg (−), CPM/DLK1low (+) and CPM/DLK1high (++) fractions as indicated in (C). n = 1 in each group (each experiment contains two technical replicates).
(E) RT-PCR analysis shows the expression of Cpm during mouse liver development (E11.5, E14.5, E17.5, Neonate, 4 weeks, 8 weeks). The product sizes of Cpm and Gapdh are 430 bp and 600 bp, respectively. Amplification of Gapdh was used as an internal control.
(F) Relative Cpm expression in liver progenitor cells (LPC), Hep (mature hepatocytes), and Chol (mature cholangiocytes). The results are shown as the mean ± SEM of four independent experiments. (each experiment contains two technical replicates) ∗∗∗p < 0.001.
(G) Correlation between the expression of CPM and the hepatic maturation during liver development.
Next, we performed FCM analysis/cell sorting to confirm the expression of CPM in mouse hepatoblasts. CPM was coexpressed with a hepatoblast marker, DLK, and CPM+ cells isolated from E14.5 fetal liver also highly expressed LPC markers such as alpha-1-fetoprotein (Afp) and albumin (Alb) (Figures 1C and 1D). Cpm was highly expressed in fetal liver, but its expression was dramatically decreased after birth (Figure 1E) and was undetectable in mature hepatocytes and cholangiocytes (Figure 1F). Collectively, these data demonstrated that CPM is strongly expressed in hepatoblasts and fetal LPCs (Figure 1G) and also suggested that CPM may be a useful marker for enrichment of LPCs from differentiating hiPSCs.
CPM Expression during Hepatic Differentiation from Human iPSCs
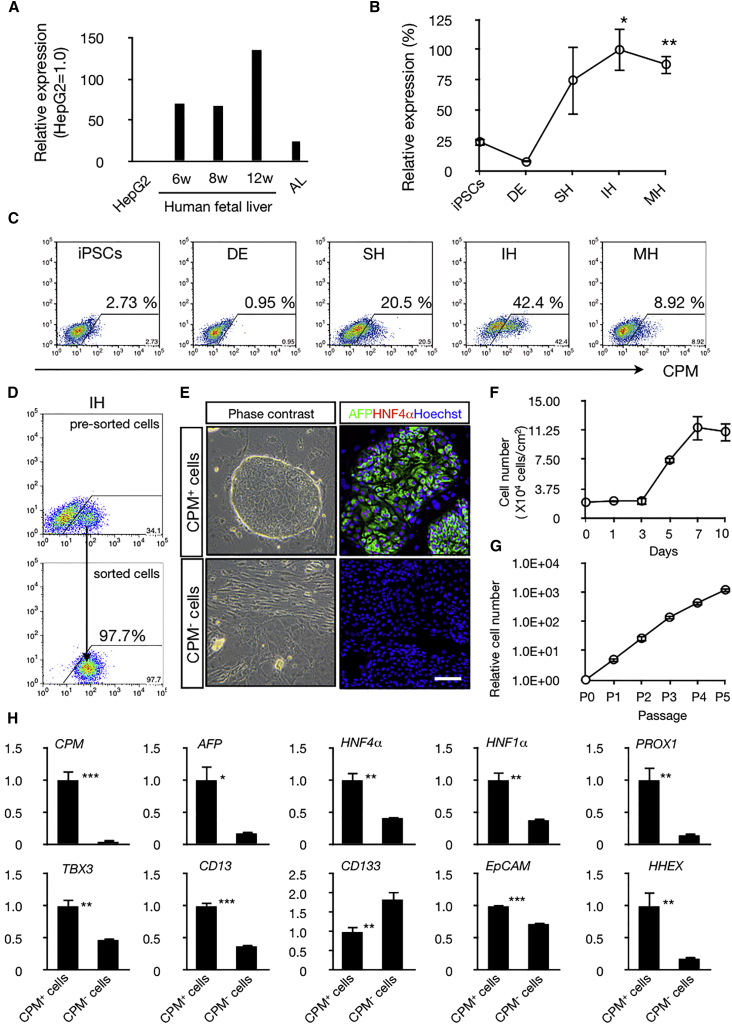
To evaluate whether CPM could be used as a marker for enrichment of LPCs from hiPSCs, we first analyzed the expression levels of CPM in human fetal liver. CPM was highly expressed in human fetal liver from 6 to 12 weeks of gestation, but was expressed at very low levels in adult liver and HepG2 cells, a human hepatocellular carcinoma cell line (Figure 2A). We then examined the CPM expression profile during hepatic differentiation from hiPSCs by qRT-PCR and FCM analysis. As described in the previously reported protocol, we induced hepatic differentiation in two hiPSC lines (454E2 and 409B2). hiPSCs showed morphological changes after induction toward the hepatic lineage (Figure S2A), with rapid downregulation of OCT4 and sequential induction of GATA4, SOX17, FOXA2, and HNF4α (Figures S2B and S2C). In this culture system, the expression level of CPM was undetectable in undifferentiated hiPSCs, but upregulated together with hepatic progenitor markers such as FOXA2 and HNF4α during differentiation (Figures 2B, S2B, and S2C). FCM analysis revealed that 20% of specified hepatic cells were CPM+, with this population increasing to ∼40% after differentiation to immature hepatocytes. CPM expression decreased along with subsequent hepatic maturation (Figure 2C). Collectively, these data showed that CPM is a specific cell surface marker for human iPSC-derived LPCs.
Figure 2.
Enrichment of the hiPSC-Derived LPCs Based on the Expression of CPM
(A) Relative CPM expression in HepG2 cells and liver tissues (adult liver, gestational ages: 6 weeks, 8 weeks, 12 weeks). n = 1 in each group (each experiment contains two technical replicates).
(B and C) CPM mRNA and protein expression were analyzed by qRT-PCR (B) and FCM (C). iPSCs, iPS cells; DE, definitive endoderm; SH, specified hepatic; IH, immature hepatocytes; MH, mature hepatocytes. For (B), error bar represents the mean ± SEM of three independent experiments. ∗p < 0.05 between iPSCs and IH, ∗∗p < 0.01 between iPSCs and MH. See also Figures S2B and S2C.
(D) FCM analysis of CPM expression was performed in pre- and post-sorted cells.
(E) Morphology of the CPM+ cells (left upper) and CPM− cells (left lower) on MEF feeder cells. Cells were cultured for 4 days. Immunohistochemical staining for AFP (green) and HNF4α (red) in CPM+ cells (right upper) and CPM− cells (right lower). Nuclei were visualized by Hoechst 33342 staining (blue). Scale bar, 100 μm.
(F) Growth curve of CPM+ cells. Each value was determined in triplicate. Error bar represents the mean ± SEM of three independent experiments.
(G) Relative cell number after several passages. Error bar represents the mean ± SEM of four independent experiments.
(H) Expression of hepatoblast markers in CPM+ cells compared with CPM− cells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. The results are shown as the mean ± SEM of eight independent experiments. (each experiment contains two technical replicates).
Enrichment of hiPSC-Derived LPCs Based on the Expression of CPM
To further characterize the CPM+ cells derived from hiPSCs, we used a magnetic cell sorter to isolate the cells of interest and established a culture system to expand them. The purity of enriched CPM+ cells was over 97% after isolation by single step sorting using a magnetic cell sorter (Figure 2D). CPM+ cells formed compact colonies on MEF feeder cells and exhibited epithelial-like morphology, whereas no such colonies were formed from CPM− cells (Figure 2E). CPM+ cells exhibited a high proliferative capacity and grew to confluence by day 7 after seeding (Figure 2F). These cells continued to expand after several passages in vitro (Figure 2G). Immunocytochemistry demonstrated that all CPM+ cells strongly expressed AFP and HNF4α, a key transcription factor for LPCs (Figure 2E). These cells could also be cultured even after several passages (Figure S2D) or cryopreservation (Figure S2E). In addition to AFP and HNF4α, hepatoblast markers HNF1α, PROX1, TBX3, CD13, EpCAM, and HHEX were significantly enriched in CPM+ cells compared with CPM− cells (Figure 2H). In contrast, CD133, a cholangiocyte marker, was significantly expressed in CPM− cells. All these data strongly suggested that CPM is a useful cell surface marker to enrich for LPCs in hiPSC-derived immature hepatic cells. Similar results were obtained from a different hiPSC line (409B2) (Figure S2F), indicating that the efficiency of our method for generating CPM+ LPCs is not cell line-dependent.
Differentiation of Hepatocytes and Cholangiocytes from CPM+ LPCs
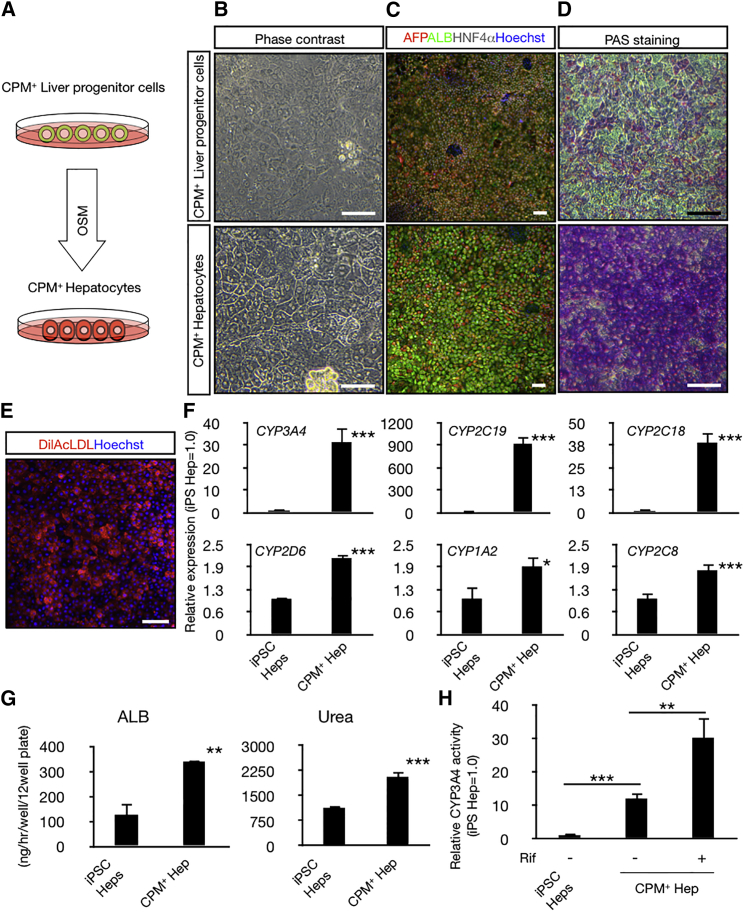
It is well established that LPCs differentiate into both hepatocytes and cholangiocytes. Therefore, we evaluated the differentiation potential of CPM+ LPCs. After expansion of the CPM+ population by serial passages on MEF feeder cells, these cells were then differentiated into hepatocytes by addition of Oncostatin M (Figure 3A). As shown in Figure 3B, the hepatocytes from CPM+ LPCs showed typical human primary hepatocyte morphology, with binuclear cells delineated distinctly by bright borders. In addition, these cells exhibited high level expression of ALB, accumulation of glycogen and uptake of Dil-Ac-LDL (Figures 3C–3E), indicative of mature hepatocytes. FCM analysis showed that almost all differentiated cells expressed ALB (Figure S3A). The levels of hepatic gene expression such as CYP3A4, CYP2C19, CYP2C18, CYP2D6, CYP1A2, and CYP2C8 in hepatocyte-like cells derived from CPM+ LPCs were much higher than those derived from hiPSCs by the conventional differentiation protocol (Figure 3F). Furthermore, hepatocytes derived from CPM+ LPCs secreted high amounts of ALB and urea into the culture medium (Figure 3G) and exhibited high CYP3A4 activity (Figure 3H). In addition, CYP3A4 activity was significantly induced in response to rifampicin treatment, which is a well-known CYP3A4 inducer (Figure 3H). Collectively, hepatocytes derived from CPM+ cells exhibited higher metabolic activity compared to those derived from hiPSCs using a conventional protocol.
Figure 3.
Differentiation of Hepatocytes from hiPSC-Derived CPM+ LPCs
(A) Schematic image of hepatocyte-like cell differentiation from CPM+ LPCs.
(B–D) The morphology of CPM+ LPCs (upper) and CPM+ hepatocyte-like cells (lower) was investigated by microcopy. (B) Phase contrast images. CPM+ LPCs exhibit light cytoplasm and indistinct cell borders. CPM+ hepatocyte-like cells exhibit cobblestone-like morphology with binucleation. (C) Immunohistochemistry for AFP (red), ALB (green) and HNF4α (gray). Nuclei were counterstained with Hoechst 33342 (blue). (D) PAS staining showed accumulation of glycogen. Scale bars, 100 μm.
(E) Uptake of DilAcLDL (red) in CPM+ hepatocyte-like cells. Scale bar, 100 μm.
(F) qRT-PCR analysis of various CYP450s mRNA levels. The results are shown as the mean ± SEM of six independent experiments. (each experiment contains two technical replicates). iPSC-Heps was used as a control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) Secretion of ALB and urea. The results are shown as the mean ± SEM of seven independent experiments. iPSC-Heps was used as a control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(H) Relative CYP3A4 activity. The results are shown as the mean ± SEM of at least three independent experiments. Treatment with 10 μM of rifampicin (Rif) for 72 hr. iPSC-Heps was used as a control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
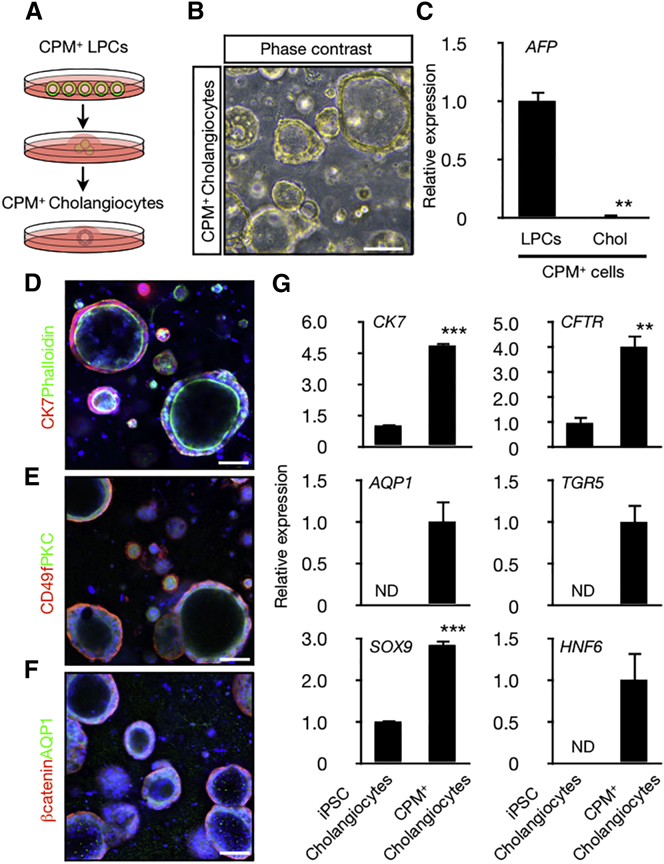
Furthermore, CPM+ LPCs were converted to cholangiocytes, which formed cysts with the luminal structure in vitro after culturing in a gel consisting of collagen and Matrigel (Figures 4A, 4B, S4A, and S4B). The expression of AFP was dramatically decreased after differentiation (Figure 4C). Moreover, immunohistochemistry showed that F-actin, PKC, and AQP1 localized to the apical membrane, whereas CD49f was detected in the basolateral membrane in cystic cells (Figures 4D–4F), thus showing the proper apico-basal structure. In this culture system, cells derived from CPM+ LPCs highly expressed cholangiocyte marker genes, such as CK7, CFTR, AQP1, TGR5, SOX9, and HNF6, compared with iPSC-derived cholangiocytes (without CPM purification process) (Figure 4G). These results indicate that CPM+ LPCs can differentiate into both hepatocyte-like cells and cholangiocyte-like cells.
Figure 4.
Differentiation of Cholangiocytes from hiPSC-Derived CPM+ LPCs
(A) Schematic image of cholangiocyte-like cells differentiation from CPM+ LPCs.
(B) Phase contrast image. CPM+ LPCs form cysts in collagen/Matrigel after 7 days of culture. See also Figures S4A and S4B.
(C) Expression of AFP in CPM+ LPCs and CPM+ cholangiocytes. The results are shown as the mean ± SEM of four independent experiments (each experiment contains two technical replicates). ∗∗p < 0.01.
(D–F) Immunofluorescence staining for cholangiocyte markers in CPM+ cholangiocyte-like cells. Localization of (D) CK7 (red), F-actin (green), (E) CD49f (red), PKC (green), (F) β-catenin (red), AQP1 (green). Nuclei were visualized by Hoechst 33342 staining (blue). Scale bars, 100 μm.
(G) Gene expression profile of CPM+ cholangiocyte-like cells compared with iPSC cholangiocyte-like cells. The results are shown as the mean ± SEM of four independent experiments (each experiment contains two technical replicates). ND, not detected. ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Generation of mature hepatocytes and cholangiocytes from iPS/ES cells requires time-consuming multiple processes with expensive cytokines. Therefore, it would be beneficial to derive expandable precursor cells to simplify the procedure. Because LPCs are able to proliferate extensively in vitro and differentiate to both hepatocytes and cholangiocytes (Suzuki et al., 2000, Tanimizu et al., 2003, Tanimizu et al., 2007), they are ideal cells to use in developing an efficient protocol for large-scale production of mature liver cells. In order to isolate LPCs, we first tested the expression of DLK1 and CXCR4, which are well known to be expressed in hepatoblasts. However, neither of these markers was appropriate for enriching LPCs from hiPSC-derived cells, as described above. We therefore searched for another marker and showed that CPM is a cell surface antigen expressed on hepatoblasts in mouse fetal liver between E11.5 and E17.5, although its expression is dramatically downregulated in mature hepatocytes and cholangiocytes. CPM is a member of the carboxypeptidase family, expressed on the cell surface to catalyze the release of C-terminal arginine or lysine residues of peptides and proteins (Skidgel et al., 1989). While its role in hepatoblasts is currently unknown, we found that the expression of CPM was gradually upregulated along with hepatic differentiation from hiPSC-like liver development. The simple method of single step cell sorting based on CPM expression made it possible to enrich the LPC fraction after induction of hiPSCs to the immature hepatocyte stage. These CPM+ cells exhibited a high proliferative potential and the expanded cells could be cryopreserved. Moreover, they expressed various liver progenitor markers (Figure 2H). While most CPM+ cells expressed HNF4α (Figure 2E), expression of midgut/hindgut markers such as CDX2 and PDX1 was also detected by RT-PCR (data not shown), suggesting that CPM+ cells may contain non-liver progenitors. However, after induction of hepatic differentiation, almost all cells became hepatocytes as shown by their morphology and ALB expression (Figures 3B, 3C, and S3A). If such non-hepatic progenitors were present in the CPM+ cells, they did not affect hepatocyte differentiation.
CPM+ LPCs were able to differentiate in a single step culture to either hepatocyte-like cells or cholangiocyte-like cells depending on the culture condition. Furthermore, hepatocytes derived from CPM+ cells exhibited a significantly higher level of metabolic activity compared to the hiPSC-derived hepatocytes using a conventional differentiation protocol. Importantly, these hepatocyte-like cells remain phenotypically stable for more than 2 weeks (Figure S3B). Thus, CPM+ LPCs derived from hiPSCs will be useful for developing a reliable long-term hepatocyte culture system, and this simplified purification method will contribute to basic and clinical research related to liver diseases. Although CPM+ hepatocytes highly expressed mature hepatic genes involved in glucogenesis (G6PC, PCK1) and the urea cycling (CPS1), they exhibited variable levels of CYP expression compared with cultured primary human hepatocytes (Figure S3C). It is well known that the capacity to metabolize drugs is variable due to genetic polymorphisms in CYPs (Ingelman-Sundberg et al., 2007). Hepatocyte-like cells differentiated from iPSCs are highly variable due to retention of donor-specific metabolic capacity (Takayama et al., 2014), suggesting that the expression of CYPs in CPM+ Hepatocytes may be affected by a donor’s genetic background.
Because freshly isolated hepatocytes rapidly lose their functions, it is a major challenge to generate fully functional hepatocytes from pluripotent stem cells. While hepatocytes derived from CPM+ LPCs expressed high levels of metabolic activity, the levels of some proteins are not as high as primary human hepatocytes and there is still room for improvement. During embryogenesis, hepatoblasts differentiate into mature hepatocytes through interactions with non-parenchymal cells. As non-parenchymal cells are in direct contact with hepatoblasts and also produce various cytokines to induce hepatic maturation, co-culture of CPM+ hepatocytes with non-parenchymal cells may be an effective way to generate more mature hepatocytes from hiPSCs, and we are currently investigating this possibility.
Experimental Procedures
Cell Culture
Two hiPSC lines (454E2 and 409B2) were provided by RIKEN Cell Bank (Okita et al., 2011). These cells were maintained on mitomycin C-treated (Wako Pure Chemical Industries) mouse embryonic fibroblast (MEF) feeder cells, and hepatic differentiation of hiPSCs was induced using the four-step protocol previously reported, with minor modifications (Si-Tayeb et al., 2010).
Isolation and Expansion of LPCs Derived from hiPSCs
After induction of hiPSCs to the immature hepatocyte stage, cells were sorted using a MoFlo XDP cell sorter (Beckman Coulter) or autoMACS Pro Separator (Miltenyi Biotech) into CPM+ and CPM− populations. To expand hiPSC-derived CPM+ cells, we modified the published method (Chen et al., 2007, Huch et al., 2013, Yanagida et al., 2013) as follows: sorted cells were cultured on mitomycin c-treated MEF feeder cells (2.0 × 104 cells/cm2) in DMEM-F12 (Sigma-Aldrich) supplemented with 10% FBS (JRH Biosciences), penicillin-streptomycin-glutamine, ITS, N-2 supplement, MEM non-essential amino acids solution, L-glutamine (Life Technologies), ascorbic acid (1 mM), nicotinamide (10 mM), N-acetylcysteine (0.2 mM) (Sigma-Aldrich), dexamethasone (1 × 10−7 M), HGF (20 ng/ml), EGF (10 ng/ml) (PeproTech), Y-27632 (5 μM) (Wako), and A83-01 (2.5 μM) (Tocris).
Differentiation of Hepatocytes and Cholangiocytes from CPM+ LPCs
To induce hepatic maturation of CPM+ LPCs, confluent cells were incubated in Hepatocyte Basal Medium (Lonza) supplemented with HCM SingleQuots (excluding EGF) and Oncostatin M (20 ng/ml) (PeproTech) for 5–10 days as described previously (Si-Tayeb et al., 2010). Induction of cholangiocyte differentiation was performed using the three-dimensional gel culture method previously reported (Tanimizu et al., 2007, Yanagida et al., 2013) with minor modifications. After expansion of CPM+ LPCs, the resulting cells were collected and re-suspended in a gel composed of 2:3 mixture of growth factor reduced Matrigel (Corning) and collagen type I (Nitta Gelatin) at a density of 1 × 105 cells/50 μl. Cell suspensions were then added to a 24-well plate (Corning) and incubated for 2 hr at 37°C until solidification. The cells were then cultured in the presence of R-spondin-1 (40 ng/ml) and WNT-3a (40 ng/ml) (PeproTech) for 7 days.
Additional details of experimental procedures are available in the Supplemental Information.
Author Contributions
T.K. designed the study, performed experiments, analyzed data, and wrote the manuscript. Y.K., K.S., A.K., Y.M., and E.C. performed experiments and analyzed data. M.T. designed the study, performed experiments, and analyzed data. A.M. designed the study, analyzed data, and wrote the manuscript.
Acknowledgments
We thank Drs. Yasuyuki Sakai (CIBiS, Institute of Industrial Science, The University of Tokyo) and Takahiro Ochiya (Division of Molecular and Cellular Medicine, National Cancer Center Research Institute) for helpful discussions, and Dr. Cindy Kok for critical review of the manuscript. This study was supported by CREST program of Japan Science and Technology Agency, and Grants-in-Aid for Scientific Research of Japan Society for the Promotion of Science.
Published: September 10, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.08.008.
Contributor Information
Taketomo Kido, Email: kido@iam.u-tokyo.ac.jp.
Atsushi Miyajima, Email: miyajima@iam.u-tokyo.ac.jp.
Supplemental Information
References
- Chen Q., Kon J., Ooe H., Sasaki K., Mitaka T. Selective proliferation of rat hepatocyte progenitor cells in serum-free culture. Nat. Protoc. 2007;2:1197–1205. doi: 10.1038/nprot.2007.118. [DOI] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M., Sim S.C., Gomez A., Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Miyajima A., Tanaka M., Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Ogawa S., Surapisitchat J., Virtanen C., Ogawa M., Niapour M., Sugamori K.S., Wang S., Tamblyn L., Guillemette C., Hoffmann E. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 2013;140:3285–3296. doi: 10.1242/dev.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidgel R.A., Davis R.M., Tan F. Human carboxypeptidase M. Purification and characterization of a membrane-bound carboxypeptidase that cleaves peptide hormones. J. Biol. Chem. 1989;264:2236–2241. [PubMed] [Google Scholar]
- Suzuki A., Zheng Y., Kondo R., Kusakabe M., Takada Y., Fukao K., Nakauchi H., Taniguchi H. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230–1239. doi: 10.1053/jhep.2000.20349. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Tanaka M., Watanabe N., Saito S., Nonaka H., Miyajima A. p75 Neurotrophin receptor is a marker for precursors of stellate cells and portal fibroblasts in mouse fetal liver. Gastroenterology. 2008;135:270–281.e3. doi: 10.1053/j.gastro.2008.03.075. [DOI] [PubMed] [Google Scholar]
- Takayama K., Inamura M., Kawabata K., Sugawara M., Kikuchi K., Higuchi M., Nagamoto Y., Watanabe H., Tashiro K., Sakurai F. Generation of metabolically functioning hepatocytes from human pluripotent stem cells by FOXA2 and HNF1α transduction. J. Hepatol. 2012;57:628–636. doi: 10.1016/j.jhep.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Takayama K., Nagamoto Y., Mimura N., Tashiro K., Sakurai F., Tachibana M., Hayakawa T., Kawabata K., Mizuguchi H. Long-term self-renewal of human ES/iPS-derived hepatoblast-like cells on human laminin 111-coated dishes. Stem Cell Reports. 2013;1:322–335. doi: 10.1016/j.stemcr.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Morisaki Y., Kuno S., Nagamoto Y., Harada K., Furukawa N., Ohtaka M., Nishimura K., Imagawa K., Sakurai F. Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes. Proc. Natl. Acad. Sci. USA. 2014;111:16772–16777. doi: 10.1073/pnas.1413481111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin O.J., Kinzel D., Cox B.J., Bell C.E., Rossant J., Lickert H. Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics. 2008;9:511. doi: 10.1186/1471-2164-9-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Okabe M., Suzuki K., Kamiya Y., Tsukahara Y., Saito S., Miyajima A. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mech. Dev. 2009;126:665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- Tanimizu N., Nishikawa M., Saito H., Tsujimura T., Miyajima A. Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J. Cell Sci. 2003;116:1775–1786. doi: 10.1242/jcs.00388. [DOI] [PubMed] [Google Scholar]
- Tanimizu N., Miyajima A., Mostov K.E. Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Mol. Biol. Cell. 2007;18:1472–1479. doi: 10.1091/mbc.E06-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida A., Ito K., Chikada H., Nakauchi H., Kamiya A. An in vitro expansion system for generation of human iPS cell-derived hepatic progenitor-like cells exhibiting a bipotent differentiation potential. PLoS ONE. 2013;8:e67541. doi: 10.1371/journal.pone.0067541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S., Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Chen S., Cai J., Guo Y., Song Z., Che J., Liu C., Wu C., Ding M., Deng H. Derivation and characterization of hepatic progenitor cells from human embryonic stem cells. PLoS ONE. 2009;4:e6468. doi: 10.1371/journal.pone.0006468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.