Abstract
White-nose syndrome (WNS), an emerging disease of hibernating bats, has rapidly spread across eastern North America killing millions of bats. Pseudogymnoascus destructans (Pd), the sole etiologic agent of WNS, is widespread and persistent in bat hibernacula. Control of Pd in the affected sites is urgently needed to break the transmission cycle while minimizing any adverse impact on the native organisms. We isolated a novel strain of Trichoderma polysporum (Tp) from one of the caves at the epicenter of WNS zoonotic. Detailed experimental studies revealed: (1) Tp WPM 39143 was highly adapted to grow at temperatures simulating the cave environment (6°C-15°C), (2) Tp WPM 39143 restricted Pd colony growth in dual culture challenges, (3) Tp WPM 39143 caused four logs reduction of Pd colony forming units and genome copies in autoclaved soil samples from one of the WNS affected caves, (4) Tp WPM 39143 extract showed specific fungicidal activity against Pd in disk diffusion assay, but not against closely related fungus P. pannorum (Pp), (5) Tp WPM 39143 extract retained inhibitory activity after exposure to high temperatures, light and proteinase K, and (6) Inhibitory metabolites in Tp WPM 39143 extract comprised of water-soluble, high polarity compounds. These results suggest that Tp WPM 39143 is a promising candidate for further evaluation as a biocontrol agent of Pd in WNS affected sites.
Introduction
White-nose Syndrome (WNS), caused by Pseudogymnoascus destructans (Pd), has devastated bat populations across eastern North America for almost a decade [1–7]. A number of bat species are affected with the most vulnerable being little brown bats (Myotis lucifugus), Indiana bats (M. sodalis), northern long-eared bats (M. septentrionalis), and tricolored bats (Perimyotis subflavus) [5, 8]. Pd invades the skin of hibernating bats including muzzles, ears, and wings. The affected animals are noticeably emaciated, and many fail to survive hibernation [9–11].
The underlying mechanisms of Pd infectivity and its precise ecological niche are largely unknown. Historically, Pd appears to be well established in European caves, and European bats harboring the fungus exhibit pathology similar to that seen in North American bats, with no recorded mass mortality [12–14]. To date, all investigations of Pd indicate that it is well adapted to cold conditions and has a clonal population in the US [15, 16]. Several DNA and culture based studies have revealed wide distribution and persistence of Pd in sediment and swab samples from bat hibernacula [12, 17–19]. Other published cave fungal surveys indicate that Pd could survive and persist in bat hibernacula for prolonged periods, and this has a dramatic impact on both disease management and epidemiology [19]. Recently, a mycobiome study found a diversity of fungi inhabiting WNS caves and mines and signs of local adaptation by Pd [20]. Further studies are needed to understand the pathogenesis of WNS and to develop control strategies that include alleviation or remediation of Pd to break the transmission cycle, especially by reducing the fungal burden available for new infections. Recently, some bacteria have been proposed as biocontrol agents, including naturally occurring bacteria [21] or volatiles produced by bacteria [22, 23].
There is a vast volume of literature on the biocontrol potential of Trichoderma spp. and several highly effective preparations incorporating these fungi are now approved for commercial use for the biocontrol of agriculture pests in the US and other parts of the world [24, 25]. The most important biocontrol agents against plant pathogenic fungi are Trichoderma virens von Arx, Beih [26, 27], T. harzianum Rifai [28, 29], T. viride Pers:Fr [30], and T. atroviride sc1 [31]. Optimum growth temperatures differ among Trichoderma spp. and previous studies have largely focused on mesophilic groups (22°C~35°C) [24, 32–34]. It has been found that Trichoderma spp. did not protect germinating seeds from soil-borne diseases caused by cold-tolerant phytopathogenic fungi during cold and wet autumn and spring seasons [35, 36]. To date, there is relatively little information available about psychrotolerant Trichoderma spp. and their biocontrol potentials. Trichoderma polysporum was shown to have biocontrol activity outside of caves against a variety of other fungi including Armilaria gallica, Fomus annosus, and Ceratocystis paradoxa [37–39].
The present study describes a psychrotolerant strain of Trichoderma polysporum (Tp) WPM 39143, which was isolated from a cave at the epicenter of the WNS zoonotic. Since T. polysporum WPM 39143 grew relatively well at low temperatures common in the cave environment [40], the objective of this investigation was to determine if it exerted inhibitory activity against Pd in laboratory media and soil matrices.
Materials and Methods
Fungal strains and media
Trichoderma polysporum (Tp), recently isolated from air sample 39143 from William Preserve Mine (WPM), NY [20], Trichoderma harzianum (Th) Rifai strain T22 purchased from BioWorks, Inc. Victor, NY, USA, T. atroviride (Ta) obtained from ARS fungal collection, Cornell University, NY, USA, Pseudogymnoascus destructans (Pd) M1379 and Pseudogymnoascus pannorum (Pp) M1372 [41] were part of this investigation. All fungi were maintained on potato dextrose agar (PDA, Difco) slants at 15°C. Fungal cultures were also stored in 15% glycerol at -70°C. Other media used were Sabauraud dextrose agar (SDA) with or without cycloheximide (400 μg/ml), rice fermentation medium (100 g rice in 150 ml of distilled water) [42], and yeast extract peptone dextrose (YPD) agar. All the experiments were performed in the biosafety cabinet 2.
Phylogenetic analysis of Tp WPM 39143
Genomic DNA from Tp WPM 39143 was extracted using thermolysis phenol extraction protocol as described in a published study [43]. In brief, approximately 5x5 mm of fungal culture was removed from the agar surface, placed in 300 μl of modified genomic DNA extraction buffer (100 mM Tris [pH 8.0], 10 mM EDTA, 2% SDS, 1.4 M NaCl, 1% CTAB, 0.4 μg/ml proteinase K), and the mixture incubated at 65°C for 1 h followed by chloroform:isoamyl alcohol (24:1) extraction, precipitation with isopropanol and washing with 70% ethanol. The precipitated DNA was centrifuged at 12,000 RPM and the resulting pellet was dried and finally dissolved in 50 μl of Tris-EDTA (TE) buffer. The extracted DNA was amplified for the internal transcribed spacer (ITS) and D1/D2 regions of the ribosomal gene with primer set ITS1-ITS4 and NL1-NL4, respectively [44]. The PCR was performed with proof reading KlenTaq DNA polymerase (Sigma-Aldrich, St. Louis, MO, USA) with initial denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 2 min and the final extension at 68°C for 10 min. The PCR amplicons were sequenced, assembled, and edited using Sequencher 4.6 software (Gene Codes Corp., Ann Arbor, MI, USA) and BLAST searched against two databases: GenBank (www.ncbi.nlm.nih.gov/) and CBS-KNAW (www.cbs.knaw.nl/).
Multiple alignments of ITS and D1/D2 sequences of Tp WPM 39143 and selected GenBank sequences were done using the CLUSTALX 1.81 [45] and MAFFT programs [46]. A phylogenetic analysis of the aligned sequences was done using the neighbor-joining (NJ) method with 1,000 bootstrap replicates using MEGA 5.1 [47]. The Dictionary of the Fungi and UniProt (http://www.uniprot.org/taxonomy/) served as the source of taxonomic references for fungal species [48].
Growth optima of Tp WPM 39143
In order to assess the biocontrol potential, we first compared the growth temperature range of Tp WPM 39143 with that of two well -characterized biocontrol fungi Th and Ta. These strains were point inoculated on the center of PDA and YPD agar plates, and cultures were incubated at 6, 10, 15, and 22°C. The diameter of the fungal colony was measured at 9 and 20 days post-incubation.
Dual culture challenge studies of Pd and Tp WPM 39143
The dual challenge experiments were performed on the laboratory medium (PDA) and soil matrices. Approximately 15 μl of Pd conidial suspension (107 conidia/ml) was inoculated close to the edge on one side of a PDA agar plate. Following incubation for 10 days at 15°C, approximately 15 μl of conidial suspension (105 cells/ml) of Tp WPM 39143 was placed on the opposite side of the plates. Similarly, a conidial suspension of Th (105 cells/ml) was also placed on the opposite side of the Pd culture plate in a separate experiment. The interactions between Pd and Tp or Th were assessed at 2 and 4 weeks post-inoculation at 15°C. The Pd culture alone served as a control.
For the dual challenge experiment simulating a natural setting, one soil sample (dark black in color with fine granular consistency) collected from Aeolus Cave on September 18, 2013, was used. In brief, the soil sample was weighed, and 3 grams placed into sterile 10-ml screw capped Econo Glass VialsTM (PerkinElmer, Waltham, MA, USA). Vials were autoclaved at 15 psi for 30 min. The pH of the autoclaved samples was determined by pulverizing the soil sample with 10 ml of sterilized distilled water and placing a drop of this suspension on to pH indicator strips ranging from pH 0–6 and pH 5–10. A loopful of autoclaved soil sample was streaked onto YPD agar plates, and the plates were incubated at 15°C for one week to confirm sterility.
Three vials, each containing 3 g of soil sample, were inoculated with 100 μl of Pd conidial suspension (3×105 conidia/ml) to get final conidia counts of 1 x 104 conidia/g of soil. The contents of the vials were thoroughly mixed with a sterile spatula, and these vials were incubated for one week at 15°C. Following incubation, each Pd-containing vial was seeded with either 100 μl of Tp WPM 39143 or Th conidial suspension (3×105 conidia/ml) to get final conidia counts of 1 x 104 conidia/g of soil. One Pd-containing vial was also inoculated with 100 μl of sterile distilled water, which served as a control. All the vials were incubated at 15°C for an additional 35 days.
Recovery of Pd from dual culture challenge
Following incubation, approximately 1 g of soil sample was removed from each vial, transferred to an autoclaved mortar and pestle and mixed gently to get a homogenous mixture. The samples were weighed, and approximately 100 mg aliquots of homogenous soil sample were transferred into six 2-ml screw cap vials. Three vials each were processed for Pd recovery by culture-dependent (CD) and culture-independent (CI) methods as shown in the flowchart (S1 Fig).
For the CD method, the content of each vial was mixed with 500 μl of sterile distilled water, vortexed vigorously for 2 minutes and the tubes were left in an up-right position for 5 minutes to allow settling of soil particles. Approximately, 200 μl of supernatant from each vial was removed and transferred to a new sterile vial. Ten-fold dilutions of the supernatant were prepared, and 100 μl of each dilution was inoculated on to SDA plates with or without cycloheximide in triplicates for the recovery of Pd, Tp WPM 39143, and Th. The culture medium selection was based upon our preliminary experiments wherein Pd was found to be resistant to cycloheximide while Tp WPM 39143 and Th showed 100% growth inhibition (details not shown). All the plates were incubated at 15°C for 7 to 30 days. The Pd colonies were counted, and results were expressed as log colony-forming unit (CFU) per gram of soil. The percent Pd growth inhibition by Tp WPM 39143 or Th was calculated as: 1-(Pd CFU experiment/Pd CFU control) x 100.
For the CI method, the remaining three vials, each containing 100 mg of soil sample with Pd alone or Pd with Tp WPM 39143 or Th, were subjected to genomic DNA (gDNA) extraction using SoilMasterTM DNA Extraction Kit (Epicentre, Chicago, IL, USA). The autoclaved pre-treated soil sample was also processed for DNA extraction to assess if it contained any Pd gDNA. The extracted DNA from all sample types was suspended in 30 μl of Tris-EDTA (TE) buffer and Pd gDNA in these samples were quantitated using real-time PCR assays targeting alpha-L-rhamnosidase gene (ALR) and intergenic spacer region (IGS) of the rRNA gene complex in an IQ5 real-time PCR detection system (Bio-Rad, Hercules, CA), as described previously [49, 50]. In brief, each reaction mixture contained 1× LightCycler Fast Start DNA master hybridization probe mix (Roche Applied Science, Indianapolis, IN, USA), 4 mM MgCl2, 1 μM concentration of each primer, 0.25 μM concentration of each probe, and 2 μl of soil DNA in a final volume of 20 μl. The soil sample was also checked for PCR inhibitors by spiking with 1 ng of Pd gDNA. The PCR cycling conditions were 10 min at 95°C followed by 15 s at 95°C and 60 s at 60°C for 40 cycles. Each sample was tested in triplicate, and the results were averaged to obtain the cycle threshold (Ct)—i.e., the point at which sample fluorescence rises above the background level. A Ct value of >40 was considered negative while a Ct value of ≤40 was considered positive. Each sample was also run with a non-DNA template (NTC) as a negative control. The dilution series of gDNA from a pure culture of Pd (M1379) was used for the generation of standard curve. The average Ct counts equivalent to DNA quantity was extrapolated from the standard curve, and then Pd genome copies was calculated using formula:
(copies = DNA quantity (ng)x(6.022x1023)/genome length (bp)x(1x109)x650 in which the 6.022x1023 molecules/mole is Avogadro's number, average weight of a base pair (bp) is 650 Daltons and Pd genome length is 30.49 Mb. More details of the formula used are available online (http://cels.uri.edu/gsc/cndna.html). The Pd genome copies obtained was calculated per gram of soil. Since IGS rRNA gene complex in fungi is a multi-copy gene [51, 52] and IGS PCR was 100 fold more sensitive than the single copy ALR PCR [50], the IGS was arbitrarily calculated as 100 copies per Pd genome. The autoclaved pre-treated soil sample was weakly positive for Pd gDNA (mean Ct = 38.58) by IGS real-time PCR assay.
Extraction of crude extracts from Tp WPM 39143
Tp WPM 39143 and the well-known biocontrol fungi, Th and Ta, were grown on PDA agar at 15°C for 30 days. Fungal growth was gently scraped with a plastic loop, and a spore/hyphal suspension was harvested in sterile distilled water. Approximately 5 ml of spore/hyphal suspension (OD600 = 1.0) was seeded into six 500-ml flasks containing 100 g of autoclaved rice fermentation medium, and cultures were then incubated at 22°C for 30 days. Following incubation, the fermented rice substrate was extracted with methanol (3 x 400 ml) by 4 h incubation at 22°C. The methanol-fungal slurry was centrifuged at 8,000 RPM for 10 min. The fungal pellet was removed, and the supernatant evaporated under vacuum. The dry pellet (approximately 7~8 g) was suspended into 30 ml of methanol, which served as a crude extract.
A portion of the crude extract was assessed for stability by exposure to high temperature (45°C), regular light and 10 μl of proteinase K (37° and 45°C) for a total of 20 h each.
HPLC fractionation of Tp WPM 39143 crude extract
The Tp WPM 39143 crude extract was analyzed for the presence of different metabolites by high-performance liquid chromatography (HPLC). In brief, 20 μl of crude extract from Tp WPM 39143 was placed into HPLC column (Agilent Zorbax SB-C18 column, 5 μm, 4.6×250 mm, 1 ml min-1) and different fractions were eluted using gradients starting with 5% CH3OH in H2O to 100% CH3OH for 55 min followed by 100% CH3OH for 5 min (12 fractions). The crude extract in the column was further eluted using gradients starting with 100% CH3OH to 90% acetonitrile in H2O for 10 min (2 fractions) and final washing with 5% CH3OH in H2O for 5 min (1 fraction). A total of 15 fractions were collected. This experiment was repeated 10 times and fractions from each run were pooled, concentrated 100 fold, and then each fraction was assessed for anti-Pd activity using antimicrobial disc diffusion bioassay.
Antimicrobial disc diffusion bioassay
The antimicrobial disc diffusion bioassay was performed as described in a previous publication [53]. In brief, conidial suspensions of Pd and Pp were prepared by gently scraping fungal growth from the agar surface and then passing through a 27-G needle. Conidia were counted microscopically using a hemocytometer, and approximately 100 μl of conidial suspension (108 conidia/ml) was spread on YPD agar plates. The 6 mm autoclaved filter discs (Whatman no. 1) were carefully placed on seeded plates and 15 μl of untreated or treated crude extract or extract purified through HPLC column was placed onto filer discs. Plates were incubated for 5–7 days at 15°C, and the zone of fungal growth inhibition around the disc was measured.
Statistical Analysis
Data in figures are presented as mean ± SD. All statistical analyses were performed using GraphPad Prism software (GraphPad, San Diego, CA, USA). The comparison of mean value of multiple groups was performed using a two-way ANOVA and comparison of two groups was performed using a two-tailed unpaired t-test. Values were accepted as significant if p ≤ 0.05.
Nucleotide Sequence Accession Numbers
All nucleotide sequences of Tp WPM 39143 (Hypocrea pachybasioide) were deposited in GenBank under accession numbers: ITS sequence, KJ494575; 28S rRNA gene partial sequence, KJ494576.
Results
Phylogenetic analysis of Tp WPM 39143
BLAST search of the ITS sequences of the ribosomal gene of Tp WPM 39143 strain showed close homology with T. polysporum strain CBS 119319 (accession no. FJ860796). Further, phylogenetic analyses using a Neighbor-Joining tree revealed that Tp strain WPM 39143 was closely related to Hypocrea pachybasioide CBS 119319 (teleomorph of Trichoderma polysporum) followed by T. piluliferum. In contrast, Tp WPM 39143 was distantly related to the well-known biocontrol fungi Th and Ta (Fig 1). Phylogenetic analysis of the D1/D2 region of the 28S ribosomal gene revealed a similar placement of respective genera (data not shown).
Fig 1. Phylogenetic analysis of Tp WPM 39143.
The ITS nucleotide sequence of Tp WPM 39143 was aligned with similar sequences from 27 taxa of Trichoderma/Hypocrea species available in the GenBank. The neighbor-joining method was used to construct the phylogenetic tree. The bootstrap scores are based on 1,000 reiterations. Fusarium oxysporum CBS 129.24 and Neotyphodium uncinatum NBRC 32642 were used as outgroup.
Growth characteristics of Tp WPM 39143
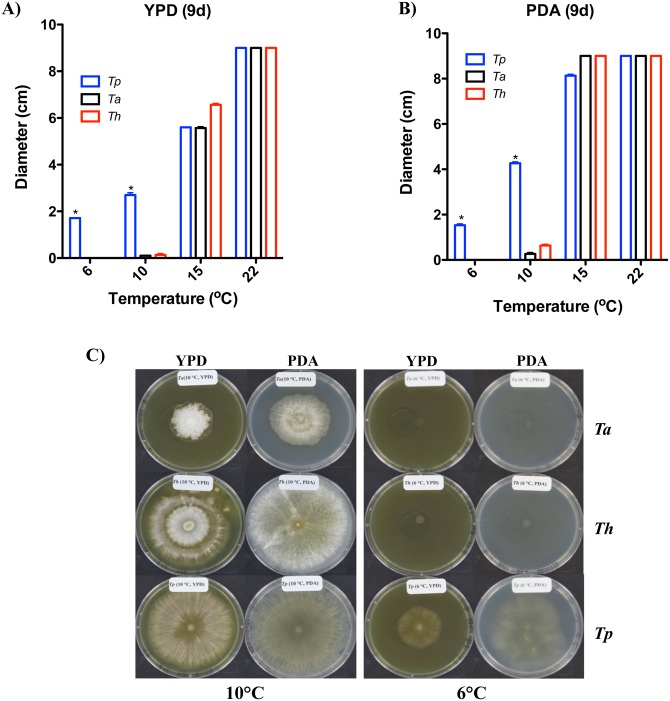
In order to examine the biocontrol potential of Tp WPM 39143, we first tested its growth at different temperatures in comparison to the well-known biocontrol strains Th and Ta. Our results indicated that Tp WPM 39143 grew fairly well at 6°C while Th and Ta did not grow at all at 6°C after 9 days post-incubation (Fig 2A and 2B). Even prolonged incubation of 20 days did not support the growth of either Th or Ta at 6°C while restricted growth of Ta was observed at 10°C (Fig 2C). Tp 39143 grew better at ≥10°C but growth was also evident at 6°C, a temperature more prevalent in the caves and mines [40].
Fig 2. Growth comparisons of Tp WPM 39143 at different temperatures.
Tp WPM 39143 along with Th and Ta strains were point inoculated on the center of PDA (A) and YPD (B) agar plates and incubated at different temperatures (6°C-22°C) for 9 days and colony diameter was measured. The Tp WPM 39143 growth was significantly rapid (asterisk denotes p<0.001) as compared to Ta or Th at lower temperatures of 6°C and 10°C. Further incubation (20 days) revealed good growth of Th and restricted growth of Ta at 10°C, but no growth of Th or Ta at 6°C (C).
Dual culture challenge studies
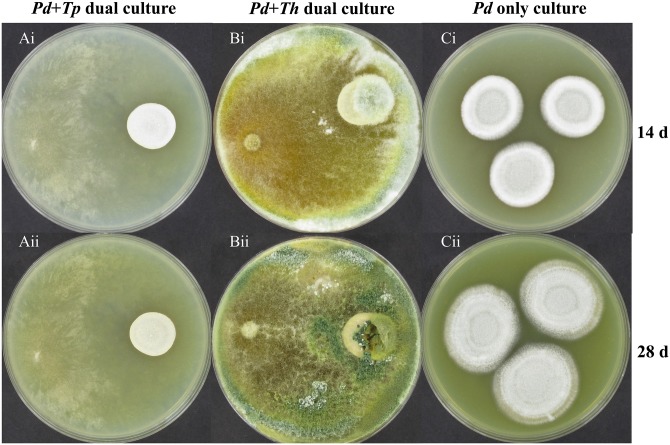
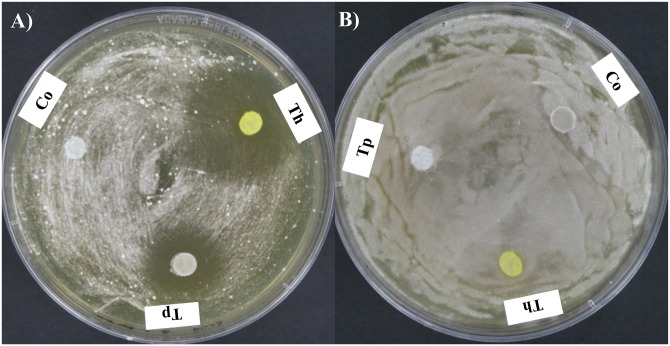
In the dual culture challenge experiments on PDA agar, the Pd hyphal extension (diameter) was 1.93 ± 0.1 cm in the presence of Tp WPM 39143 (Fig 3Ai) while it was 2.82 ± 0.2 cm in the absence of Tp WPM 39143 at 14 days post-incubation (Fig 3Ci). No extension of Pd hyphal growth was observed in the presence of Tp WPM 39143 (Fig 3Aii) while it steadily increased to 3.90 ± 0.2.cm at 28 days post-incubation (Fig 3Cii). Of particular interest, Pd colonies in the presence of Tp WPM 39143 appeared white and restricted as compared to fluffy and pigmented when grown alone (Fig 3Ci and 3Cii). A similar trend was observed for Pd grown in the presence of Th (Fig 3Bi and 3Bii).
Fig 3. Dual culture challenge on PDA agar medium.
Approximately 15 μl spore suspension of Pd (107/ml) was inoculated near the edge of the PDA plate. Following incubation at 15°C for 10 days, 15 μl of Tp WPM 39143 or Th conidial suspension (105 cells/ml) was inoculated on the opposite edge of the culture plate and the interaction between these two fungi was assessed at 14 and 28 days post-incubation. Pd colonies were white and restricted in the presence of Tp WPM 39143 or Th as compared to fluffy and pigmented when grown alone.
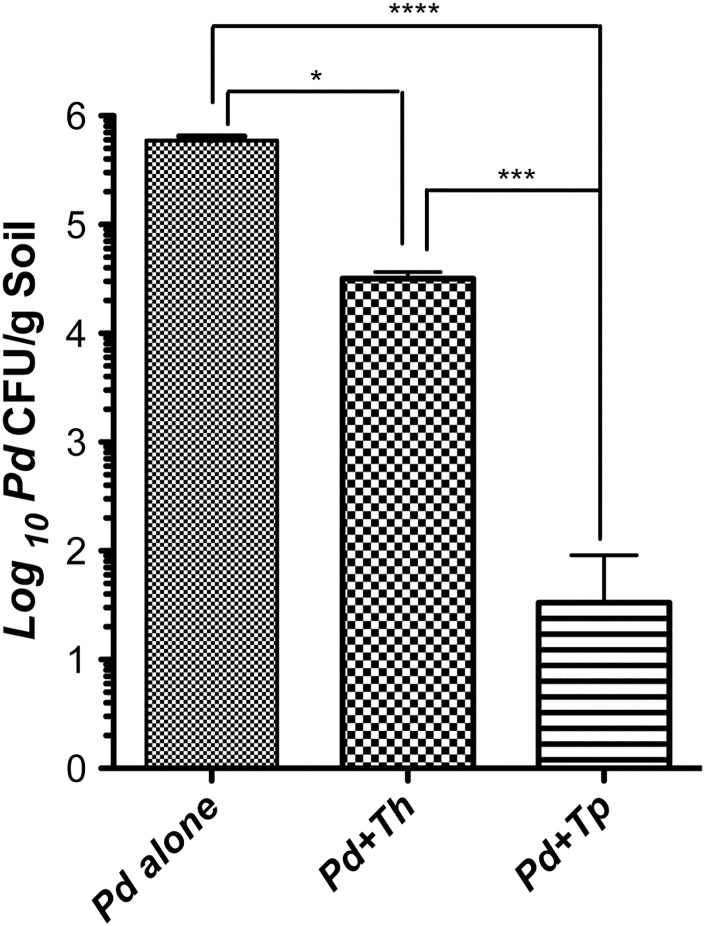
Next, we determined if Tp WPM 39143 induced growth inhibition was sustainable in autoclaved soil sample mimicking the natural environment. This information is crucial if Tp WPM 39143 is to be considered as a biocontrol agent of Pd in caves and mines. The Th induced growth inhibition was also determined for comparison. The autoclaved soil sample supported good growth of Pd (106 CFU/g soil) and Tp or Th (approximately 108 CFU/g soil). When compared to the control soil sample harboring Pd alone, the recovery of Pd from soil samples harboring Tp WPM 39143 was reduced by approximately 4.0-logs (99.98% inhibition). In contrast, the recovery of Pd from soil samples harboring Th was reduced by 1.7-log (89% inhibition) (Fig 4). A similar trend was observed for Pd genome copies in soil samples by real-time PCR assay. As compared to the recovery of Pd genome copies from control soil sample harboring Pd alone, several logs reduction of Pd genome copies was observed in soil samples harboring Tp WPM 39143, and only one log reduction of Pd genome copies was observed in soil samples harboring Th (Fig 5). These results confirmed that Tp WPM 39143 had potent inhibitory activity against Pd, and the inhibitory activity was sustainable under a simulated natural setting.
Fig 4. Pd CFU recovery from dual culture challenge in soil (culture-dependent method).
Autoclaved soil sample was first inoculated with 100 μl of Pd conidia (104/g) and following incubation for 7 days at 15°C, one soil sample each containing Pd, was inoculated with 100 μl of Tp WPM 39143 or Th conidia (104/g) or 100 μl of water alone. Dual culture challenge samples and control samples were incubated at 15°C for another 35 days, and three aliquots of 100 mg soil from each treatment group were processed for the recovery of Pd in culture. Approximately, 4-logs reduction of Pd CFU by Tp WPM 39143 (p<0.0001) compared to 1.7-logs reduction of Pd CFU by Th (p<0.05) was observed.
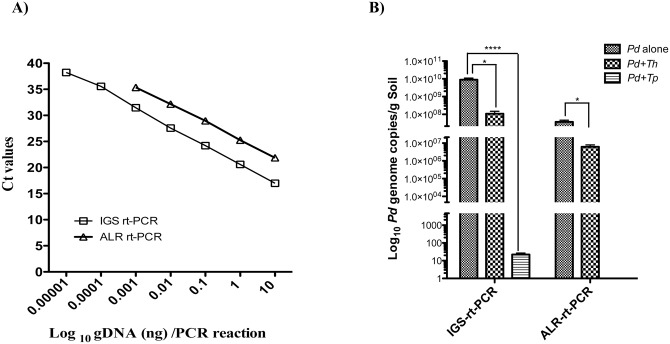
Fig 5. Pd genome copies recovery from dual culture challenge in soil (culture-independent method).
A, Standard curves of Pd intergenic spacer region (IGS) and alpha L-Rhamnosidase (ALR) gene by real-time PCR assays. The purified genomic DNA from Pd was used for the generation of standard curve against IGS and ALR targets, which was used to extrapolate Pd genome copies in the soil samples with or without biocontrol agents. All data points represent the mean Ct value of amplification reactions done in triplicates with error bars denoting standard deviation. The assay was linear over 6 orders of magnitude for IGS gene (y = –3.5886x+42.131, R2 = 0.9982) and over 4 orders of magnitude for ALR gene (y = –3.398x+45.716, R2 = 0.9994). B, Pd genome copies recovery by real-time PCR assays. Autoclaved soil sample was first inoculated with 100 μl of Pd conidia (104/g) and following incubation for 7 days at 15°C, one each soil sample containing Pd was inoculated with 100 μl of Tp WPM 39143 or Th conidia (104/g) or 100 μl of water alone. The dual challenge samples were incubated at 15°C for additional 35 days and three aliquots of 100 mg each soil sample from each treatment group were processed for Pd gDNA extraction followed by IGS and ALR real-time PCR assays. The mean Ct counts equivalent to DNA was extrapolated from the standard curve, and Pd genome copies were calculated based on the formula described in Materials and Methods. Substantial reduction of Pd genome copies observed in samples challenged with Tp WPM 39143 (p<0.0001) as compared to samples challenged with Th (p<0.05).
Inhibitory activity of Tp WPM 39143 extract
In order to identify active ingredients for biocontrol, preliminary experiments were done with crude extract of Tp WPM 39143 against Pd. We also included crude extracts from well-known biocontrol fungi Th and Ta for comparison. Pseudogymnoascus pannorum (Pp), a species closely related to Pd and abundant in caves and mines [20] was also included in the challenge experiment as a target to compare species-specific biocontrol activities. The crude extract from Tp WPM 39143 showed inhibitory activity against Pd as judged by the presence of clear zone surrounding the disc (Fig 6A) but not against Pp (Fig 6B). Similarly, the Tp WPM 39143 crude extract was not inhibitory against fungi common in caves and mines including Aspergillus species, Penicillium species, Mucor species, Fusarium species, Cladosporium species, Alternaria species and Paecilomyces species (data not shown). The Th crude extract also showed more pronounced inhibitory activity against Pd but not against Pp (Fig 6A and 6B). The Ta crude extract did not inhibit either Pd or Pp (data not shown). These results indicated that the inhibitory metabolites in Tp or Th crude extracts might be specific against Pd.
Fig 6. Inhibition of Pd by Tp WPM 39143 and Th extracts.
YPD agar plates were streaked with 108 spore suspensions of Pd or Pp. Sterile filter discs (6 mm) were placed on the surface of the inoculated plate and saturated with 15 μl of Tp WPM 39143 or Th crude extract. Plates were incubated at 15°C for 10 days. Both Tp WPM 39143 and Th extract showed inhibitory activity against Pd (A) but not against Pp (B). The fermentation medium without metabolites, which went through similar extraction process as medium containing metabolites served as a negative control (NCo).
Characterization of Tp WPM 39143 inhibitory metabolites
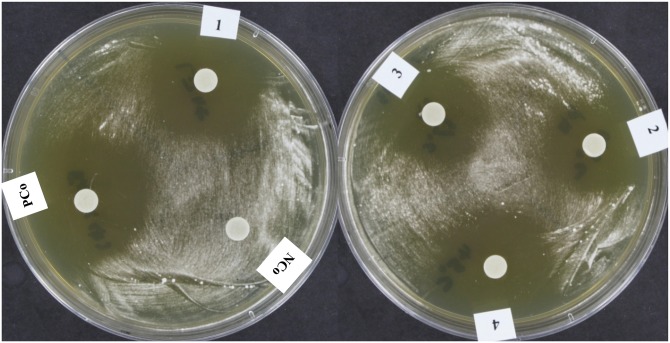
Since Tp WPM 39143 grew in the laboratory at cave and mine temperatures (6°C-15°C) and exhibited Pd inhibition, we further assessed its inhibitory potential by determining the stability of metabolites at different temperatures, light exposure, and proteinase K treatment. The treated extracts inhibited Pd growth as efficiently as non-treated extract indicating that Tp WPM 39143 extract is stable at high temperatures and under regular light (Fig 7).
Fig 7. Stability assessment of Pd inhibitory activity in Tp WPM 39143 crude extract.
YPD agar plates were streaked with 100 μl of Pd conidial suspension (108) and sterile filter discs (6 mm) were placed on agar plates and saturated with 15 μl of treated or untreated Tp WPM 39143 extract. Plates were incubated at 15°C for 10 days and were photographed for inhibition zone around discs. Abbreviations: PCo, positive control (untreated Tp WPM 39143 extract), NCo, negative control (fermentation medium alone); 1, Tp WPM 39143 extract exposed to regular light for 20 h; 2, Tp WPM 39143 extract exposed to proteinase K at 37°C for 20 h; 3, Tp WPM 39143 extract exposed to proteinase K at 45°C for 20 h; 4, Tp WPM 39143 extract exposed to 45°C for 20 h.
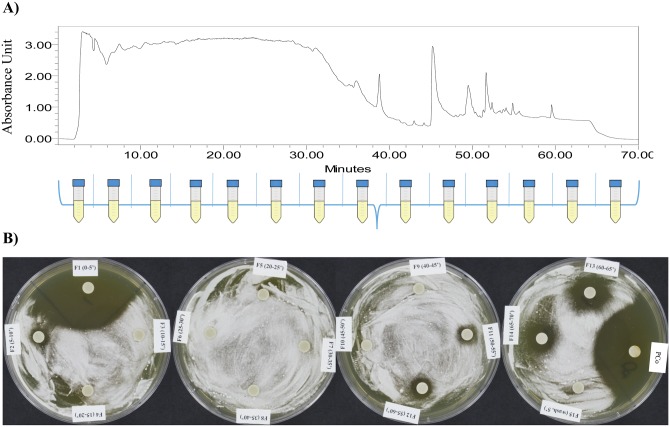
Since Tp WPM 39143 extract was not altered by proteinase k treatment indicating that the inhibitory metabolite(s) in Tp WPM 39143 extract are presumably not proteins, but a mixture of compounds (Fig 7). Therefore, we explored the chemical nature of inhibitory metabolites in Tp WPM 39143 crude extract. The Tp WPM 39143 crude extract was fractionated using high performance liquid chromatography (HPLC) and a total of 15 fractions were collected (Fig 8A). The first two sub-fractions (F1, 0–5 min and F2, 5–10 min-red arrow) and the last four sub-fractions (F11, F12, F13, and F14 –yellow arrow) displayed inhibitory activity against Pd while other fractions did not (Fig 8B). These results indicated that there are two major subclasses of compounds in Tp WPM 39143 extract, high polarity compounds with higher inhibitory activity to Pd and low polarity compounds with lower inhibitory activity to Pd.
Fig 8. Inhibitory activity of HPLC fractions obtained from Tp WPM 39143 extract.
A) Tp WPM 39143 crude extract was fractionated by HPLC (details in methods). Total of 15 sub-fractions were collected at the intervals of 5 min. B) Disc diffusion assay to measure inhibition of Pd growth by various HPLC sub-fractions. F1, 0–5 min sub-fraction; F2, 5–10 min sub-fraction; F3, 10–15 min sub-fraction; …………. F14, 65–70 min sub-fraction; F15, 70–75 min, 5% CH3OH in H2O elution time. Abbreviation: PCo, positive control (untreated Tp WPM 39143 crude extract).
Discussion
The major findings of this study were: the isolation of novel Tp WPM 39143 strain from a WNS affected cave, growth of the novel strain at temperatures simulating cave and mine environments, and potent and specific inhibitory activity of Tp WPM 39143 against Pd in laboratory media and autoclaved soil. Biocontrol agents are already accepted as an environmentally friendly alternative to chemicals for plant disease management in agriculture [54, 55]. Recently, a zooplankton Daphnia magna was suggested as a promising biocontrol agent for Batrachochytrium dendrobatitis, a chytrid fungus responsible for deadly chytridiomycosis in amphibians [56, 57]. Our findings of a novel, psychotolerant Tp WPM 39143 now sets the stage for potential biological decontamination of Pd at WNS affected sites.
There were no published studies prior to a report from our laboratory that documented Tp from caves and mines [20]. We isolated Tp from only William Preserve Mine, and that too from 3 of the 25 samples investigated. The rare isolation indicated that it is probably not present in large numbers as are some other fungi or has a restricted niche. Since, we did not observe any inhibition of at least some of the major fungal genera by Tp apart from Pd, it follows that Tp could serve as a biocontrol agent for the inhibition or eradication of Pd from caves and mines. The excellent growth of Tp observed in autoclaved soil sample from Aeolus Cave suggests that the organic and inorganic contents of the soil favor the growth of Tp. Additionally, our limited studies documented the fungicidal activity of Tp against Pd in spiked autoclaved soil sample. These results further confirmed the potent biocontrol activity of Tp against Pd and the biocontrol activity was sustainable under simulated natural setting. Therefore, the stage is set for future experiments to demonstrate if Tp can grow well in the presence of other microbes in natural settings and if it can sustain inhibitory activity against Pd.
Trichoderma spp. are prominent among the most effective mycoparasites. Several species such as T. harzianum, T. polysporum, T. viride and T. virens are currently in commercial production for the control of plant pathogenic fungi in agriculture and horticulture [24, 58]. A number of studies have documented that psychrophilic range limits the biocontrol potential of Trichoderma spp. and only a few cold adapted strains with biocontrol potential have been reported, including T. aureoviride, T. harzianum, and T. viride [35, 59]. Although T. polysporum has been reported from cold environments including the Arctic and Antarctic, none of the reported strains are known for their potent biocontrol activity against other fungi [60, 61]. Thus, the discovery and characterization of Tp WPM 39143 establishes it as a potentially unique biocontrol agent against the psychrophilic, zoopathogen Pd.
Trichoderma polysporum secondary metabolites include cyclosporin [62, 63], trichosporin [64], peptaibols [65], anthraquinones [38], trichodermin [24], and minor cyclonerodiol derivatives [66]. Among them, trichosporin, cyclosporine, peptaibols, and cyclonerodiol derivatives have antifungal activities [62, 63, 66]. Many of these metabolites are amphipathic [67, 68]. It is relevant to recall that water soluble, high polarity compounds with specific inhibitory activity against Pd were identified in our preliminary analyses. The next logical experiments will be to define the fungicidal molecule (s) in Tp extracts active against Pd.
In conclusion, we have identified a novel biocontrol strain Tp WPM 39143 from a cave at the epicenter of the WNS zoonotic. Trichoderma polysporum WPM 39143 produced potent inhibitory compound(s) that impeded the growth of Pd in both laboratory media and soil matrices. These results could be used to design a viable strategy for the biological decontamination of Pd in ‘cave or mine in a lab’ environment or under field conditions.
Supporting Information
Flow chart representing culture-dependent (CD) and culture-independent (CI) methods for the recovery of Pd in the presence or absence of biocontrol fungi, Tp WPM 39143 or Th.
(TIF)
Acknowledgments
We thank Wadsworth Center Biochemistry Core for preliminary HPLC analysis, Applied Genomic Technology Core for sequencing and Media & Tissue Culture Core for the preparation of various media and reagents. We also thank Sunanda S. Rajkumar and Holland DeFiglio for the initial experimental contributions. We thank five anonymous reviewers for their valuable critique of our original submission.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was partly supported with funds from Fish and Wildlife Service (F12AP01167), the National Science Foundation (000600793) to V.C. and S.C., and Wadsworth Center State Purpose Activities. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Veilleux JP. Current status of white-nose syndrome in the Northeastern United States. Bat Res News. 2008;49:15–7. [Google Scholar]
- 2. Turner GG. Update of white nose syndrome in bats, September 2009. Bat Res News. 2009;50:47–53. [Google Scholar]
- 3. Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, et al. Bat white-nose syndrome: an emerging fungal pathogen? Science. 2009;323(5911):227 10.1126/science.1163874 [DOI] [PubMed] [Google Scholar]
- 4. Chaturvedi V, Springer DJ, Behr MJ, Ramani R, Li X, Peck MK, et al. Morphological and molecular characterizations of psychrophilic fungus Geomyces destructans from New York bats with White Nose Syndrome (WNS). PLoS One. 2010;5(5):e10783 10.1371/journal.pone.0010783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, et al. An emerging disease causes regional population collapse of a common North American bat species. Science. 2010;329(5992):679–82. 10.1126/science.1188594 [DOI] [PubMed] [Google Scholar]
- 6. Blehert DS. Fungal disease and the developing story of bat white-nose syndrome. PLoS Pathogen. 2012;8:e1002779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O'Regan SM, MagorI K, Pulliam JT, Zokan MA, Kaul RB, Barton HD, et al. Multi-scale model of epidemic fade-out: Will local extirpation events inhibit the spread of white-nose syndrome? Ecol App 2015;25:621–33. [DOI] [PubMed] [Google Scholar]
- 8. Puechmaille SJ, Frick WF, Kunz TH, Racey PA, Voigt CC, Wibbelt G, et al. White-nose syndrome: is this emerging disease a threat to European bats? Trends Ecol Evol. 2011;26(11):570–6. 10.1016/j.tree.2011.06.013 . [DOI] [PubMed] [Google Scholar]
- 9. Cryan PM, Meteyer CU, Boyles JG, Blehert DS. Wing pathology of white-nose syndrome in bats suggests life-threatening disruption of physiology. BMC biology. 2010;8:135 10.1186/1741-7007-8-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reeder DM, Frank CL, Turner GG, Meteyer CU, Kurta A, Britzke ER, et al. Frequent arousal from hibernation linked to severity of infection and mortality in bats with white-nose syndrome. PLoS One. 2012;7:e38920 10.1371/journal.pone.0038920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Warnecke L, Turner JM, Bollinger TK, Misra V, Cryan PM, Wibbelt G, et al. Pathophysiology of white-nose syndrome in bats: a mechanistic model linking wing damage to mortality. Biol Lett. 2013;9:20130177 10.1098/rsbl.2013.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puechmaille SJ, Wibbelt G, Korn V, Fuller H, Forget F, Muhldorfer K, et al. Pan-European distribution of white-nose syndrome fungus (Geomyces destructans) not associated with mass mortality. PLoS One. 2011;6(4):e19167 10.1371/journal.pone.0019167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bandouchova H, Bartonicka T, Berkova H, Brichta J, Cerny J, Kovacova V, et al. Pseudogymnoascus destructans: evidence of virulent skin invasion for bats under natural conditions, Europe. Transbound Emerg Dis. 2015;62(1):1–5. 10.1111/tbed.12282 . [DOI] [PubMed] [Google Scholar]
- 14. Leopardi S, Blake D, Puechmaille SJ. White-Nose Syndrome fungus introduced from Europe to North America. Curr Biol. 2015;25(6):R217–9. 10.1016/j.cub.2015.01.047 . [DOI] [PubMed] [Google Scholar]
- 15. Rajkumar SS, Rudd RJ, Okoniewski JC, Xu J, Chaturvedi S, Chaturvedi V. Dispersal of a single, clonal genotype of Geomyces destructans among New York bats with geomycosis (White Nose Syndrome). Emerg Infect Dis. 2011;17:1273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ren P, Haman KH, Last LA, Rajkumar SS, Keel MK, Chaturvedi V. Clonal Spread of Geomyces destructans among Bats, Midwestern and Southern United States. Emerging infectious diseases. 2012;18:883–5. 10.3201/eid1805.111711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lindner DL, Gargas A, Lorch JM, Banik MT, Glaeser J, Kunz TH, et al. DNA-based detection of the fungal pathogen Geomyces destructans in soils from bat hibernacula. Mycologia. 2010;103(2):241–6. Epub 2010/10/19. 10–262 [pii] 10.3852/10-262 . [DOI] [PubMed] [Google Scholar]
- 18. Shuey MM, Drees KP, Lindner DL, Keim P, Foster JT. Highly sensitive quantitative PCR for the detection and differentiation of Pseudogymnoascus destructans and other Pseudogymnoascus species. Appl Environ Microbiol. 2014;80(5):1726–31. 10.1128/AEM.02897-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorch JM, Muller LK, Russell RE, O'Connor M, Lindner DL, Blehert DS. Distribution and environmental persistence of the causative agent of white-nose syndrome, Geomyces destructans, in bat hibernacula of the Eastern United States. App Environ Microbiol. 2013;79:1293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang T, Victor TR, Rajkumar SS, Li X, Okoniewski JC, Hicks AC, et al. Mycobiome of the bat white nose syndrome affected caves and mines reveals diversity of fungi and local adaptation by the fungal pathogen Pseudogymnoascus (Geomyces) destructans . PLoS One. 2014;9(9):e108714 Epub 2014/09/30. 10.1371/journal.pone.0108714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoyt JR, Cheng TL, Langwig KE, Hee MM, Frick WF, Kilpatrick AM. Bacteria isolated from bats inhibit the growth of Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PLoS One. 2015;10(4):e0121329 10.1371/journal.pone.0121329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cornelison CT, Gabriel KT, Barlament C, Crow SA Jr. Inhibition of Pseudogymnoascus destructans growth from conidia and mycelial extension by bacterially produced volatile organic compounds. Mycopathologia. 2014;177(1–2):1–10. 10.1007/s11046-013-9716-2 . [DOI] [PubMed] [Google Scholar]
- 23. Cornelison CT, Keel MK, Gabriel KT, Barlament CK, Tucker TA, Pierce GE, et al. A preliminary report on the contact-independent antagonism of Pseudogymnoascus destructans by Rhodococcus rhodochrous strain DAP96253. BMC Microbiol. 2014;14:246–52. 10.1186/s12866-014-0246-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nature Rev Microbiol. 2004;2:43–56. [DOI] [PubMed] [Google Scholar]
- 25. Punja ZK, Utkhede RS. Using fungi and yeasts to manage vegetable crop diseases. Trends in Biotechnology. 2003;21:400–7. [DOI] [PubMed] [Google Scholar]
- 26. Howell CR. Understanding the Mechanisms Employed by Trichoderma virens to Effect Biological Control of Cotton Diseases. Phytopathology. 2006;96(2):178–80. 10.1094/PHYTO-96-0178 . [DOI] [PubMed] [Google Scholar]
- 27. Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E, et al. Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol 2000;66:1890–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cloyd RA, Dickinson A, Kemp KE. Effect of diatomaceous earth and Trichoderma harzianum T-22 (Rifai strain KLR-AG2) on the fungus gnat Bradysia sp. nr. coprophila (Diptera: Sciaridae). J Econ Entomol. 2007;100:1353–9. [DOI] [PubMed] [Google Scholar]
- 29. Yedidia II, Benhamou N, Chet II. Induction of defense responses in cucumber plants (Cucumis sativus L.) By the biocontrol agent trichoderma harzianum. Appl Environ Microbiol. 1999;65(3):1061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papavizas GC. Trichoderma and Gliocladium: Biology, Ecology, and Potential for Biocontrol. Annual Review of Phytopathology 1985;23:23–54. [Google Scholar]
- 31. Longa CM, Savazzini F, Tosi S, Elad Y, Pertot I. Evaluating the survival and environmental fate of the biocontrol agent Trichoderma atroviride SC1 in vineyards in northern Italy. J Appl Microbiol. 2009;106(5):1549–57. 10.1111/j.1365-2672.2008.04117.x . [DOI] [PubMed] [Google Scholar]
- 32. Benitez T, Rincon AM, Limon MC, Codon AC. Biocontrol mechanisms of Trichoderma strains. Int Microbiol. 2004;7(4):249–60. . [PubMed] [Google Scholar]
- 33. Mukherjee PK, Raghu K. Effect of temperature on antagonistic and biocontrol potential of Trichoderma sp. on Sclerotium rolfsii . Mycopathologia. 1997;139(3):151–5. 10.1023/A:1006868009184 . [DOI] [PubMed] [Google Scholar]
- 34. Gupta V, Sharma AK. Assesment of optimum temperature of Trichoderma harzianum by monitoring radial growth and population dynamics in different compost manures under different temperatures. Octa J Biosci. 2013;1:151–7. [Google Scholar]
- 35. Kredics L, Antal Z, Manczinger L, Szekeres A, Kevei F, E. N. Influence of environmental parameters on Trichoderma strains with biocontrol potential. Food Tech Biotechnol. 2003;41:37–42. [Google Scholar]
- 36. Antala Z, Manczingerb L, Szakacsc G, Tengerdyd RP, Ferenczy L. Colony growth, in vitro antagonism and secretion of extracellular enzymes in cold-tolerant strains of Trichoderma species. Mycol Res. 2000;104:545–9. [Google Scholar]
- 37. Dumas MT, Boyonoski NW. Scanning electron microscopy of mycoparasitism of Armillaria rhizomorphs by species of Trichoderma . Eur J For Path. 1992;22:379–83. [Google Scholar]
- 38. Donnelly DMX, Sheridan MH. Anthraquinones from Trichoderma polysporum . Phytochemistry. 1986;25:2303–4. [Google Scholar]
- 39. Eziashi EI, Uma NU, Adekunle AA, Airede CE, Odigie EE. Evaluation of lyophilized and non lyophilized toxins from Trichoderma species for the control of Ceratocystis paradoxa . African J Agricul Res. 2010;5:1733–8. [Google Scholar]
- 40. Perry RW. A review of factors affecting cave climates for hibernating bats in temperate North America. Environ Rev. 2013;21:28–39. [Google Scholar]
- 41. Chibucos MC, Crabtree J, Nagaraj S, Chaturvedi S, Chaturvedi V. Draft Genome Sequences of Human Pathogenic Fungus Geomyces pannorum Sensu Lato and Bat White Nose Syndrome Pathogen Geomyces (Pseudogymnoascus) destructans . Genome Announcement. 2013;1:e01045–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang QX, Li SF, Zhao F, Dai HQ, Bao Lea. Chemical constituents from endophytic fungus Fusarium oxysporum . Fitoterapia. 2011;82:777–81. 10.1016/j.fitote.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 43. Möller EM, Bahnweg G, Sandermann H, Geiger HH. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992;20:6115–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. White TJ, Bruns T, Lee SR, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors: Academic Press Inc, New York; 1990. [Google Scholar]
- 45. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. Epub 2009/04/21. 10.1007/978-1-59745-251-9_3 . [DOI] [PubMed] [Google Scholar]
- 47. Tamura K, Peterson D, Peterson N, Stecher G, Nei Mea. MEGA5:molecular evolutionary genetic analysis using maximum likelihood evolutionary distance, and maximum parsimony methids. Molecular biology and evolution. 2011;28:2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kirk PM, Cannon PF, Minter DW, Stalpers JA. Ainsworth & Bisby's Dictionary of the Fungi: CAB International, Wallingford, United kingdom; 2008. [Google Scholar]
- 49. Chaturvedi S, Rudd RJ, Davis A, Victor TR, Li X, Appler KA, et al. Rapid Real-Time PCR Assay for Culture and Tissue Identification of Geomyces destructans, the Etiologic Agent of Bat Geomycosis (White Nose Syndrome). Mycopathologia. 2011;172:247–56. 10.1007/s11046-011-9435-5 [DOI] [PubMed] [Google Scholar]
- 50. Muller LK, Lorch JM, Lindner DL, O'Connor M, Gargas Aea. Bat white-nose syndrome: a real-time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans . Mycologia. 2012;105:253–9. 10.3852/12-242 [DOI] [PubMed] [Google Scholar]
- 51. Ganley AR, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17(2):184–91. 10.1101/gr.5457707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101(19):7329–34. 10.1073/pnas.0401648101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Driscoll AJ, Bhat N, Karron RA, O'Brien KL, Murdoch DR. Disk diffusion bioassays for the detection of antibiotic activity in body fluids: applications for the Pneumonia Etiology Research for Child Health project. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;54 Suppl 2:S159–64. 10.1093/cid/cir1061 . [DOI] [PubMed] [Google Scholar]
- 54. Alabouvette C, Olivain C, Steinberg C. Biological control of plant diseases: the European situation. Eur J Plant Pathology. 2006;114:329–41. [Google Scholar]
- 55. Fravel DR. Commercialization and implementation of biocontrol. Annu Rev Phytopathol. 2005;43:337–59. 10.1146/annurev.phyto.43.032904.092924 . [DOI] [PubMed] [Google Scholar]
- 56. Searle CL, Mendelson JR 3rd, Green LE, Duffy MA. Daphnia predation on the amphibian chytrid fungus and its impacts on disease risk in tadpoles. Ecol Evol. 2013;3(12):4129–38. 10.1002/ece3.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buck JC, Truong L, Blaustein AR. Predation by zooplankton on Batrachochytrium dendrobatidis: biological control of the deadly amphibian chytrid fungus? Biodiver and Conserv. 2011;20:3549–53. [Google Scholar]
- 58. Howell CR. Mechanisms Employed by Trichoderma Species in the Biological Control of Plant Diseases: The History and Evolution of Current Concepts. Plant Dis. 2003;87:4–10. [DOI] [PubMed] [Google Scholar]
- 59. Hudec K. Influence of temperature on Trichoderma harzianum antagonistic activity against Microdochium nivale, Fusarium culmorum and F. equiseti under "in vitro" conditions. Acta Fytotechnica et Zootechnica 2000;3:98–100. [Google Scholar]
- 60. Yamazaki Y, Tozo M, Hoshino T, Kida K, Sakamoto Tea. Characterization of Trichoderma polysporum from Spitsbergen, Svalbard archipelago, Norway, with species identity, pathogenicity to moss, and polygalacturonase activity. Fungal Ecol. 2011;4:15–21. [Google Scholar]
- 61. Tozo M, Newsham KK. Snow moulds in polar environments. Fungal Ecol. 2012;5:395–402. [Google Scholar]
- 62. Ruegger A, Kuhn M, Lichti H, Loosli HR, Huguenin R, Quiquerez C, et al. [Cyclosporin A, a Peptide Metabolite from Trichoderma polysporum (Link ex Pers.) Rifai, with a remarkable immunosuppressive activity]. Helv Chim Acta. 1976;59(4):1075–92. 10.1002/hlca.19760590412 . [DOI] [PubMed] [Google Scholar]
- 63. Traber R, Kuhn M, Loosli HR, Pache W, von Wartburg A. [New cyclopeptides from Trichoderma polysporum (Link ex Pers.) Rifai: cyclosporins B, D and E (author's transl)]. Helv Chim Acta. 1977;60(5):1568–78. 10.1002/hlca.19770600513 . [DOI] [PubMed] [Google Scholar]
- 64. Fujita T, Iida A, Uesato S, Takaishi Y, Shingu T, Saito M, et al. Structural elucidation of trichosporin-B-Ia, IIIa, IIId and V from Trichoderma polysporum . J Antibiot (Tokyo). 1988;41(6):814–8. . [DOI] [PubMed] [Google Scholar]
- 65. New AP, Eckers C, Haskins NJ, Neville WA, Elson Sea. Structure of polysporin A-D, four new pentaibolsisolated from Trichoderma polysporum. Tretrahedron Lett. 1996;37:3039–42. [Google Scholar]
- 66. Fujita T, Takahashi y, Takeda K, Fujiyama T, Nishi T. Fungal metabolites. Structural elucidation of minor metabolites, valinotricin, cyclonerodiol oxide, and epicyclonerodiol oxide from Trichoderma polysporum . Chem Pharm Bull (Tokyo). 1984;32:4419–25. [Google Scholar]
- 67. Toniolo C, Crisma M, Formaggio F, Peggion C, Epand RF, Epand RM. Lipopeptaibols, a novel family of membrane active, antimicrobial peptides. Cell Mol Life Sci. 2001;58(9):1179–88. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Szekeres A, Leitgeb B, Kredics L, Antal Z, Hatvani L, Manczinger L, et al. Peptaibols and related peptaibiotics of Trichoderma. A review. Acta Microbiol Immunol Hung. 2005;52(2):137–68. 10.1556/AMicr.52.2005.2.2 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart representing culture-dependent (CD) and culture-independent (CI) methods for the recovery of Pd in the presence or absence of biocontrol fungi, Tp WPM 39143 or Th.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.