Summary
The discovery of induced pluripotent stem cells (iPSCs) and the concurrent development of protocols for their cell-type-specific differentiation have revolutionized our approach to cell therapy. It has now become critical to address the challenges related to the generation of iPSCs under current good manufacturing practice (cGMP) compliant conditions, including tissue sourcing, manufacturing, testing, and storage. Furthermore, regarding the technical challenges, it is very important to keep the costs of manufacturing and testing reasonable and solve logistic hurdles that permit the global distribution of these products. Here we describe our efforts to develop a process for the manufacturing of iPSC master cell banks (MCBs) under cGMPs and announce the availability of such banks.
Highlights
-
•
Generation of clinical-grade iPSCs manufactured under a cGMP-compliant process
-
•
A robust and reproducible iPSC culture system
-
•
A completely defined, serum-free, and feeder-free process
-
•
A comprehensive characterization, including gene expression profiles of cGMP iPSCs
Rao, Fellner, and colleagues report the development of a cGMP-compliant process for the manufacturing of iPSC master cell banks and announce the availability of such banks for the development of clinical cell therapy applications. This manuscript highlights the generation of a clinical-grade human iPSC line using a fully cGMP-compliant process starting from tissue sourcing, a robust and reproducible hiPSC generation process, and well established testing.
Introduction
Induced pluripotent stem cell (iPSC)-based therapy is a newly developing field and builds on several key technical advances that have enabled the widespread use of embryonic stem cell (ESC)-based technology (Ellerström et al., 2006, Rao, 2008, Rao and Condic, 2008, Chen et al., 2012) for drug discovery and basic biology. Companies such as Geron, Asteris, Ocata (formerly known as Advanced Cell Technology), Biotime, Viacyte, and J&J have developed products from ESCs, several have initiated early-stage clinical trials (Carpenter and Rao, 2015), and several patients have been treated with no deleterious side effects (Schwartz et al., 2012). These results have led companies such as Healios and Megakaryon to initiate plans to generate products using iPSCs. Recently, a study involving one patient treated with retinal pigment epithelium (RPE) cells derived from iPSCs was carried out using cells manufactured in a current good laboratory practice (cGLP) environment using autologous cells (http://www.dddmag.com/articles/2014/10/japan-starts-world-first-stem-cell-trial-plans-more).
These groups have demonstrated to the Food and Drug Administration (FDA) that products derived from pluripotent stem cells (PSCs) can be manufactured without a demonstrable risk of contaminating undifferentiated cells. Although current good manufacturing practice (cGMP) compliant cells have been generated from ESCs (Crook et al., 2007, Tannenbaum et al., 2012), most of the cells were derived under non-cGMP conditions and then qualified for cGMP by additional testing. The cells were exposed to xenogeneic agents and feeder cells, and/or, in some cases, donor consent would not permit their use as a commercial product. To our knowledge, no fully cGMP-compliant cell line has been generated where the entire manufacturing process, from tissue sourcing to cell expansion and banking processes as well as documentation, raw materials, staff training, cell therapy facility, and quality control (QC) testing, was validated.
Developing a cGMP-compliant manufacturing protocol or using integration-free methods and xenogeneic-free material in a cGMP-compliant facility will not be sufficient to ensure clinically relevant products, nor will adding certification or training complete the process. Conformation to regulations governing the acquisition of human donor tissue will need to be ensured (in the United States according to FDA 21 CFR 1271 Human Cells, Tissues, and Cellular and Tissue-based Products). Reference or control material will need to be developed to generate convincing data on in-process testing, lot-to-lot variability, and release assays. The assays themselves will need to be developed and qualified or validated (depending on the clinical trial phase of application). Moreover, cGMP manufacturing requirements that are incompatible with cell manufacture need to be modified, including developing specific guidance for sterility/aseptic processes for patient-specific cells. Attention will need to be paid to the different interpretations of ethical issues, patent law, and the special property rights issues that arise for cells that may make gametes (Andrews et al., 2014). In addition to being in compliance with FDA regulations, one will need to comply with requirements that are imposed by institutional review boards (IRBs), the Health Insurance Portability and Privacy Act (HIPPA), and the Office for Human Research Protection (OHRP). Furthermore, given that iPSC-derived cells may be distributed internationally, the cell manufacturing process will need to adhere to additional country-specific guidelines as well. Developers will also have to devise a strategy for international distribution in countries where regulations are still being formulated (http://c.ymcdn.com/sites/www.celltherapysociety.org/resource/resmgr/2014AnnualMeeting/ISCT2014-AcademicProgram_Web.pdf).
Given that a global effort has been initiated to develop donor banks of human leukocyte antigen (HLA)-matched iPSC banks that will serve as an extensive library for selecting close to optimal matches for patients (Turner et al., 2013, Fairchild, 2015, Solomon et al., 2015), it is clear that the need for a large number of cGMP-compliant lines exists, and paying attention to international regulations and not just United States regulations will be critical.
As a custom manufacturing organization (CMO) offering cGMP manufacturing services around a variety of cell therapy products, Lonza attempted to develop clinically compliant processes to generate cGMP-compliant human iPSC lines. In this manuscript, we report the development of such human iPSC lines that primarily address United States regulations. We show that sourcing from cord blood when using an optimized and reproducible integration-free method allows one to develop a bank of well characterized and tested pluripotent cells that can be used for differentiation into clinically relevant cells, holding potential for clinical applications.
Results
Tissue Acquisition
Tissue for the derivation of cellular products must be obtained by following the appropriate guidelines. Figure 1 illustrates some of the main activities associated with sourcing donor tissue for use in cell therapy manufacturing. These activities include defining tissue requirements, working with a tissue recovery agency, establishing forms and standard operating procedures, recovering tissue, and donor eligibility determination. For this iPSC MCB, the starting material was an umbilical cord blood unit. Critical elements of the process included informed consent, which covered the intended use of the donated tissue; the benefits, if any, to the donor or others; the risks and potential discomfort of the donation; confidentiality of donor information; and a statement demonstrating voluntary participation of the donor in the program, obtained from the mother. Notably, the informed consent form developed for this study was reviewed and approved by the hospital IRB in which the cord blood tissue collection work took place.
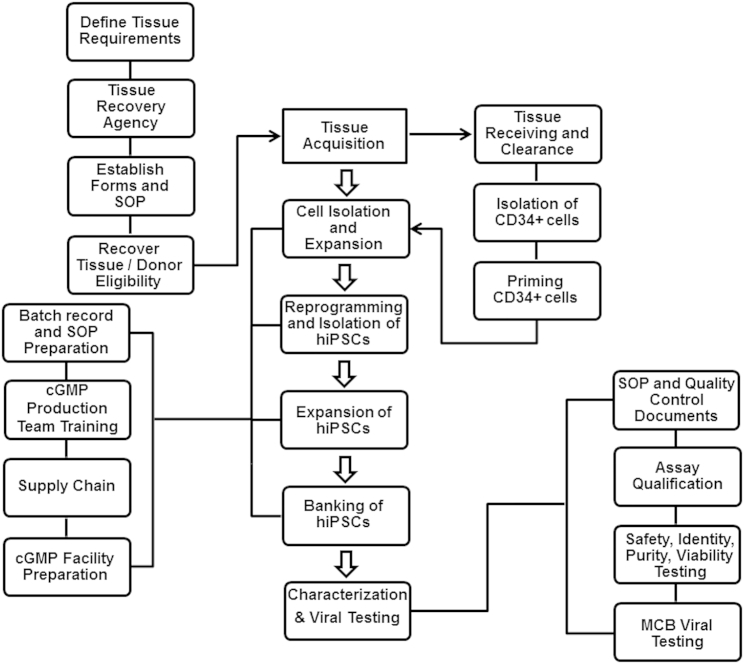
Figure 1.
An Overview of the Manufacturing of Human Induced Pluripotent Stem Cells under cGMPs
A tissue acquisition program was established, focusing on defining tissue requirements, working with a tissue recovery agency, establishing forms and standard operating procedures, recovering tissue, and donor eligibility determination. The manufacturing started with the isolation of CD34+ cells from a fresh cord blood unit and continued to priming, expansion, and then reprogramming of CD34+ cells. After generation of iPSCs and expansion, the cells were banked and eventually tested. Every step of the manufacturing process was documented and performed according to the batch records and standard procedures. Following characterization of the final bank, the results were reviewed by the quality assurance group to release the GMP iPSC lot.
Another critical part of the donor tissue sourcing process was donor eligibility determination, which was based on the results of donor screening and testing. This activity involved review of the medical records and a physical exam. Additionally, clinical laboratory testing was performed to test for relevant communicable disease agents or diseases (RCDADs). The RCDADs listed in the regulations are HIV, hepatitis B virus (HBV), hepatitis C virus (HCV), transmissible spongiform encephalopathies (TSEs), human T-lymphotropic virus (HTLV), Treponema pallidum, Chlamydia trachomatis, and Neisseria gonorrhoea (see FDA 21 CFR 1271 regulations at http://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Tissue/ucm073964.htm). Also, other communicable diseases are considered relevant but not listed, such as West Nile virus, sepsis, and Vaccinia virus.
The collection of cord blood tissue was performed according to defined procedures by trained tissue recovery specialists in a hospital setting using aseptic techniques in accordance with good tissue practices (GTP) regulations as stated in 21 CFR 1271. The tissue was packaged and shipped to the manufacturing site (Lonza Walkersville) using validated procedures (American Association of Blood Banks [AABB]- and Foundation for the Accreditation of Cellular Therapy [FACT]-accredited). When received, the tissue was stored appropriately and transferred to an assigned cell therapy area according to standard protocols. In the interim, the quality assurance (QA) group reviewed the initial documents, which included the date of tissue collection, shipping and handling conditions, and tissue volume/size. When approved, the tissue entered the manufacturing process. Because the process utilized a fresh cord blood unit, the initial cell isolation and priming steps were performed under quarantine conditions until the results of the screening and testing were received. When the data were received, they were reviewed by a qualified person (Lonza’s medical director) and the QA group. After sign-off, the tissue was released from quarantine status.
Development of a cGMP-Compatible Process
iPSC MCBs have to be generated using a robust, reproducible, and cGMP-compliant manufacturing process, applying best practices for cell culture, documentation, and quality control (Stacey et al., 2013). Our early iPSC generation studies focused on the development of a reliable and efficient process that resulted in a sufficient number of high-quality iPSC clones and could be successfully transitioned into the GMP manufacturing suites. The reprogramming process was evaluated based on the number of generated iPSC colonies, morphology and quality of iPSC colonies, and expression of pluripotency markers using flow cytometry and immunostaining. A defined and feeder-free cell reprogramming and cell culture system was developed to avoid the issues of lot-to-lot variability, regulatory and safety concerns of co-culture, and scalability challenges. Important parameters evaluated and optimized in the process development phase included the reprogramming method, nucleofection parameters, reprogramming enhancers, cell culture medium, matrix and subculture reagents, expansion and subculture protocols, and characterization of human iPSCs. Figure 2 illustrates the critical manufacturing steps of cord blood (CB)-derived CD34+ cells.
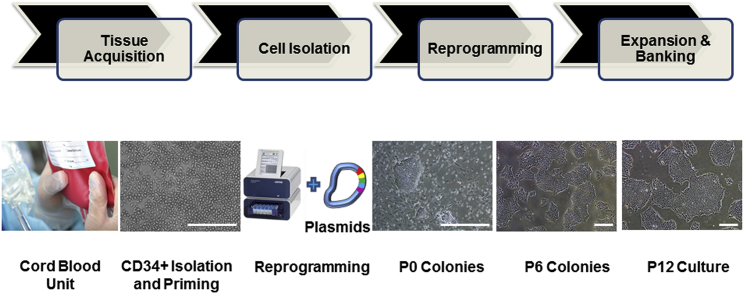
Figure 2.
Key Steps of the Human iPSC Manufacturing Process
The manufacturing of human iPSCs under defined and cGMP conditions include tissue acquisition to obtain a fresh cord blood unit, CD34+ cells isolation from the cord blood, reprogramming of CD34+ cells into iPSCs using the 4D nucleofector system and episomal-based technology, and expansion and banking of iPSCs. The photomicrographs represent CD34+ cells isolated from cord blood and expanded in culture (CD34+ Isolation and Priming), iPSC colonies on day 10 post-nucleofection (P0 Colonies), and iPSCs at passage 6 (P6 Colonies) passage 12 (P12 Culture). Scale bars, 500 μm, except in the CD34+ Isolation and Priming image (250 μm).
CB-derived CD34+ cells were chosen as the starting material because of several practical and technical considerations, as described previously (Rao et al., 2012). Selecting CD34+ cells also eliminated the risk of reprogramming T or B cells. During the process development phase, cryopreserved vials of magnetic bead-isolated CD34+ cells were used. This isolation process resulted in a population of 80% or more CD34+ cells. The cryopreserved CD34+ cells were then thawed and primed for a period of 4 days in a medium supporting the expansion of CD34+ cells. The quality of the cells, level of expansion, and purity of CD34+ cells were evaluated based on the morphology; cell counts on days 0, 1 (after splitting the cells), and 4 (prior to nucleofection); and flow cytometry analysis on days 0 and 4. On average, the priming step yielded a 4- to 8-fold expansion of CD34+ cells and allowed the evaluation of multiple reprogramming conditions.
Given the safety concerns surrounding the use of integrating viral vectors (Okita et al., 2011, Goh et al., 2013), a non-integrative episomal-based technology initially developed by Chou et al. (2011) was used for reprogramming based on a single transfection of two plasmids: pEB-C5 and pEB-Tg (Chou et al., 2011). CD34+ cells were transfected with the two plasmids using a cGMP-compliant electroporation system. Because all of the commercially available substrates we tested did not meet our success criteria, we developed a proprietary matrix in-house (L7 human pluripotent stem cell [hPSC] matrix, unpublished data). This matrix, in combination with a feeder-free cell culture medium (Lonza L7 hPSC Medium, see Supplemental Experimental Procedures for additional information), optimally supported the expansion of human pluripotent stem cells (unpublished data) and allowed us to develop a feeder-independent system for the generation of human iPSCs. However, we observed a significantly lower reprogramming efficiency compared with reprogramming using a feeder system. Therefore, further modifications were incorporated in the reprogramming process, including the use of hypoxic culture conditions (3%–5% O2) and a transforming growth factor β (TGF-β) inhibitor (A83-01), a selective inhibitor of the TGF-β type I receptor ALK5, which has been shown to enhance reprogramming efficiency (Ichida et al., 2009). We also incorporated Alhydrogel (i.e., an aluminum hydroxide wet gel suspension), which enhanced integration-free reprogramming under defined and feeder-free conditions (Figure S1). The mechanism of action of aluminum hydroxide is still under investigation, although it may act by the recently described “transflammation mechanism,” similar to Poly I:C, which functions through cell surface Toll-like receptors (TLRs) to promote reprogramming (Lee et al., 2012). Finally, a chemically defined, non-enzymatic passaging solution (i.e., Lonza L7 hPSC Passaging Solution) based on sodium citrate was used for serial subculture of iPSCs (Nie et al., 2014). Combining these improvements in a complete system allowed us to generate iPSCs reliably and reproducibly, as assessed by testing (Figure S2).
Raw Material Handling and Supply Chain Considerations for cGMP Manufacturing
An important component of establishing a cGMP manufacturing process includes establishing a complete list of raw materials as well as establishing relevant quality control and bioassay tests, primary and secondary vendor qualifications, raw material part setup, and inventory management (including storage conditions, tracking expiration, reorder points, and material flow to and within GMP manufacturing suites) (Stacey et al., 2013). Following establishment of the process, a bill of material (BOM) was created to evaluate the criticality of the materials for the process and assess vendors’ qualifications based on the United States regulatory requirements. In our case, vendor evaluation was performed by supply chain and quality assurance, focusing on accepted or approved vendors depending on the criticality of the materials. Approved vendors were evaluated using a written vendor questionnaire along with verification of specific tests shown on the certificate of analysis (COA) (see Supplemental Experimental Procedures for evaluation of vendors for reprogramming plasmids). Accepted vendors involved with QC test reagents or general supplies were evaluated based on obtaining generic information around the materials with appropriate COAs as well as relevant material shipment documents. Another important aspect of supply chain activities was setting up client/project-specific part numbers in the Enterprise Resource Planning (ERP) system. As part of setting up each material in the system applications and products (SAPs), all relevant information, including the COA, material origin information, and shipping conditions, were collected and included on the raw material specification sheet (RMSS). The RMSS was then reviewed and approved by the subject matter expert and QA group. A specific part number was assigned to each raw material at the end of the part setup process. SAP was used throughout the period of manufacturing process to facilitate the handling, ordering, tracking, and availability of the materials from suppliers to the warehouse and from the warehouse to the cGMP suite.
Transferring the Manufacturing Process to the cGMP Suite
Following the optimization of human iPSC generation and completing the process development phase, but prior to manufacturing in a cGMP manufacturing suite, training runs and engineering runs were performed. The purpose of training runs is training of cell therapy technicians on the process prior to execution of the actual process in a manufacturing suite as well as development of draft master batch records (MBRs). The training runs were carried out in a process development/research laboratory. Engineering runs, commonly referred to as validation runs, served as the final evaluation of all prerequisites related to manufacturing of iPSCs utilizing approved (or near-approved) production records and standard operating procedures (SOPs). The objective of the engineering runs was to identify gaps throughout the entire manufacturing process. Engineering runs were carried out in the cGMP manufacturing suite, with products (including intermediate products and final products) being tested and evaluated using cGMP testing protocols.
It should be noted that training, engineering, and cGMP manufacturing runs were all conducted using the same optimized iPSC manufacturing process established during process development (PD) runs, but they differ in the process used for CD34+ cells isolation as described below.
Phase I: Training Runs
Two training runs were initiated 20 days apart to generate iPSC lines from cryopreserved research and development (R&D)-grade, CB-derived CD34+ cells (derived from two different donors) using the defined and cGMP-compatible process and cell culture system established during the process optimization phase. Two reprogramming methods were used on each sample: episomal plasmids plus TGF-β inhibitor (method A) and episomal plasmids plus TGF-β inhibitor and 2% Alhydrogel (method B). On days 10–12 post-nucleofection, six hiPSC colonies were isolated for each method and donor for further expansion and characterization. The remaining cell culture plate was stained with alkaline phosphatase (AP). In training run 1, AP staining exhibited 110 potential iPSC colonies for method A and 182 colonies for method B. In training run 2, 54 iPSC colonies were generated using method A and 39 colonies using method B. Following serial sub-culturing of the isolated human iPSCs for four to six passages, all iPSC colonies were cryopreserved. Two iPSC clones per donor were then thawed after initial evaluation based on the quality of each clone with respect to morphology, growth, and level of spontaneous differentiation and serially sub-cultured before banking and characterization. As illustrated in Figure S3, the iPSC clones (LiPSC-TR1.1) generated during training run 1 proliferated and expanded under cGMP-compatible conditions, stained positively for pluripotent stem cell markers (OCT4, NANOG, TRA-1-60, TRA-1-81, and SSEA4), exhibited potential to differentiate into all three germ layers (ectoderm, endoderm, and mesoderm), matched the genetic identity of starting material using short tandem repeat (STR) analysis, and had a normal karyotype at passage 16. Surprisingly, karyotype analysis of both the CD34+ cells used as starting material for reprogramming and iPSCs generated during training run 2 demonstrated an abnormal female karyotype with paracentric inversion on the long arm of chromosome 7 in all 20 cells. We concluded that the karyotypic abnormality was already present in the donor cells rather than introduced during the reprogramming process. At the end of the training runs, the draft MBRs as well as quality control and testing documents, including the sampling plan, were updated.
Phase II: Engineering Runs
For the engineering runs, we elected to use CD34+ cells freshly isolated from CB for manufacturing cGMP iPSCs. Because cryopreserved CD34+ cells had been used for PD runs and training runs, we performed a gap analysis to re-evaluate the CD34+ cell isolation step. The gap analysis focused on pretreatment of the cord blood tissue prior to cell separation using Miltenyi Biotec’s CliniMACS system, improving the purity and yield of CD34+ cells, and CD34+ cell format (cryopreserved versus fresh unfrozen cells). Based on this gap analysis study, a proprietary multi-step CD34+ cell isolation process was established. Using the CliniMACS system in the CD34+ cell isolation process, we achieved a purity of >60% CD34+ cells and a yield of about 40% CD34+ isolation, with a minimum of 2.5 × 106 CD34+ cells required to start the priming step. A total of two engineering runs were performed using two R&D-grade tissues undergoing CD34+ cell isolation, followed by a 4-day priming step. Following the reprogramming of methods A and B as described earlier, nine iPSC colonies were isolated per method and donor. The selected human iPSC colonies were subcultured until passages 5–7 and cryopreserved. Each of the iPSC clones was evaluated based on cell morphology and plasmid clearance. The plasmid clearance assay was developed based on a qPCR assay (residual qPCR) to quantitatively detect residual EBNA/OriP sequences originating from both pEB-C5 and pEB-Tg (see Supplemental Experimental Procedures). Based on the results of the in-process controls, two to three clones per donor were thawed, expanded further, and banked. We found that the two clones generated during engineering run 1 and expanded further to passage 19 were not clear of the plasmids, which could indicate integration of plasmid DNA. In contrast, three iPSC clones (clones A, B, and G) generated during engineering run 2 exhibited an undetermined level of cycle threshold (Ct) value using qPCR assay at passages 10–13, confirming clearance. All three iPSC clones demonstrated (1) the ability to recover after cryopreservation, (2) expression of pluripotent stem cell markers, (3) pluripotency using embryoid body (EB) formation, (4) a genetic identity matching the starting material, and (5) normal karyotype following banking (Figure 3 shows the characterization results for LiPSC-ER2.2). The iPSCs generated during engineering run 2 also passed standard safety assays for sterility, mycoplasma, and endotoxin tests. Based on the results of the engineering runs, the MBRs were updated to include the CD34+ cell isolation process and in-process controls.
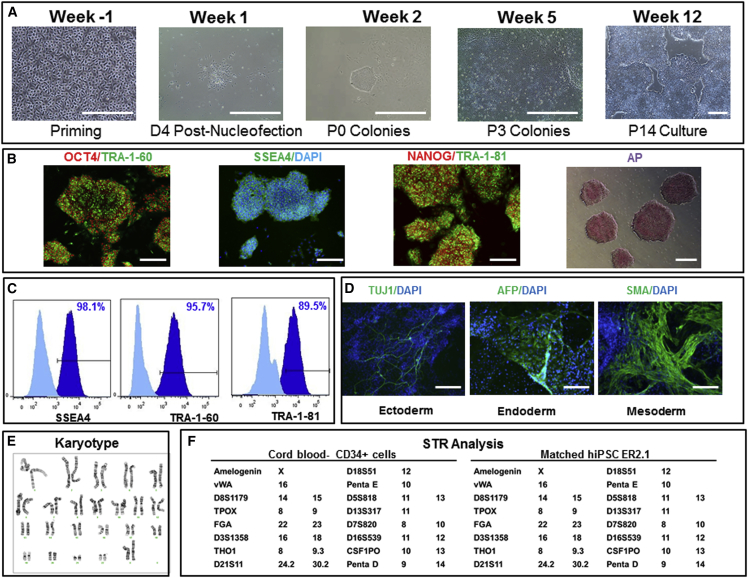
Figure 3.
Generation, Expansion, and Characterization of Human iPSCs: Engineering Runs—LiPSC-ER2.2
(A) Priming of CD34+ cells isolated from cord blood unit and expanded in culture on day 4 prior to nucleofection (Priming), the iPSC colony emerged on day 4 post nucleofection (D4 Post-Nucleofection), iPSC colonies on day 11 post nucleofection (P0 colonies), iPSCs at passage 3 (P3 colonies), and iPSCs at passage 14 (P14 culture). Scale bars, 500 μm, except in the Priming image (250 μm).
(B) iPSCs stained positively with OCT4, TRA-1-60, SSEA4, NANOG, TRA-1-81, and AP. Scale bars, 250 μm, except in the AP image (500 μm).
(C) iPSCs expressing the pluripotent stem cell surface markers SSEA4, TRA-1-60, and TRA-1-81 (dark blue). Light blue indicates the isotype control.
(D) iPSCs differentiated into embryoid bodies and readily expressing the markers for early ectoderm (TUJ1), endoderm (AFP), and mesoderm (SMA). Scale bars, 125 μm.
(E) The iPSCs demonstrated a normal karyotype after 14 passages.
(F) STR analysis showed that the iPSCs matched the starting CD34+ donor sample.
Phase III: Manufacturing Runs
After completing the tissue acquisition and recovery procedures described earlier, a fresh cord blood unit was sourced from one donor, packaged, and shipped to the manufacturing site according to the validated procedures. When received, the cGMP-compliant tissue was quarantined while initial documents associated with the tissue, including the tissue collection date (i.e., August 14, 2014), shipping and handling conditions (room temperature), and tissue volume/size (total weight of 188 g and a net cord blood volume of 156 ml) were reviewed. The manufacturing process was initiated within 36 hr of tissue recovery. The CD34+ cell isolation process resulted in the isolation of 1.74 × 107 total viable cells, 89.10% of which were CD34+ cells passing the acceptance criteria for the initial CD34+ cell isolation. Priming of the cells for 4 days resulted in a more than 3-fold expansion and increase in CD34+ cell purity (94.9% CD34+ cells) (Figure 4A). Karyotype analysis of the sample taken on day 4 demonstrated that the enriched CD34+ cells were karyotypically normal. The sterility test result was negative, confirming that the CD34+ cell isolation and priming steps were carried out in compliance with appropriate aseptic techniques. The cells were then transfected with the pEB-C5 and pEB-Tg plasmids using two nucleofection methods (one million enriched CD34+ cells per method). After 18 days in the L7 cell culture system, method A, which included TGF-β inhibitor only, resulted in the generation of nine hESC-like colonies, three of which (clones A, D, and I) survived and actively proliferated in culture after the colony isolation step. Notably, the number of colonies observed for both reprogramming procedures during GMP run 1 was significantly lower than the number of colonies observed during PD runs, training runs, and engineering runs. Donor-to-donor variability likely explains the low number of colonies observed for method A (nine potential hiPSC colonies) and no colony observed for method B (Mack et al., 2011). Despite this observation, we continued with the manufacturing process according to established GMP protocols and batch records, given the high quality of surviving iPSCs (Figure 4B) and successful completion of in-process testing (including sterility and karyotype). Clone A (product no. CTI-1134, lot no. 50-001-02A) was cryopreserved at passage 5, and clone D (product no. CTI-1134, lot no. 50-001-02D) and clone I (product no. CTI-1134, lot no. 50-001-02I) were cryopreserved at passage 6. In-process residual plasmid clearance showed that all three iPSC clones had undetermined levels of plasmid at the cryopreservation point. In parallel to manufacturing, screening and testing of the donor cells was performed by an independent laboratory registered with the FDA. The test results showed that the donor met the eligibility criteria. The donor tested negative for HIV-1, HIV-2, hepatitis B, HCV, HTLV, cytomegalovirus (CMV), syphilis, and West Nile virus. Upon confirmation of donor eligibility, the starting tissue was released from quarantine status.
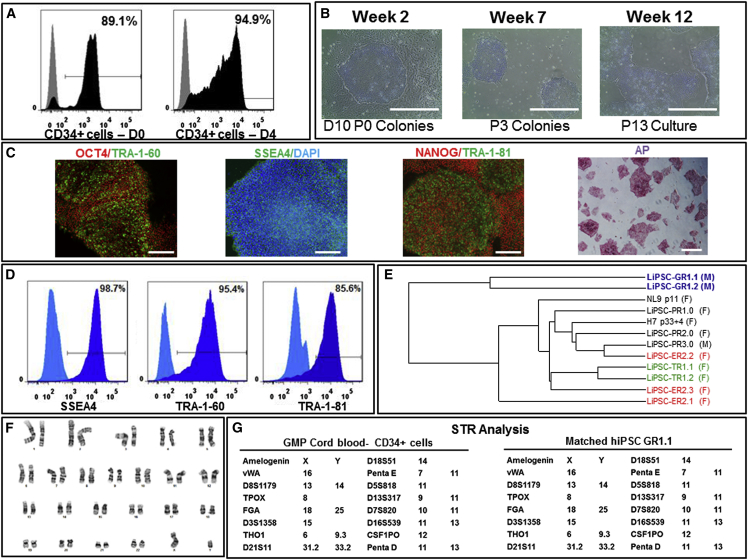
Figure 4.
cGMP Manufacturing of Human iPSCs: GMP Runs—LiPSC-GR1.1
(A) CD34+ cells isolated from afresh cord blood unit expanded and further purified from days 0–4 in the priming step. Black represents the CD34+ cell population, and gray represents the isotype control.
(B) An iPSC colony on day 10 post-nucleofection (D10 P0 Colonies), iPSCs at passage 3 (P3 Colonies), and iPSCs at passage 13 (P13 Culture). Scale bars, 500 μm.
(C) iPSCs stained positively with OCT4, TRA-1-60, SSEA4, NANOG, TRA-1-81, and AP. Scale bars, 250 μm, except in the AP image (500 μm).
(D) iPSCs expressing the pluripotent stem cell surface markers SSEA4, TRA-1-60, and TRA-1-81 (dark blue). Light blue indicates the isotype control.
(E) A dendrogram developed through whole gene expression analysis, confirming the clustering of the iPSC lines generated in this work and lines published previously. The colored lines indicate iPSC clones generated from the same donor. F and M indicate iPSC generation from female and male donors, respectively.
(F) The iPSCs demonstrated a normal karyotype after 14 passages in culture.
(G) STR analysis showed that the iPSCs matched the starting CD34+ donor sample.
Following the confirmation of donor eligibility and in-process evaluation of the cryopreserved intermediate iPSC clones, two iPSC clones, lot no. 50-001-02A (hereafter labeled LiPSC-GR1.1) and lot no. 50-001-02D (hereafter labeled LiPSC-GR1.2), were selected, thawed, serially subcultured, and finally banked. Each bank received a unique part number for identification and was tested for plasmid clearance one passage after thawing. Evaluation of the lots was conducted at each passage until plasmid clearance was confirmed for two consecutive passages (LiPSC-GR1.1 at passages 7 and 8 and LiPSC-GR1.2 at passages 8 and 9). After confirming plasmid clearance, the human iPSCs were expanded further. LiPSC-GR1.1 and LiPSC-GR1.2 were cryopreserved at passage levels P14 and P13, respectively. For each line, between 150–200 vials (2 × 106 cells/vial) were cryopreserved. Although a well-mixed cell suspension was used during the filling process, each cryovial was labeled with a number indicating the order of filling. Following the cryopreservation, cryovials from the start, middle, and end of the filling process were used for QC testing to ensure that the relatively long filling process did not adversely affect the quality of the cells.
QC Testing and Release Assays
In addition to developing a process to generate iPSCs, it was critical to establish appropriate final product release testing to ensure the identity, safety, purity, and viability of the final products (Table S1). In the absence of specific guidelines for characterization of iPSCs, the decision was made to include an assay as a release assay or for information only (FIO) based on the criticality of the assay (i.e., indicating safety, identity, or purity).
Upon evaluation of pluripotent stem cells using standard/qualified assays and other test methods, the human iPSCs manufactured under cGMP conditions fulfilled the main characteristics of pluripotent stem cells (Figures 4B–4D) and passed standard safety assays, including the plasmid clearance, karyotype (Figure 4F), STR (Figure 4G), sterility, mycoplasma, and endotoxin tests.
Gene Expression Profiling of iPSC Lines
Whole genome expression analysis was performed on iPSC lines generated during process development, training runs, engineering runs, and GMP manufacturing runs (GEO: GSE72078). Hierarchical clustering of all lines and overall correlation co-efficiency (R2 value) between each line are shown in Figure 4E and Table S2. The correlation co-efficiency between each line manufactured in this study is greater than 0.9. We further compared the gene expression profile of these lines with several iPSC lines generated and characterized previously (Pei et al., 2015, Shaltouki et al., 2015) and found them to be closely correlated with the R2 values of more than 0.9 for all lines crossed and compared (Table S2). No significant difference in gene expression profiles at a global level was observed among the lines generated in this work and lines generated previously using similar or different reprogramming methodologies (Shaltouki et al., 2015).
In addition to the overall gene expression comparison, we developed a panel of approximately 250 markers, including markers of pluripotency, gender, imprint, endoderm, mesoderm, and ectoderm that can serve as quality control for cells to be used and analyzed their expression in these lines (data not shown). As expected, several pluripotency markers, including Oct4, Nanog, and Sox2, were highly expressed in all iPSC samples. No difference was observed between the male or female lines, and no change in the expression of imprinted genes was seen. Overall, our gene expression profiling of each line was similar and similar to previously reported iPSC and ESC lines (Momčilović et al., 2014).
Validation of Neural Differentiation and Gene Targeting
To determine whether the manufactured iPSC lines can be differentiated into a neural lineage, we generated neural stem cells (NSCs) from LiPSC-TR1.2 using a well-established protocol (Swistowski et al., 2010). We observed no difference in the timeline of NSC formation between LiPSC-TR1.2 and XCL1 (XCell Science), a well characterized iPSC line reprogrammed by the identical method (Shaltouki et al., 2015). Figure 5A, a–c, shows a homogeneous population of NSCs that uniformly expressed SOX1 and NESTIN, markers of NSCs.
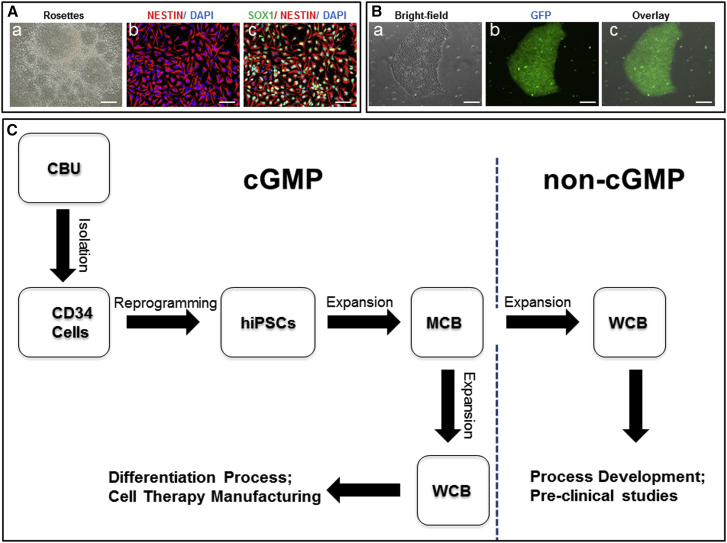
Figure 5.
Validation of Neural Differentiation and Gene Targeting and Preparation of the Human iPSC MCB and WCBs
(A and B) Use of iPSCs (LiPSC-TR1.2) generated using a cGMP-compatible process in pre-clinical studies for neural differentiation (A) and genetic engineering (B).
(A, a–c) Neural rosettes formed via EBs were isolated manually and expanded to a homogenous NSC population. Immunocytochemical analysis showed positive expression of the NSC markers SOX1 and NESTIN. Scale bars, 200 μm, except in b and c (100 μm).
(B, a–c) TALEN-mediated homologous recombination targeting the safe harbor site AAVS1 on chromosome 19. A representative example of a GFP-positive clone is shown. GFP was driven by the constitutively active CAG promoter. Scale bars, 200 μm.
(C) Human iPSCs generated under cGMP conditions can be used as MCB seed stocks to create working cell banks under both the GMP setting (for manufacturing specialized cell therapy products) and in a non-GMP environment (to carry out research studies for multiple cell therapy applications).
To demonstrate that the iPSC lines manufactured under cGMP-compliant conditions can be genetically modified, we tested one line, LiPSC-TR1.2, using a platform we developed for the rapid generation of targeted clones in iPSCs via TALEN and zinc finger nuclease (ZFN) (Pei et al., 2015). As a proof of concept, we used a TALEN construct comprised of a GFP reporter driven by the constitutively active CAG promoter targeting the AAVS1 site. As expected, a similar targeting efficiency was observed for LiPSC-TR1.2 and XCL1 (Pei et al., 2015). A representative image of a GFP-expressing clone is shown in Figure 5B, a–c.
Preparing a Seedstock and Matched Research-Grade Stock
Upon completion of the cGMP manufacturing process, an MCB of approximately 100 vials, with each vial containing at least 2 × 106 human iPSCs, was produced. The cells passed all established release criteria for iPSCs, standard QC assays, and viral testing (listed in Table S3). The MCBs are stored in vapor-phase liquid nitrogen in a warehouse following established protocols and QA oversight. These MCBs fulfill all criteria described by Stacey et al. (2013) and the references therein, including manufacturing and banking under standard GMP guidelines, traceability, high-quality (pertaining to robustness) and standard QC testing, pluripotent stem cell characteristics, and well documented storage. As illustrated in Figure 5C, iPSC MCBs can be used for developing both GMP-compliant and research-grade working cell banks (WCBs). The matched research-grade WCBs can be used by research laboratories to perform pre-clinical studies (similar to the neural differentiation and gene-targeting studies described earlier for LiPSC-TR1.2) or to establish/optimize directed differentiation protocols. These studies allow researchers to focus on generating functional and clinically relevant cell types such as dopaminergic neurons, retinal pigment epithelial cells, or insulin-producing cells for cell transplantation therapies. Indeed, we developed a research-grade WCB by thawing one MCB vial and limited expansion to prepare about 50 cryopreserved vials, which were further tested for karyotype stability. A WCB can be also generated under GMP manufacturing conditions to generate a clinical product. Considering the limited number of MCB stocks, establishing these WCBs will allow the sustainable replenishment of high-quality and valuable iPSCs generated under cGMP manufacturing guidelines (Stacey et al., 2013, Andrews et al., 2015).
Discussion
We have described a step-by-step cGMP-compliant process to generate clinically compliant cell lines. The results of bioassay/QC tests and manufacturing runs confirm that the process is robust and reproducible. Moreover, the iPSCs generated in each phase (i.e., process development, training run, engineering run, and GMP manufacturing run) fulfill pluripotent stem cell characteristics using standard assays from validated suppliers. Additionally, we have shown that we can generate reporter sub-clones for pre-clinical studies. Furthermore, these experiments confirm that the lines produced in this manner can be used for safe harbor gene editing to repair genetic defects and that such engineered lines could be manufactured using a similar clinically compliant process.
Because iPSC could be used as starting material for multiple products given the pluripotent nature of the cell line, we felt that a larger seed stock and MCB should be planned. In the HLA haplobank model that has been proposed (Turner et al., 2013, Solomon et al., 2015), the same line may be used for multiple individuals in addition to multiple indications. We expect both proponents of autologous and allogeneic iPSC-based therapy to utilize a seed stock strategy. Specifically for autologous users, their back-up cell stock can be preserved for future needs, whereas, for allogeneic products, it can be used from a MCB.
From one of the GMP manufacturing run clones, we prepared a MCB and a research-grade WCB as well. Although essentially identical, this research-grade WCB is significantly less expensive (because the cGMP regulation is not applied) and can be used for evaluating the lines and the differentiation protocols that may be used to generate a clinical product. Lonza has filed a drug master file (DMF) with the FDA that can be accessed via reference in a future investigational new drug (IND) submission. Although we used cord blood (Rao et al., 2012), our process is agnostic to the starting material, and the episomal plasmid-based electroporation protocol has been used successfully with a variety of tissue samples in multiple laboratories (Chou et al., 2011 Chou et al., 2015, Okita et al., 2011, Goh et al., 2013, Schlaeger et al., 2015).
There were three levels of concerns we had to address during the derivation process. One was related to the process of consent as discussed by Lowenthal et al., 2012. Was the process of obtaining tissue compliant with the new guidance provided by the FDA and compatible with the final donor consent rule? Had we maintained the appropriate level of confidentiality with HIPPA regulations, and could we re-approach the donor should we need further questions addressed? The second issue was keeping a backup tissue sample and multiple clones of iPSC lines from the same donor. In the end, we decided against this for the first line we generated because this line will likely be used for developing processes of differentiation and for pre-clinical studies, but, for actual therapy, each group will prefer generating its own lines. We expect, however, that, if they opt to use the described process and SOPs, then their cost will be significantly lower, and that the FDA will accept much of the pre-clinical data that have been generated with this line. The third issue was exposure to serum and xeno material in the manufacturing process. Our process, as described, does not include serum or feeders, and all incoming raw materials were assessed for their quality. Both primary and secondary suppliers were evaluated. Our process, although xeno-free, is not animal origin-free, but we have recently established a completely animal origin-free process (data not shown). All materials used in the process are commercially available, and, in the future, the animal origin-free media will be made commercially available as well. We note, however, that using xeno-free material is not an obligatory requirement in the United States for use of a biologic material in a human.
Other, more routine issues were instrumentation validation, materials suppliers, and the development of release assays. Although there is a recommendation from investigators as to which additional tests should be considered, several assays are required. In general, most tests need to be validated, and the tests must be performed by a certified vendor. Specific attention was given to the safety assays and evaluation of pluripotent markers expression using flow cytometry. As described earlier, plasmid clearance, karyotype, mycoplasma, sterility, endotoxin level, and flow cytometry assays were performed using a qualified or validated method or service provider. In addition to the required tests, additional tests, such as embryoid body formation, gene array analysis, comparative genomic hybridization + SNP microarray analysis, post-thaw evaluation using alkaline phosphatase, and colony morphology, were performed as optional tests or FIO, which is mainly due to the subjective nature of these assays or the lack of a qualified/validated service provider.
We worked with Dr. Zeng and colleagues to develop a safe harbor reporter line using methods we have described before. Our rationale here was two-fold: to obtain a reporter line that would not go silent after differentiation and could be used for pre-clinical studies as an isogenic labeled control and to demonstrate that gene targeting is possible and can be implemented at the seed bank stage (i.e., downstream of the derivation and expansion process). We expect that this will make getting approval of an engineered sub-clone as a product much easier in the future.
We note that several alternate paths could be followed to obtain a clinically compliant line (Malik and Rao, 2013, Schlaeger et al., 2015). Because these processes will use different starting materials, different reagents, and, presumably, a different site of manufacture, each of those groups will have to develop their own 510k or DMF package as part of the IND application. Investigators could choose to access our process in its entirety and reference our DMF. Alternatively, they could modify our process, which would significantly reduce their development time. Our time range to develop a process for manufacturing iPSC MCBs under GMP and performing the manufacturing runs was more than 2 years.
Although we followed all of the rules, we hesitate to use the phrase cGMP-compliant because this is a determination that will ultimately be made by the regulatory authorities when the entire IND package is provided to the FDA. We also note that the approval of the use of this line as part of an IND process does not presume that this will be deemed cGMP-compatible in other regulatory domains such as Europe, Japan, or China. We will take the DMF that we have prepared to these other authorities to develop a gap analysis. We are aware that Europe has a well-defined consultation process for this purpose (Ancans, 2012).
Our goal in making a matched research-grade iPSC line from the cells generated through manufacturing runs was to make a standardized line available that could be used to evaluate the directed differentiation process. We understand that use of this line by different organizations may still require incorporating a new risk assessment and validation to address the local reagents and conditions. However, this scenario will determine whether the processes will work or need to be standardized further. If it works with the matched research sample, then it will work with the cGMP-manufactured seed stock and, perhaps, with the lines made using the same process. If there was variability, then we would know by comparing the results between the matched research-grade line and lines made during the process development phase. We believe that this will result in a significant time and cost reduction and, perhaps even more importantly, a reduction in risk as individual investigators move their projects forward.
In summary, we believe that this is a complete report of a process for the sourcing, derivation, and development of a clinically compliant iPSC MCB that is widely available for use and evaluation. Additional lines can be generated using this reproducible process, and the process can be readily adapted to meet the European and Japanese guidelines.
Experimental Procedures
Generation of Human Induced Pluripotent Stem Cells
Cryopreserved human umbilical cord blood (hUCB) CD34+ cells (Lonza, 2C-101) were thawed and expanded in a priming medium described in the Supplemental Experimental Procedures. 1 × 106 hUCB CD34+ cells were reprogramed using episomal plasmids encoding Oct4, Sox2, Klf4, c-Myc, and Lin28 and pEB-Tg (Chen et al., 2011, Dowey et al., 2012) using the 4D nucleofector system and P3 solution kit (Lonza, V4XP-3012). Details regarding isolation and expansion of iPSC colonies are provided in the Supplemental Experimental Procedures.
Characterization of Human Induced Pluripotent Stem Cells
Human iPSCs were characterized using in-process and final characterization methods, including flow cytometry and immunocytochemistry techniques evaluating the expression of pluripotent stem cells markers, karyotype, and STR analyses, embryoid body formation, whole genome expression, qPCR for the detection of residual plasmids, and standard safety assays, as described in the Supplemental Experimental Procedures.
Detailed procedures are provided in the Supplemental Experimental Procedures and are available upon request or by accessing the DMF.
Author Contributions
M.S.R., T.F., and B.A.B. wrote the manuscript, designed process development and optimization studies, and provided supervision and intellectual guidance. B.A.B., X.T., B.H.N., A.B., and T.D. carried out the cGMP manufacturing and quality control assays. X.Z. and G.S. contributed to gene engineering, NSC differentiation, and data analysis. K.W. and D.P.K. provided intellectual and financial support and helped with writing the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgments
The authors thank the staff and colleagues at Lonza and the Buck Institute and members of Dr. Rao’s group who provided intellectual input and practical advice and access to specialized expertise. See Supplemental Information for a detailed list of individuals who supported this work.
Published: September 24, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, three figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.08.015.
Contributor Information
Thomas Fellner, Email: thomas.fellner@lonza.com.
Mahendra S. Rao, Email: rao1789@gmail.com.
Accession Numbers
The accession number for the data reported in this paper is GEO: GSE72078.
Supplemental Information
References
- Ancans J. Cell therapy medicinal product regulatory framework in Europe and its application for MSC-based therapy development. Front. Immunol. 2012;3:253. doi: 10.3389/fimmu.2012.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.W., Cavagnaro J., Deans R., Feigal E., Horowitz E., Keating A., Rao M., Turner M., Wilmut I., Yamanaka S. Harmonizing standards for producing clinical-grade therapies from pluripotent stem cells. Nat. Biotechnol. 2014;32:724–726. doi: 10.1038/nbt.2973. [DOI] [PubMed] [Google Scholar]
- Andrews P.W., Baker D., Benvinisty N., Miranda B., Bruce K., Brüstle O., Choi M., Choi Y.M., Crook J.M., de Sousa P.A. Points to consider in the development of seed stocks of pluripotent stem cells for clinical applications: International Stem Cell Banking Initiative (ISCBI) Regen. Med. 2015;10(2, Suppl):1–44. doi: 10.2217/rme.14.93. [DOI] [PubMed] [Google Scholar]
- Carpenter M.K., Rao M.S. Concise review: making and using clinically compliant pluripotent stem cell lines. Stem Cells Transl. Med. 2015;4:381–388. doi: 10.5966/sctm.2014-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.C., Couture S.M., Ye J., Lin Z., Hua G., Huang H.-I.P., Wu J., Hsu D., Carpenter M.K., Couture L.A. Scalable GMP compliant suspension culture system for human ES cells. Stem Cell Res. (Amst.) 2012;8:388–402. doi: 10.1016/j.scr.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Chou B.K., Mali P., Huang X., Ye Z., Dowey S.N., Resar L.M., Zou C., Zhang Y.A., Tong J., Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou B.K., Gu H., Gao Y., Dowey S.N., Wang Y., Shi J., Li Y., Ye Z., Cheng T., Cheng L. A facile method to establish human induced pluripotent stem cells from adult blood cells under feeder-free and xeno-free culture conditions: a clinically compliant approach. Stem Cells Transl. Med. 2015;4:320–332. doi: 10.5966/sctm.2014-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook J.M., Peura T.T., Kravets L., Bosman A.G., Buzzard J.J., Horne R., Hentze H., Dunn N.R., Zweigerdt R., Chua F. The generation of six clinical-grade human embryonic stem cell lines. Cell Stem Cell. 2007;1:490–494. doi: 10.1016/j.stem.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Dowey S.N., Huang X., Chou B.K., Ye Z., Cheng L. Generation of integration-free human induced pluripotent stem cells from postnatal blood mononuclear cells by plasmid vector expression. Nat. Protoc. 2012;7:2013–2021. doi: 10.1038/nprot.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerström C., Strehl R., Moya K., Andersson K., Bergh C., Lundin K., Hyllner J., Semb H. Derivation of a xeno-free human embryonic stem cell line. Stem Cells. 2006;24:2170–2176. doi: 10.1634/stemcells.2006-0130. [DOI] [PubMed] [Google Scholar]
- Fairchild P.J. Taming the lion: the challenge of immunity in regenerative medicine. Regen. Med. 2015;10:227–229. doi: 10.2217/rme.15.15. [DOI] [PubMed] [Google Scholar]
- Goh P.A., Caxaria S., Casper C., Rosales C., Warner T.T., Coffey P.J., Nathwani A.C. A systematic evaluation of integration free reprogramming methods for deriving clinically relevant patient specific induced pluripotent stem (iPS) cells. PLoS ONE. 2013;8:e81622. doi: 10.1371/journal.pone.0081622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of tgf-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Sayed N., Hunter A., Au K.F., Wong W.H., Mocarski E.S., Pera R.R., Yakubov E., Cooke J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal J., Lipnick S., Rao M., Hull S.C. Specimen collection for induced pluripotent stem cell research: harmonizing the approach to informed consent. Stem Cells Transl. Med. 2012;1:409–421. doi: 10.5966/sctm.2012-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack A.A., Kroboth S., Rajesh D., Wang W.B. Generation of induced pluripotent stem cells from CD34+ cells across blood drawn from multiple donors with non-integrating episomal vectors. PLoS ONE. 2011;6:e27956. doi: 10.1371/journal.pone.0027956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N., Rao M.S. A review of the methods for human iPSC derivation. Methods Mol. Biol. 2013;997:23–33. doi: 10.1007/978-1-62703-348-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momčilović O., Liu Q., Swistowski A., Russo-Tait T., Zhao Y., Rao M.S., Zeng X. Genome wide profiling of dopaminergic neurons derived from human embryonic and induced pluripotent stem cells. Stem Cells Dev. 2014;23:406–420. doi: 10.1089/scd.2013.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y., Walsh P., Clarke D.L., Rowley J.A., Fellner T. Scalable passaging of adherent human pluripotent stem cells. PLoS ONE. 2014;9:e88012. doi: 10.1371/journal.pone.0088012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Pei Y., Sierra G., Sivapatham R., Swistowski A., Rao M.S., Zeng X. A platform for rapid generation of single and multiplexed reporters in human iPSC lines. Sci. Rep. 2015;5:9205. doi: 10.1038/srep09205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. Scalable human ES culture for therapeutic use: propagation, differentiation, genetic modification and regulatory issues. Gene Ther. 2008;15:82–88. doi: 10.1038/sj.gt.3303061. [DOI] [PubMed] [Google Scholar]
- Rao M., Condic M.L. Alternative sources of pluripotent stem cells: scientific solutions to an ethical dilemma. Stem Cells Dev. 2008;17:1–10. doi: 10.1089/scd.2008.0013. [DOI] [PubMed] [Google Scholar]
- Rao M., Ahrlund-Richter L., Kaufman D.S. Concise review: Cord blood banking, transplantation and induced pluripotent stem cell: success and opportunities. Stem Cells. 2012;30:55–60. doi: 10.1002/stem.770. [DOI] [PubMed] [Google Scholar]
- Schlaeger T.M., Daheron L., Brickler T.R., Entwisle S., Chan K., Cianci A., DeVine A., Ettenger A., Fitzgerald K., Godfrey M. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.D., Hubschman J.P., Heilwell G., Franco-Cardenas V., Pan C.K., Ostrick R.M., Mickunas E., Gay R., Klimanskaya I., Lanza R. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- Shaltouki A., Sivapatham R., Pei Y., Gerencser A.A., Momčilović O., Rao M.S., Zeng X. Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Reports. 2015;4:847–859. doi: 10.1016/j.stemcr.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Pitossi F., Rao M.S. Banking on iPSC--is it doable and is it worthwhile. Stem Cell Rev. 2015;11:1–10. doi: 10.1007/s12015-014-9574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G.N., Crook J.M., Hei D., Ludwig T. Banking human induced pluripotent stem cells: lessons learned from embryonic stem cells? Cell Stem Cell. 2013;13:385–388. doi: 10.1016/j.stem.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Swistowski A., Peng J., Liu Q., Mali P., Rao M.S., Cheng L., Zeng X. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannenbaum S.E., Turetsky T.T., Singer O., Aizenman E., Kirshberg S., Ilouz N., Gil Y., Berman-Zaken Y., Perlman T.S., Geva N. Derivation of xeno-free and GMP-grade human embryonic stem cells--platforms for future clinical applications. PLoS ONE. 2012;7:e35325. doi: 10.1371/journal.pone.0035325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Leslie S., Martin N.G., Peschanski M., Rao M., Taylor C.J., Trounson A., Turner D., Yamanaka S., Wilmut I. Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell. 2013;13:382–384. doi: 10.1016/j.stem.2013.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.