Abstract
Most imprinted genes are concerned with embryonic development, especially placental development. Here, we identified a placenta-specific imprinted gene Qpct. Our results show that Qpct is widely expressed during early embryonic development and can be detected in the telecephalon, midbrain, and rhombencephalon at E9.5–E11.5. Moreover, Qpct is strikingly expressed in the brain, lung and liver in E15.5. Expression signals for Qpct achieved a peak at E15.5 during placental development and were only detected in the labyrinth layer in E15.5 placenta. ChIP assay results suggest that the modification of histone H3K4me3 can result in maternal activating of Qpct.
Keywords: expression patterns, H3K4me3, imprinted gene, Qpct, placenta
INTRODUCTION
It is well-known that the generation of mammals requires homologous genes from both parents. However, a large number of genes, termed imprinted genes, exhibit parent-of-origin-specific expression patterns. Until recently, about 150 imprinted genes have been characterized in the murine genome. Most of the imprinted genes are well-conserved in mammals (Plasschaert and Bartolomei, 2014). The majority of identified imprinted genes in the mouse influenced embryonic development stages (Lim and Ferguson-Smith, 2010; Schulz et al., 2010). For instance, the imprinting control region of Meg1/Grb10 can regulate the pre- and postnatal embryonic weight of the mouse (Shiura et al., 2009). Some related imprinted genes are concerned with the regulation of placental growth. For example, Igf2 contributes to placental growth, while Igf2r inhibits placental growth (Coan et al., 2005; Nafee et al., 2008; Wang et al., 2013). Grb10 expressed in the placenta limit placental size and efficiency (Charalambous et al., 2010).
Qpct encodes glutaminyl-peptide cyclotransferase. In the mouse, Qpct is located on chromosome 17 (chr17:79451246-79489583) and encodes glutaminyl cyclase, which is a Zn++-dependent acyltransferase with a mixed α-helix and β- sheet structure. It has been isolated from animals, plants, and bacteria (Busby et al., 1987; Messer and Ottesen, 1964). The cyclase converts N-terminal glutaminyl residues into pyroglutamate, and plays a role in stabilizing these proteins (Fischer and Spiess, 1987; Hartlage-Rubsamen et al., 2009; Schilling et al., 2008). QC is the alias of Qpct and its mRNA expressed level is the highest in the brain. And the formation of QC is conductive to obtain a greater understanding learning, and memory (Atwood et al., 2005; Iourov et al., 2014), which is vital for treating Alzheimer’s disease. In addition, QC is co-localized with Golgi apparatus, endoplasmic reticulum, and secretory granules (Hartlage-Rubsamen et al., 2009). Furthermore, in the latest studies, it has been found that Qpct has a close relationship with some cancers, such as melanoma (Gillis, 2006), papillary thyroid carcinoma (da Silveira Mitteldorf et al., 2011; Jarzab et al., 2005), and renal cell carcinoma (Morris et al., 2011).
DNA methylation, histone modification, and RNA-associated silencing can lead to genomic imprinting (Egger et al., 2004). It has been reported that Lys4 methylation of histones is connected with transcriptional activity (Carr et al., 2007).As a repressive mark, H3K9me3 was associated with methylated maternal and paternal alleles of the H19/Igf2 imprinting control region (Singh and Szabo, 2012). Besides H3K9me3, the methylation of histone H3K27 correlated with the allele-specific expression of the major-type Grb10 transcript (Yamasaki-Ishizaki et al., 2007). Many studies have confirmed that this abnormal imprinting was closely related to disorders in growth and placental function (Trasler, 2006). Therefore, a study of regulatory mechanism of imprinted genes’ in the placenta becomes important. Qpct has been predicted as a potential imprinted gene with DNA sequence features (Brideau et al., 2010).
In this study, we redefined Qpct to be a placenta-specific maternally expressed gene. Meanwhile, we explored the expression of Qpct in the mouse embryo and placenta using in situ hybridization (ISH), real-time quantitative PCR (qRT-PCR) and immunohistochemistry. We further investigated the imprinting regulatory mechanism of Qpct by methylation analysis and chromatin immunopreciptation (ChIP) assays, and found that the modification of histone H3K4me3 can result in maternal activating of Qpct. Furthermore, Qpct could provide a target in the treatment of disease related to the development of the placenta and embryo.
MATERIALS AND METHODS
Animals and preparation of embryos
Male and female mice (C57BL/6J) were obtained from Beijing Laboratory Animal Research Center (China). The presence of a vaginal plug the next morning was defined as 0.5 day post-conception. The day of conception was designated as embryonic day (E). The embryos were obtained and used for qRT-PCR and ISH analyses. All the experiments were approved and carried out according to the “Rules for Experimental Animals” published by the Chinese Government.
Imprinting analysis
DNA was extracted from BDF1 (C57BL/6♀ × DBA/2♂) mice using proteinase K (Roche, Germany) digestion, followed by phenol-chloroform and ethanol. The total RNA of brain, tongue, heart, lung, liver, and placenta and chorionic plate from BDF1 (C57BL/6♀ × DBA/2♂) and DBF1 (DBA/2♀ × C57BL/6♂) embryos at day E15.5 was isolated using TRIzol (Invitrogen, USA). RNA was treated with DNaseI (USA), and oligo-dT-primed reverse transcription (RT) reactions were carried out using 2 μg total RNA and a Superscript™ III RNase H-Reverse Transcriptase kit (Invitrogen). The specific primers (sense, 5′-ACC ATT GAC AATCTCAAC-3′ and antisense, 5′-CAC TAT ACC ATA CAG CAT C-3′) were designed to amplify a small region containing the SNP. The PCR conditions were as follows: amplification at 95°C, for 30 s, 50°C, for 30 s, and 72°C, for 32 s for 30 cycles. The PCR reaction products were purified using the Axygen PCR Purification kit (Axygen) and sequenced.
In-situ hybridization
To prepare Qpct RNA probes, a mouse cDNA fragment was subcloned into pBluescript® II KS (+) phagemids (Strata-gene, USA) T-vector using a DNA Ligation Kit (TaKaRa, China) according to the manufacturer’s protocol, using the same primers as in the imprinting analysis. A digoxigenin (DIG)-UTP labeled RNA probe was generated using a DIG RNA labeling kit (Roche Molecular Biochemicals, Germany). For whole- mount in-situ hybridization (WISH) analysis, embryos at E9.5, E10.5, and E11.5 were fixed with 4% paraformaldehyde in phosphate buffered saline and incubated at 4°C overnight. For ISH, fixed embryos and placentas (E15.5) were dehydrated and subsequently embedded in paraffin. Sections (8 μm) of the embryos were placed on slides, and WISH and ISH were performed according to protocols described previously (Moorman et al., 2001; Wilkinson and Nieto, 1993).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from embryonic organs and placentas according to standard protocols using TRIzol reagent (Invitrogen). The cDNAs were synthesized using a Superscript™ III RNase H-Reverse Transcriptase kit (Invitrogen). The cDNAs were subsequently used for quantitative analysis of gene expression by qRT-PCR with a Perfect Real-Time SYBR® Premix Ex TaqTM Kit (TaKaRa) using a ABI PRISM 7500 Real-Time PCR System. The sequences for the Qpct-specific primer were: sense, 5′-CCT GTC TCT GAC AGC TGG GAA-3′ and anti-sense, 5′-TTT TGC CAC ATT TCA GAA ATG CTA GTG-3′. The Gapdh gene (sense, 5′-GTC GTG GAG TCT ACT GGT GTC-3′; antisense, 5′-GAG CCC TTC CAC AAT GCC AAA-3′) and the β-actin gene (sense, 5′-AAA TGG TGA AGG TCG GTG TGA-3′; antisense, 5′-CCG TTG AAT TTG CCG TGA GTG-3′) were used as a loading control. Cycle threshold values were normalized against the β-actin gene. Experiments were carried out in triplicate for each sample.
Immnocytochemistry
To detect Qpct experssion in mouse placenta at the protein level, the E12.5, E13.5, E15.5 and E18.5 placental paraffin sections were dehydrated with graded ethanol and xylene and washed with 1× PBS. Then antigen retrieval was conducted by using 10 mM sodium citrate buffer (pH6.0) for 10min. The sections were blocked with 5% BSA in PBS for 1 h. Anti-Qpct antibody (cat. HPA008406, Sigma, USA) was diluted 1:500 in blocking buffer and incubated at 4°C overnight. Next that, the sections were incubated with Alexa Flour488(Invitrogen) labeled anti-rabbit secondary antibody (1:1000 dilution) at 37°C for 1 h, washed with PBS and counterstained with DAPI for 10min at RT. The signals were imaged with using a fluorescence microscope (Olympus IX71; Olympus, Japan).
DNA methylation and chromatin immunoprecipitations (ChIP) assay
For methylation analysis of oocytes, sperm, and placentas, 160 oocytes were directly subjected to proteinase K digestion and bisulfite treated, DNA of sperm was isolated from paternal mature sperms and (C57BL/6 × DBA and DBA × C57BL/6), and placental DNA was isolated from E15.5 F1-generation placenta. Then the DNA (sperm, placenta and chorionic plate) and treated oocyte samples were analyzed using an EZ DNA methylation-Gold kit (Zymo Research, USA), according to the manufacturer’s instructions. Chromatin immunoprecipitation assays were performed using a ChIP assay kit (Upstate Biotechnology, USA) following the manufacturer’s instructions. The bisulfite-treated DNA was amplified by nested PCR using 2× GoldStar Best MasterMix (Cowin Biotech, China). The PCR products were sequenced using an ABI PRISM 3500 Genetic Analyzer (Applied Biosystems, USA). Nested PCR primer sequences were: forword, 5′-TAG GTT TTT GGG AGA GGA TTG T-3′; reverse-out, 5′-ATA ATT AAT TTA GGG GTG GAG AAG G-3′; reverse-in, 5′-AAT TCC CAA CTA TCA AAA ACA AAA C-3′. The antibody were used as following, anti-H3K4me3 and H3K4me1 (Cell Signaling Technology, USA), anti-H3K9me3 antibody (ab8898, Abcam, United Kingdom), normal rabbit IgG (Santa Cruz Biotechnology, USA).
RESULTS
Qpct is specifically expressed from the maternal allele in the placenta
The mouse Qpct gene has seven exons in the mouse genome, and its full-length DNA and mRNA span 38 kb and 1.9 kb, respectively. We retrieved a CpG island of 197 bp in length, overlapping the exon 1 and the intron 1 of Qpct from the UCSC Genome Browser (Fig. 1A). A DNA polymorphism (T/C) at exon 7 was detected between C57BL/6 and DBA. The polymorphic site was used to distinguish monoallelic and biallelic expression. The results showed that Qpct was exclusively maternal allele-specifically expressed in the placenta and chorionic plate at E15.5 (Fig. 1B).
Fig. 1.
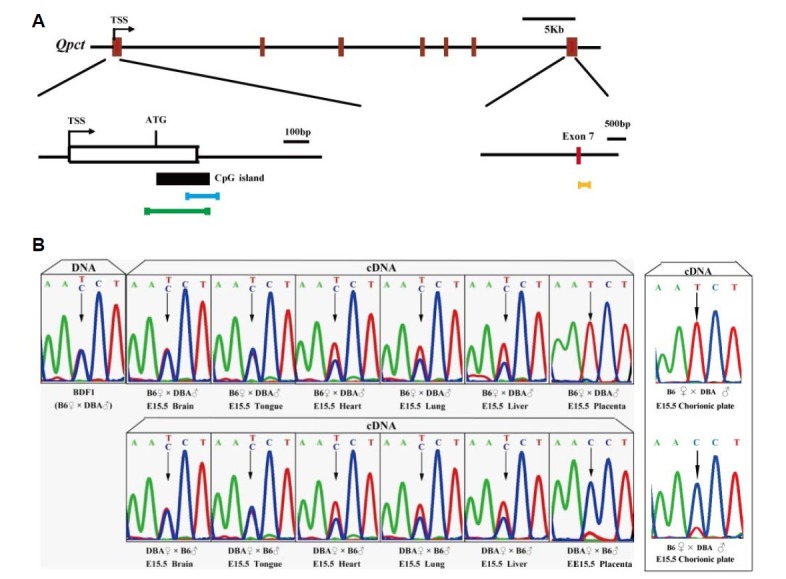
Genomic structure diagram and imprinting analysis of Qpct gene in mouse embryo and placenta. (A) Brown-red, exon; black vertical line, intron; blue horizontal line, position of methylation analysis primers; green horizontal line, position of ChIP analysis primers; yellow horizontal line, position of imprinting analysis primers; TSS, transcriptional start site. (B) Sequencing of genomic DNA from E15.5 C57BL/6 and DBA embryos, placenta and chorionic plate from hybrid F1 is heterozygous for polymorphism C/T. Sequencing of cDNA from hybrid F1 proves maternally biased expression of Qpct in E15.5 placenta and chorionic plate.
Expression of Qpct by qRT-PCR and in-situ hybridization
To determine the relative expression of Qpct at different embryonic stages in mice, qRT-PCR was carried out from E8.5 to E15.5. During mid-gestation mouse embryogenesis (E8.5–E10.5), Qpct exhibited a low expression pattern. However, its expression increased gradually after E11.5 and was highest at E15.5 (Fig. 2A). To further determine the spatiotemporal expression patterns of Qpct, WISH was performed during E9.5–E11.5 (Fig. 2B). At E9.5, strong signals were detected in the telencephalon, midbrain, and rhombencephalon (Fig. 2B, a). At E10.5, in addition to strong Qpct expression in the brain, intermediate expression signals were observed in the optic vesicle and lateral plate mesoderm (Fig. 2B, b). At E11.5, a higher expression of Qpct was found in the forebrain, midbrain, rhombencephalon, and neural tube (Fig. 2B, c–e). Furthermore, Qpct expression among various tissues at E15.5 was investigated by in-situ hybridization. Higher expressions of Qpct were found in the brain, lungs and liver while relative lower expression were found in the tongue and kidney (Fig. 2D). Similar expression patterns were further confirmed by qRT-PCR (Fig. 2C).
Fig. 2.
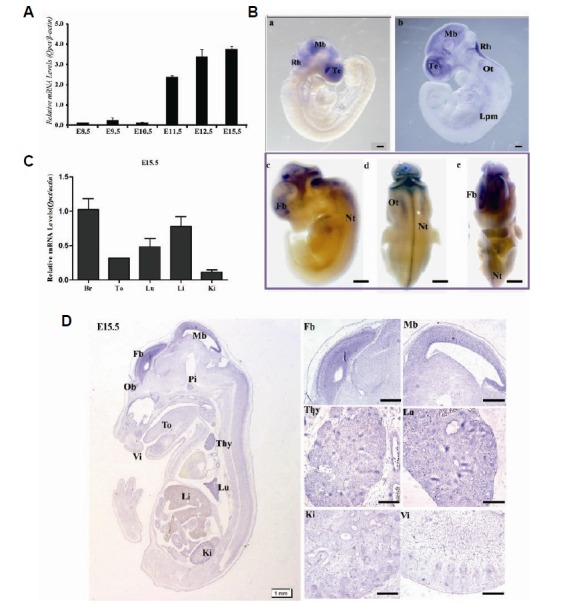
Spatiotemporal expression pattern of Qpct during mouse embryogenesis. (A, B) Expression of Qpct in E9.5, E10.5, and E11.5 embryos by whole-mount in-situ hybridization and qRT-PCR. Scale bars, 100 μm (B, a–b); 200 μm (B, c–e). Fb, forebrain; Lpm, lateral plate mesoderm; Mb, midbrain; Te, telencephalon. (C) Qpct mRNA expression level in major E15.5 tissues determined by qRT-PCR; qRT-PCR data were normalized to the level of β-actin. (D) Sagittal sections demonstrating widespread expression of Qpct in E15.5 developmental tissues by in-situ hybridization. Scale bar, 50 μm. Thy, thymus; Pi, pituitary; Lu, lung; Li, liver; Ki, kidney; Ob, olfactory bulb; Vi, vibrissae follicle, To, tongue.
Expression patterns of Qpct in placental development
Owing to the imprinting of Qpct in the placenta, we further examined the placental expression of Qpct. Expression of Qpct was higher at E12.5 and achieved a peak at E15.5, decreasing until E18.5 (Fig. 3A). The immunocytochemistry were carried out to detect the expression of Qpct. The results showed that the main localization signals were clearly observed in the decidual layer and labyrinth in E12.5 and E13.5 placentae. However, the strong expression of Qpct was found in E15.5 placenta’s labyrinth (Fig. 3B).
Fig. 3.
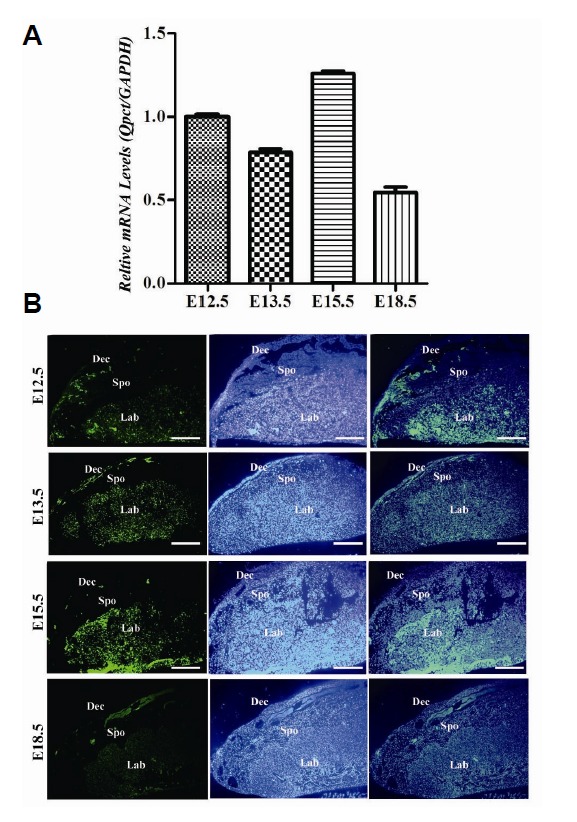
Qpct expression in the placenta. (A) Qpct mRNA expression level during early developmental stages of the placenta; qRT-PCR data were normalized to the level of GAPDH. (B) Sagittal sections of E12.5, E13.5 and E18.5, demonstrating by immunocytochemistry that the greatest expression is in the decidual layer and labyrinth. In E15.5 placenta, Qpct was strong expressed in labyrinth layer. Scale bar = 50 μm
DNA methylation and histone modification analysis of Qpct
Early studies have affirmed that epigenetic reprogramming in germ cells is of critical importance for imprinting. Therefore, we analyzed the methylation status of an existing CpG island spanning 197 bp with 16 CpG sites overlapping the exon 1 and the intron 1 of Qpct. The bisulfate sequencing was carried out in sperm, oocytes and placentas at E15.5. The results showed that the CpG sites were completely unmethylated in these tissues (Fig. 4A). Thus, we were interested whether the expression of Qpct is regulated by histone modification. The results suggested that histones H3K4me3, H3K4me1 and H3K9me3 were indeed enriched at E15.5 placentae (IBF1, BIF1). In the meantime, E15.5 liver was used as negative control (Fig. 4B). We found that only H3K4me3 was preferentially enriched on unmethylated or normally active alleles. As a marked signature, H3K4me3 was related to the expression of some imprinted genes. An SNP (G/T) at exon 1 of Qpct between C57BL/6 and ICR mice was used to distinguish maternal or paternal allelic expression. ChIP primers were designed to span the SNP (G/T) that allowed us to recognize which allele was enriched by more H3K4me3, H3K4me1 and H3K9me3 signals. The enriched DNA fragments amplified by the designed ChIP primer were collected and sequenced. The ChIP results showed that there was a higher proportion of co-enrichment of active (H3K4me3) at imprinted loci in the maternal chromosome with reciprocal crosses between ICR and C57BL/6 (Fig. 4C). These results illustrated that H3K4me3 histone modification can regulate the expression of Qpct.
Fig. 4.
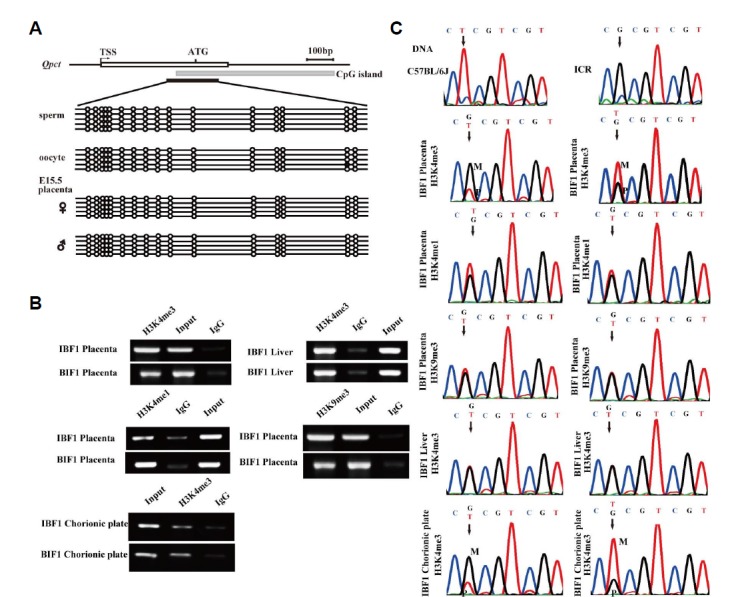
Analysis of the imprinting regulatory mechanism of Qpct. (A) Methylation analysis of Qpct in mouse sperm, oocyte, and E15.5 placenta. The methylation primers include 16 CG loci in the CpG island. (B) ChIP assays were performed in placenta and liver (E15.5 BIF1 and IBF1). The immunoprecipated DNA was analyzed by ChIP PCR primers. (C) H3K4me3, H3K4me1 and H3K9me3 histone modification analysis between C57BL/6 and ICR mouse placentae and chorionic plates. G/T (SNP) denotes the M/P peak height by dividing the percentages precipitation of the maternal and paternal alleles (‘M’ and ‘P’ indicate maternal and paternal alleles, as recognized by the SNP); BIF1 (C57BL/6 × ICR); IBF1 (ICR × C57BL/6).
DISCUSSION
Studies have reported that imprinted genes are closely linked with fetal growth, pluripotency and carcinogenesis. The majority of imprinted genes are expressed in the early developmental stages (Wang et al., 2011) .Therefore the exploration of imprinted gene regulation has become a crucial research area.
Gene regulation has been widely-used to predict potential imprinted genes with DNA sequence features in recent research (Brideau et al., 2010). However, Hiroaki Okae et al. (2012) regarded Qpct as not a maternally expressed imprinted gene. They speculate that Qpct is mistaken for an imprinted gene because of its high expressing in the decidual layer. These data were obtained from a novel approach that uses embryo transfer technology to remove maternal contamination. The imprinted status of Qpct was identified using this method. However, it is worth noting that embryo transfer experiments and invitro fertilization might change the status of imprinting. It was previously demonstrated that the H19 imprinted gene was found to be biallelically expressed because of embryos culture by in-vitro fertilization (Doherty et al., 2000). In addition, many other genes display the same phenomenon, such as Snrpn (Mann et al., 2004), Xist (McDonald et al., 1998), and its anti-sense transcript Tsix (Prissette et al., 2001). To avoid contaminating maternal cells, we performed imprinted analysis of Qpct with E15.5 mouse placenta which seldom expressed in decidual layer. Then the experiments were conducted separately in the placentae from which the decidua was removed completely and in the chorionic plates of placentae. The results showed that Qpct was a maternally allele-specific expression gene in E15.5 mouse placenta and chorionic plate. In addition, the WISH results showed that Qpct might play an important role during mid-gestation mouse embryogenesis. Moreover, Qpct was extensively expressed in the telencephalon, pituitary, neural tube, and lateral plate mesoderm, which may be closely linked to the neuroendocrine system. Immunocytochemistry results indicated that Qpct is mainly expressed in the labyrinth and decidual layers of the placenta. It is well that both layers contain plenty of fetal blood vessels and blood sinusoids. As a result, it can not be ignored that Qpct might make a great difference in nutrient substance transport in placenta.
So far, DNA methylation and histone modification have been identified as two main epigenetic modifications that regulate imprinted genes (Novakovic and Saffery, 2012; Reik and Dean, 2001). Furthermore, the imprint status of imprinted genes changed along with the variation of methylation modification (Santos et al., 2002). Therefore we assayed the CpG island near the Qpct promoter region and showed that Qpct was not regulated by DNA methylation. However, the placenta is a diversifying tissue and contains many differentiated cell types. Each layer of cells may show a different methylation status. For example, owing to different cell types, the DNA methylation of cytotrophoblasts and fibroblasts is different in the human placenta (Grigoriu et al., 2011). Therefore, it is also possible that the imprinted mechanism of Qpct is related to DNA methylation modification in placenta. Moreover, some other kinds of DNA fragments might regulate the imprinting of Qpct. In future, Qpct methylation analysis can be employed in other genomic regions.
In addition to DNA methylation, modifications of histone proteins are also important regulators in imprinted genes. Specific histone modifications are needed to direct imprinted maintenance (Li, 2002). According to a related research report, the methylation of Lys4 (H3K4) is associated with active transcriptional activity (Grewal and Elgin, 2002), while H3K9me3 and H3K27me3 are linked to inhibited transcriptional activity (Hagarman et al., 2013). In our study, to determine whether the paternal silencing and maternal expression of Qpct were due to the methylation of particular residues on histone H3, ChIP as-says were performed using H3K4me3, H3K9me3, and H3K4me1 antibodys. There was greater enrichment of histone H3K4me3 on the maternal chromosome, relative to the paternal one of Qpct. Biparental chromosomes had the same enrichment of histone modification using H3K9me3 and H3K4me1 antibody. Therefore, it is suggested that the enrichment of H3K4me3 leads to specific maternal expression of Qpct.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos.31371478, 31171383 and 31100928), the State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (No.2013DX07), the Natural Science Foundation of Heilongjiang Province (No.QC2014C016), and the Innovation and Technology Special Fund for Researchers of Haerbin City (No. 2013RFXXJ010).
REFERENCES
- Atwood C.S., Meethal S.V., Liu T., Wilson A.C., Gallego M., Smith M.A., Bowen R.L. Dysregulation of the hypothalamic-pituitary-gonadal axis with menopause and andropause promotes neurodegenerative senescence. J. Neuropathol. Exp. Neurol. 2005;64:93–103. doi: 10.1093/jnen/64.2.93. [DOI] [PubMed] [Google Scholar]
- Brideau C.M., Eilertson K.E., Hagarman J.A., Bustamante C.D., Soloway P.D. Successful computational prediction of novel imprinted genes from epigenomic features. Mol. Cell. Biol. 2010;30:3357–3370. doi: 10.1128/MCB.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby W.H., Quackenbush G.E., Humm J., Youngblood W.W., Kizer J.S. An enzyme(s) that converts glutaminylpeptides into pyroglutamyl-peptides - presence in pituitary, brain, adrenal-medulla, and lymphocytes. J. Biol. Chem. 1987;262:8532–8536. [PubMed] [Google Scholar]
- Carr M.S., Yevtodiyenko A., Schmidt C.L., Schmidt J.V. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics. 2007;89:280–290. doi: 10.1016/j.ygeno.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous M., Cowley M., Geoghegan F., Smith F.M., Radford E.J., Marlow B.P., Graham C.F., Hurst L.D., Ward A. Maternally-inherited Grb10 reduces placental size and efficiency. Dev. Biol. 2010;337:1–8. doi: 10.1016/j.ydbio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Coan P.M., Burton G.J., Ferguson-Smith A.C. Imprinted genes in the placenta--a review. Placenta. 2005;26(Suppl A):S10–20. doi: 10.1016/j.placenta.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Da Silveira Mitteldorf C.A., de Sousa-Canavez J.M., Leite K.R., Massumoto C., Camara-Lopes L.H. FN1, GALE, MET, and QPCT overexpression in papillary thyroid carcinoma: molecular analysis using frozen tissue and routine fine-needle aspiration biopsy samples. Diagn Cytopathol. 2011;39:556–561. doi: 10.1002/dc.21423. [DOI] [PubMed] [Google Scholar]
- Doherty A.S., Mann M.R.W., Tremblay K.D., Bartolomei M.S., Schultz R.M. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Fischer W.H., Spiess J. Identification of a mammalian glutaminyl cyclase converting glutaminyl into pyroglutamyl peptides. Proc. Natl. Acad. Sci. USA. 1987;84:3628–3632. doi: 10.1073/pnas.84.11.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J.S. Microarray evidence of glutaminyl cyclase gene expression in melanoma: implications for tumor antigen specific immunotherapy. J. Transl. Med. 2006;4:27. doi: 10.1186/1479-5876-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I.S., Elgin S.C. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- Grigoriu A., Ferreira J.C., Choufani S., Baczyk D., Kingdom J., Weksberg R. Cell specific patterns of methylation in the human placenta. Epigenetics. 2011;6:368–379. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagarman J.A., Motley M.P., Kristjansdottir K., Soloway P.D. Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS One. 2013;8:e53880. doi: 10.1371/journal.pone.0053880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlage-Rubsamen M., Staffa K., Waniek A., Wermann M., Hoffmann T., Cynis H., Schilling S., Demuth H.U., Rossner S. Developmental expression and subcellular localization of glutaminyl cyclase in mouse brain. Int. J. Dev. Neurosci. 2009;27:825–835. doi: 10.1016/j.ijdevneu.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Iourov I.Y., Vorsanova S.G., Kurinnaaya O.S., Kolotii A.D., Demidova I.A., Kravets V.S., Yurov Y.B. The use of molecular cytogenetic and cytogenetic techniques for the diagnosis of Prader-Willi and Angelman syndrome. Zh. Nevrol. Psikhiatr. Im. S S Korsakova. 2014;114:49–53. [PubMed] [Google Scholar]
- Jarzab B., Wiench M., Fujarewicz K., Simek K., Jarzab M., Oczko-Wojciechowska M., Wloch J., Czarniecka A., Chmielik E., Lange D., et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Lim A.L., Ferguson-Smith A.C. Genomic imprinting effects in a compromised in utero environment: implications for a healthy pregnancy. Semin. Cell Dev. Biol. 2010;21:201–208. doi: 10.1016/j.semcdb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Mann M.R.W., Lee S.S., Doherty A.S., Verona R.I., Nolen L.D., Schultz R.M., Bartolomei M.S. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- McDonald L.E., Paterson C.A., Kay G.F. Bisulfite genomic sequencing-derived methylation profile of the Xist gene throughout early mouse development. Genomics. 1998;54:379–386. doi: 10.1006/geno.1998.5570. [DOI] [PubMed] [Google Scholar]
- Messer M., Ottesen M. Isolation and properties of glutamine cyclotransferase of dried Papaya Latex. Biochim. Biophys. Acta. 1964;92:409–411. doi: 10.1016/0926-6569(64)90204-4. [DOI] [PubMed] [Google Scholar]
- Moorman A.F., Houweling A.C., de Boer P.A., Christoffels V.M. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- Morris M.R., Ricketts C.J., Gentle D., McRonald F., Carli N., Khalili H., Brown M., Kishida T., Yao M., Banks R.E., et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30:1390–1401. doi: 10.1038/onc.2010.525. [DOI] [PubMed] [Google Scholar]
- Nafee T.M., Farrell W.E., Carroll W.D., Fryer A.A., Ismail K.M. Epigenetic control of fetal gene expression. BJOG. 2008;115:158–168. doi: 10.1111/j.1471-0528.2007.01528.x. [DOI] [PubMed] [Google Scholar]
- Novakovic B., Saffery R. The ever growing complexity of placental epigenetics - Role in adverse pregnancy outcomes and fetal programming. Placenta. 2012;33:959–970. doi: 10.1016/j.placenta.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Okae H., Hiura H., Nishida Y., Funayama R., Tanaka S., Chiba H., Yaegashi N., Nakayama K., Sasaki H., Arima T. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 2012;21:548–558. doi: 10.1093/hmg/ddr488. [DOI] [PubMed] [Google Scholar]
- Plasschaert R.N., Bartolomei M.S. Genomic imprinting in development, growth, behavior and stem cells. Development. 2014;141:1805–1813. doi: 10.1242/dev.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prissette M., El-Maarri O., Arnaud D., Walter J., Avner P. Methylation profiles of DXPas34 during the onset of X-inactivation. Hum. Mol. Genet. 2001;10:31–38. doi: 10.1093/hmg/10.1.31. [DOI] [PubMed] [Google Scholar]
- Reik W., Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22:2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Santos F., Hendrich B., Reik W., Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Schilling S., Wasternack C., Demuth H.U. Glutaminyl cyclases from animals and plants: a case of functionally convergent protein evolution. Biol. Chem. 2008;389:983–991. doi: 10.1515/BC.2008.111. [DOI] [PubMed] [Google Scholar]
- Schulz R., Proudhon C., Bestor T.H., Woodfine K., Lin C.S., Lin S.P., Prissette M., Oakey R.J., Bourc’his D. The parental non-equivalence of imprinting control regions during mammalian development and evolution. PLoS Genet. 2010;6:e1001214. doi: 10.1371/journal.pgen.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiura H., Nakamura K., Hikichi T., Hino T., Oda K., Suzuki-Migishima R., Kohda T., Kaneko-ishino T., Ishino F. Paternal deletion of Meg1/Grb10 DMR causes maternalization of the Meg1/Grb10 cluster in mouse proximal Chromosome 11 leading to severe pre- and postnatal growth retardation. Hum. Mol. Genet. 2009;18:1424–1438. doi: 10.1093/hmg/ddp049. [DOI] [PubMed] [Google Scholar]
- Singh P., Szabo P.E. Chromatin immunoprecipitation to characterize the epigenetic profiles of imprinted domains. Methods Mol. Biol. 2012;925:159–172. doi: 10.1007/978-1-62703-011-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasler J.M. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod. Fert. Dev. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- Wang X., Soloway P.D., Clark A.G. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics. 2011;189:109–122. doi: 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Miller D.C., Harman R., Antczak D.F., Clark A.G. Paternally expressed genes predominate in the placenta. Proc. Natl. Acad. Sci. USA. 2013;110:10705–10710. doi: 10.1073/pnas.1308998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D.G., Nieto M.A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Yamasaki-Ishizaki Y., Kayashima T., Mapendano C.K., Soejima H., Ohta T., Masuzaki H., Kinoshita A., Urano T., Yoshiura K., Matsumoto N., et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 2007;27:732–742. doi: 10.1128/MCB.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


