Abstract
Both of the uplift of Qinghai-Tibet Plateau (QTP) and the development of East Asian monsoon system (EAMS) could have comprehensively impacted the formation and evolution of Arid Central Asia (ACA). To understand how desert plants endemic to ACA responded to these two factors, we profiled the historical population dynamics and distribution range shift of a constructive desert shrub Reaumuria soongarica (Tamaricaceae) based on species wide investigation of sequence variation of chloroplast DNA and nuclear ribosomal ITS. Phylogenetic analysis uncovered a deep divergence occurring at ca. 2.96 Mya between the western and eastern lineages of R. soongarica, and ecological niche modeling analysis strongly supported that the monsoonal climate could have fragmented its habitats in both glacial and interglacial periods and impelled its intraspecific divergence. Additionally, the population from the east monsoonal zone expanded rapidly, suggesting that the local monsoonal climate significantly impacted its population dynamics. The isolation by distance tests supported strong maternal gene flow along the direction of the East Asian winter monsoon, whose intensification induced the genetic admixture along the latitudinal populations of R. soongarica. Our results presented a new case that the development of EAMS had prominently impacted the intraspecific divergence and population dynamics of this desert plant.
Global climate fluctuations and geological events were major forces in shaping the current vegetation distribution and geographical structure of genetic diversity of species in the Northern Hemisphere1. Exploring the patterns of species responding to climatic and geological events could help us understand how species’ distributions and their population dynamics are affected under climate scenarios2. Plenty of investigations focusing on the Qinghai-Tibetan Plateau (QTP) and adjacent regions have hypothesized that the QTP uplift might have triggered the evolutionary radiations of native species, meanwhile, global climate cyclic fluctuations during the Pleistocene could have also immensely influenced their distribution ranges and genetic patterns3,4. Up to date, however, the effects of geological and climatic scenarios on species diversification had not been well dissected in the organisms native to QTP5.
Arid Central Asia (also called arid Asian interior, ACA in this paper), adjoining to the QTP and accounting for one third of the arid regions in the world, had greatly attracted geological and ecological concerns with its formation and spatial and temporal evolution, especially the processes of aridification6. It was proposed that the aridification of ACA has been profoundly aggravated by two factors: the QTP uplifts and East Asian monsoon system (EAMS). The phased uplifts of QTP originated from the collision of the Indian subcontinent with Eurasia in the Eocene, extensive uplifts during ca. 15–13 Mya (million years ago), 8–7 Mya, and the last rapid uplift during 3.6–1.2 Mya7,8,9,10,11,12. The further aridification of ACA could be associated with the development of modern EAMS during the Pliocene and Pleistocene7,8,13,14, especially with the increasing variability and strengthening of East Asian winter monsoon (EAWM) since 2.6 Mya7. The aridification of ACA and the underlying monsoonal climate could have shaped the ranges and the present genetic constitution of the biotas in the region. For example, with the monsoonal climate oscillation during the Pleistocene, the eastern deserts (Tengger and Badain Jaran deserts) ranges have shifted more significantly than that of the western deserts (Taklimakan desert)15, and these changes could have drastically shifted the plant communities14. To date, a few studies have revealed that desert plants of ACA experienced episodes of population differentiation and expansion during their evolutionary histories16,17,18. Although the EAMS had been impacting the vegetation of ACA during late Pliocene and Pleistocene7, the role of EAMS in shaping species diversity and population dynamics has not been well addressed yet. Deeper investigation on evolutionary history on these desert plants could provide a platform to discuss how EAMS impacted their habitat fragmentations and intraspecific divergences, and could also acquire the information of aridification of ACA and the evolution of EAWM in the phylogeographic view.
Reaumuria soongarica (Tamaricaceae), a Tertiary relic shrub and typical xerophyte19, is widely distributed in all the desert regions across ACA20. As a constructive and zonal species, the plant plays a crucial role in sustaining fragile desert ecosystems, including Tengger Desert, Badain Jaran Desert, Gurbantunggut Desert, Gashun Gobi, Kumtag Desert, Qaidam Basin and Taklimakan Desert (www.eflora.org). The geographical distribution range of R. soongarica is covering the whole ACA and also in situ fragmented by the QTP, which could provide an ideal model for understanding the demographic history of desert plants in response to the uplift of QTP and monsoonal climate oscillation. Previous phylogeographical investigation on one chloroplast(cp) DNA fragment of R. soongarica has revealed a significant intraspecific divergence that occurred from 1.1 to 3.5 Mya between two regional groups adjoining to QTP21. However, partly due to the insufficient informative sites and incomplete sampling, it could not adequately dissect the effects of paleogeological and/or paleoclimatic changes on the demographic history of R. soongarica. Here, based on the species wide variations on five cpDNA fragments (maternally inherited)22,23,24 and one nuclear ribosomal internal transcribed spacer (nrITS, biparentally inherited) region from 34 populations across the ACA, we profiled the population structure and dynamics with the coalescent-based program MIGRATE-N25 and reconstructed its habitat fragmentation and range shift during Last Glacial Maximum (LGM) and Last Interglacial (LIG) periods with ecological niche modeling (ENM)26. We were aiming to elucidate the roles of the EAMS in shaping the evolutionary history of this desert plant by addressing the following three questions: 1) How the geological and monsoonal climatic change affected the intraspecific divergence of R. soongarica? 2) Whether the population dynamics among the regional groups were affected by local monsoonal or global climate change? 3) How the EAMS affected the gene flow among the regional groups of R. soongarica? This work would shed light on our understanding on the demographic history of constructive species in the fragile desert ecosystems in response to the monsoonal climate change, and also supported empirical phylogeographic data to understand the spatial and temporal evolution of ACA and EAMS.
Results
Genealogies and geographical distributions of haplotypes.
Five cpDNA fragments and one nrITS fragments for a total of 272 in-group and 3 out-group individuals were sequenced. The sequences obtained from five cpDNA fragments were concatenated into a total length of 3,836 bps, with 41 chlorotypes defined by 44 polymorphic sites, including two indels (Supplementary Table S1). A total of 65 ribotypes (59 ribotypes from in-groups and 6 ribotypes from out-groups) were detected from the nrITS fragment (662 bps). Accordingly, the genealogical topology of chlorotypes and ribotypes were both clustered into two major lineages from the out-groups, and this clustering was strongly supported by the maximum likelihood (ML) trees (Fig. 1; Supplementary Fig. S1a). All chlorotypes and ribotypes from the two lineages were then projected to the population location on the geographical map (Fig. 2a,b). Most of the western populations harbored the chlorotypes and ribotypes from the western lineages, whereas most of the eastern populations were dominated by the chlorotypes and ribotypes from the eastern lineages.
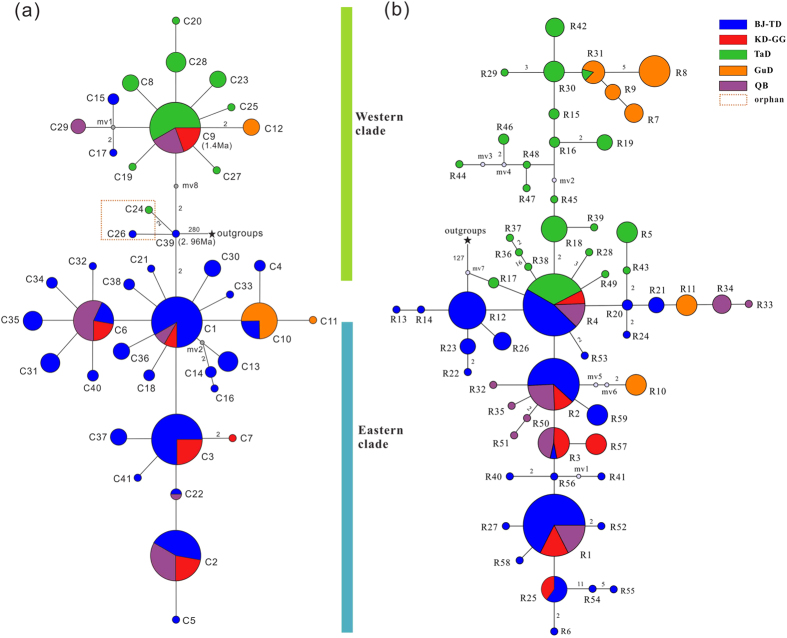
Figure 1.
Median-joining networks of (a) 41 chlorotypes (C1-C41) and (b) 59 nuclear ribotypes (R1-R59) in R. soongarica from the five groups in ACA, e.g. Taklamakan Desert (TaD), Gurbantunggut Desert (GuD), Qaidam Basin (QB), Kumtag Desert and Gashun Gobi (KD-GG), and Badain Jaran and Tengger deserts (BJ-TD). The size of the circles corresponds to the frequency of each haplotype. Small gray circles indicate unsampled or extinct haplotypes. Each number near the line represents the mutational steps interconnecting two haplotypes, and only the steps over two mutations are listed. There were three mutation steps between C39 and C9, as each mutation represents 5.35 × 105 year (1 substitution representing the divergence time of 1/[4.87 × 10−10 s/s/y × 3,836 bps) years]. Chlorotypes C39 and C9 are presented at ca. 2.96 and 1.4 Mya, respectively.
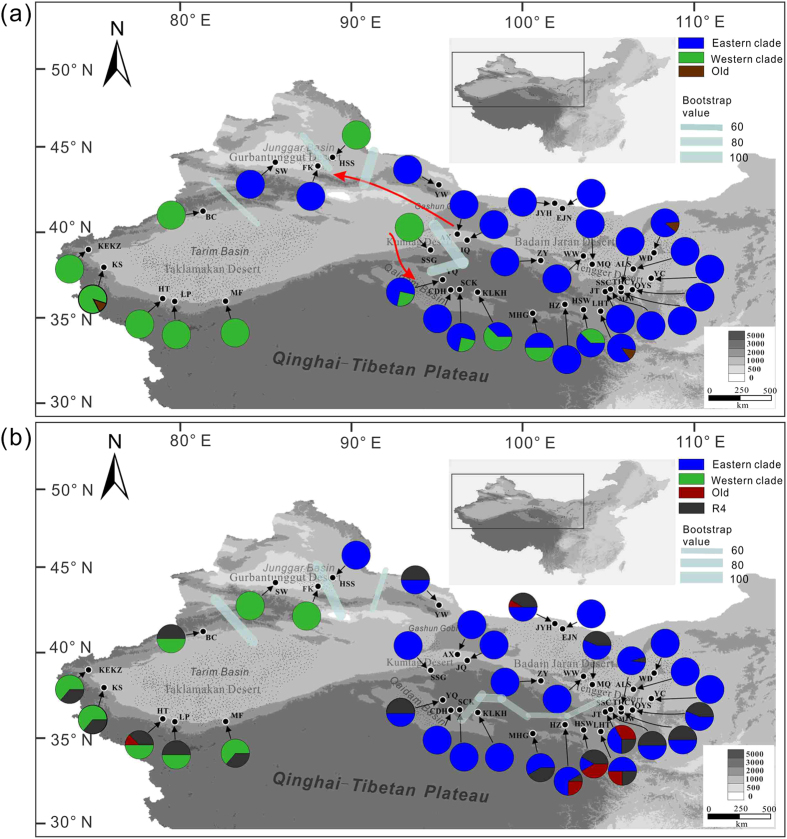
Figure 2. Geographical locations and haplotype distributions of the 34 R. soongarica populations examined in this study.
(a) cpDNA chlorotype distribution; (b) ITS ribotype distribution. Population locations are presented as small black spots with white margins; slice sizes are proportional to the frequency of the haplotypes (eastern or western clade). The colors of the pie diagrams correspond to the clades of the network genealogies in Fig. 1. “Old” refers to C26, C39 and C24. The gene flow along the East Asian winter monsoon (EAWM) and East Asian summer monsoon (EASM) directions are displayed with red arrow lines in (a). In addition, genetic barriers are superimposed on the map between populations using a gray bold line, with the width of the barrier lines representing the bootstrap values. These figures were drawn by H.X.Y. using CorelDraw X6 (Corel Corporation, Ottawa, Canada) after being originated from the software packages Diva-GIS version 7.5.0 (http://www.diva-gis.org/) loading with the Chinese map.
Chlorotypes C1 and C9, which coalesced to chlorotype C39 connecting with the out-group, could be the most recent common ancestral chlorotypes for the eastern and western clades, respectively (Fig. 1a; Supplementary Fig. S1a). Chlorotype C1 was found in most of the populations from the eastern group BJ-TD (grouping refers to Materials and Methods), whereas C9 was dominant in the western group TaD. Interestingly, chlorotypes C1 and C9 were both found in the populations from the QB and KD-GG groups. Four of the five populations from the group QB and one marginal population HSW from the group BJ-TD near the QB harbored both the western and eastern chlorotypes (Fig. 2a; Supplementary Table S2), which suggested that the maternal genetic admixture occurred along the populations of the QB “corridor”. In the KD-GG region, however, one population SSG was fixed with the western chlorotype C9, while the other three populations were fixed with the eastern chlorotypes (Fig. 2a; Supplementary Table S2), indicating that strong genetic structures were characteristics of chloroplast DNA in the group KD-GG.
Similar to the chlorotypes, most of the ribotypes could be divided into two lineages, except for the ancestral ribotype R4 being shared among all populations from the eastern and western regions (Fig. 1b), which showed incomplete divergence and/or imcomplete lineage sorting on nrITS within this species. However, we failed to detect the genetic admixture occurred on the nuclear genome in the group QB, as none of western ribotypes but ancestral ribotype R4 and the eastern ribotypes were found in all the populations from the group QB (Fig. 2b). Furthermore, we could not detect genetic structure on the nuclear genome within the group KD-GG, as the population SSG and the other three populations from the KD-GG region were all fixed with the eastern ribotypes and R4.
As noted above, all chlorotypes and ribotypes found in the group GuD were derived from both lineages (Fig. 1a,b). Interestingly, in this region, two populations (SW and FK) were dominated by the western chlorotypes and eastern ribotypes, whereas the other population, HSS, was dominated by the eastern chlorotypes and western ribotypes (Fig. 2a,b). These results indicated that R. soongarica recently colonized into the GuD region from two directions.
Divergence and diversification of chlorotype lineages.
Due to insufficiency of pollen or fossil records of Reaumuria, we carefully chose an average mutation rate of 4.87 × 10–10 substitutions per site per year (s/s/y) for chloroplast genome, which is suitable for desert shrub taxa27, to estimate the divergence time between the genealogical lineages. According to the BEAST analyses of cpDNA based on the above mutation rate, the intraspecific divergence time between the eastern and western lineages can be dated to ca. 2.96 Mya (95% HPD, 1.45–5.50 Mya, point “c” in Fig. 3), corresponding to the last peak of higher temperature during the Pliocene28 and/or the prominent uplift of the QTP10. The eastern lineage coalesced at ca. 1.72 Mya (95% HPD, 0.83–3.13 Mya point “d” in Fig. 3), considerably earlier than the western lineage at ca. 1.26 Mya (95% HPD, 0.52–2.46 Mya point “e” in Fig. 3), suggesting the asynchronous lineage diversifications of R. soongarica between the two regions (Badain Jaran and Tengger deserts and Taklamakan desert) before the Middle Pleistocene. Interestingly, in the group GuD, the presence of the west-like chlorotype C12 and the east-like C10 and C11 could be dated to later than ca. 0.37 Mya (point “f” in Fig. 3), indicating that the bi-directional colonization of R. soongarica could have occurred in the Gurbantunggut desert during the Late Pleistocene.
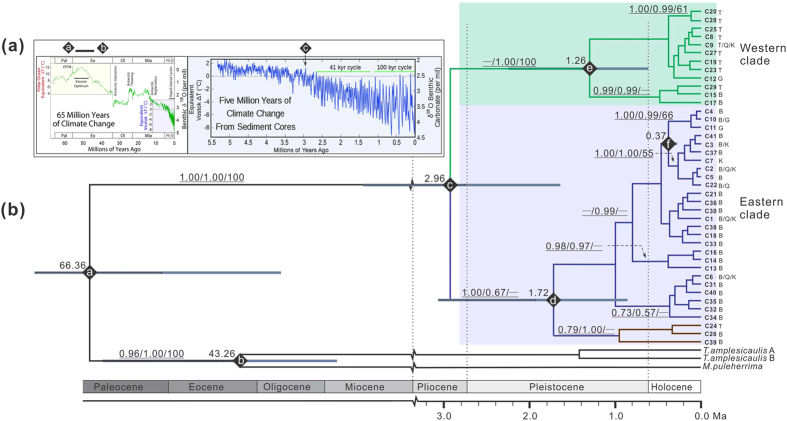
Figure 3. BEAST-generated maximum clade credibility tree of 41 chlorotypes (C1-C41) identified in R. soongarica and 3 chlorotypes identified in out-groups using the concatenated dataset with five cpDNA fragments.
(a) PETM events (Zachos et al., 2001); (b) BEAST tree with bootstrap support values (%) from 1,000 replicates (maximum likelihood analysis using PhyML software/posterior probabilities (Bayesian inference)/posterior probabilities (maximum likelihood using PAUP analysis) added above each branch with higher support. The node age (Mya) estimates are marked with letters of the alphabet; (a–f) represent the coalescence time of each major clade, and the length of the blue bars represents 95% highest posterior density. The capital letters at the end of each branch representing the original groups, e.g. Taklamakan Desert (T), Gurbantunggut Desert (G), Qaidam Basin (Q), Kumtag Desert and Gashun Gobi (K), and Badain Jaran and Tengger Desert (B).
Genetic isolation and gene flow
The overall genetic diversity (HT = 0.934) of R. soongarica on cpDNA was considerably higher than the average diversity within populations (HS = 0.527), which was concordant with the significant phylogeographical structure found on cpDNA (NST = 0.612 with P = 0.04, GST = 0.436 with P = 0.04, Table 1) and on nrITS (NST = 0.386 with P = 0.03, GST = 0.279 with P = 0.02; Supplementary Table S3). A hierarchical AMOVA indicated that the molecular variation among five groups (48.04%) was nearly two-fold higher than that within populations (24.74%, Table 1). In addition, strong significant pairwise genetic differentiation (ΦST) was found between the group TaD and the other four groups (from 0.675 to 0.819, Table 2), which supported the geographical isolation of group TaD. This finding suggested that the gene flow from other groups couldn’t have affected the population diversity within group TaD. In contrast, negligible pairwise ΦST values were found between the three latitudinal groups (ΦST (QB, KD-GG) = 0.005, ΦST (QB, GuD) = 0.084, ΦST (GuD, KD-GG) = 0.108; Table 2). This finding supported the hypothesis that frequent latitudinal gene flow on chloroplast genome occurred among these three groups during the evolutionary history of R. soongarica.
Table 1. Estimates of genetic diversity and molecular variance for chlorotype sequences.
| Regions | HS | HT | GST | NST | Source of variation | d.f. | SS | VC | PV (%) | Fixation index | θw(×10−3) | Ne(×105) | Neutrality tests |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fu’s Fs | Tajiam’s D | |||||||||||||
| TaD | 0.375 (0.086) | 0.747 (0.090) | 0.498 (0.171) | 0.501 (0.196) | Among populations | 5 | 14.46 | 0.28 | 29.20 | FST =0.292* | 0.7 | 6.67 | −1.60 (0.220) | −1.72 (0.021) |
| Within populations | 42 | 28.25 | 0.67 | 70.80 | ||||||||||
| Total | 47 | 42.71 | 0.95 | |||||||||||
| GuD | 0.083 (0.083) | 0.708 (0.175) | 0.882 (0.162) | 0.985 (0.022)* | Among populations | 2 | 90.75 | 5.67 | 99.27 | FST =0.993* | 0.6 | 5.75 | 12.41 (0.998) | 1.78 (0.962) |
| Within populations | 21 | 0.88 | 0.04 | 0.73 | ||||||||||
| Total | 23 | 91.63 | 5.71 | |||||||||||
| BJ-TD | 0.630 (0.062) | 0.916 (0.023)* | 0.313 (0.066) | 0.402 (0.058) | Among populations | 15 | 84.42 | 0.56 | 33.63 | FST =0.336* | 1.4 | 14.48 | −12.13 (<.0001) | −1.55 (0.035) |
| Within populations | 112 | 124.75 | 1.11 | 66.37 | ||||||||||
| Total | 127 | 209.17 | 1.68 | |||||||||||
| QB | 0.629 (0.045)* | 0.802 (0.056) | 0.216 (0.040)* | 0.219 (0.009)* | Among populations | 4 | 40.35 | 0.82 | 19.09 | FST =0.191* | 0.6 | 6.37 | 9.42 (0.993) | 1.89 (0.973) |
| Within populations | 35 | 122.25 | 3.49 | 80.91 | ||||||||||
| Total | 39 | 162.60 | 4.32 | |||||||||||
| KD-GG | 0.179 (0.112) | 0.997 (0.047)* | 0.821 (0.113) | 0.923 (0.078) | Among populations | 3 | 105.81 | 4.39 | 96.09 | FST =0.961* | 0. 6 | 6.67 | 6.72 (0.980) | 1.00 (0.877) |
| Within populations | 28 | 5.00 | 0.18 | 3.91 | ||||||||||
| Total | 31 | 110.81 | 4.57 | |||||||||||
| TaD, GuD, BJ-TD, KD-GG and QB versus | Among groups | 4 | 485.59 | 2.29 | 48.04 | FCT =0.524* | ||||||||
| Among populations within groups | 29 | 335.79 | 1.30 | 27.22 | FSC =0.753* | |||||||||
| Within populations | 238 | 281.13 | 1.18 | 24.74 | FST =0.480* | |||||||||
| Total | 0.527 (0.048)* | 0.934 (0.011)* | 0.436 (0.041)* | 0.612 (0.044)* | 271 | 1102.50 | 4.77 | 1.8 | 18.28 | −4.68 (0.192) | −1.27 (0.070) | |||
HS, average gene diversity within populations; HT, total gene diversity; GST, interpopulation differentiation; NST, number of substitution types; d.f., degrees of freedom; SS, sum of squares; VC, variance component; FST, correlation within populations relative to total; FSC, correlation of haplotypes within groups relative to the total; FCT, correlation within populations relative to groups; θw, theta per site; Ne, estimates of the effective population size; P-values are enclosed in the parenthesis; *P < 0.05.
Table 2. Pairwise genetic differentiation (ΦST) among five groups estimated from internal transcribed spacer (ITS) sequences (upper part) and cpDNA sequences (lower part) of R. soongarica.
| BJ-TD | KD-GG | GuD | TaD | QB | |
|---|---|---|---|---|---|
| BJ-TD | — | 0.064*** | 0.550*** | 0.337*** | 0.029* |
| KD-GG | 0.139*** | — | 0.583*** | 0.417*** | 0.089*** |
| GuD | 0.322*** | 0.108 NS | — | 0.333*** | 0.511*** |
| TaD | 0.819*** | 0.675*** | 0.638*** | — | 0.311*** |
| QB | 0.212*** | 0.005** | 0.084NS | 0.581*** | — |
*P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
Significant population structure was also found within the groups GuD and KD-GG (Hs = 0.083 and 0.179, FST = 0.993 and 0.961, respectively; Table 1). In contrast, the highest genetic diversity within populations (Hs = 0.629) and the lowest genetic differentiation (FST = 0.191) were found within the group QB, supporting the frequent maternal gene flow among the populations within this group (Fig. 2a; Table 1).
To determine whether genetic differentiation was significantly related to physical distance (isolation by distance), Mantel tests were conducted along the latitudinal and longitudinal directions both on maternally inherited cpDNA and bi-parentally inherited nrITS. Significant genetic isolation by physical distance was found along both latitudinal and longitudinal directions on nrITS (r = 0.430, P < 0.001; r = 0.794, p < 0.001, respectively), and along the longitudinal direction on cpDNA (r = 0.609, p < 0.001; Table 3). However, no significant genetic isolation was detected along the latitudinal direction on cpDNA (r = 0.262, P = 0.110; Table 3). This finding implied that the EAWM played a crucial role in enhancing the maternal gene flow, which minimized the genetic differentiation between populations along the latitudinal direction.
Table 3. Mantel tests along the monsoon directions for genetic isolation by eographicl distance on cpDNA and ITS.
| Directions | Population groups | cpDNA | ITS |
|---|---|---|---|
| Latitudinal (EAWM) | GuD, KD-GG and QB | r = 0.262 | r = 0.794 |
| p = 0.110 | p < 0.001 | ||
| Longitudinal (Westerly) | TaD, QB, KD-GG and BJ-TD | r = 0.609 | r = 0.430 |
| p < 0.001 | p < 0.001 | ||
| Southeastern (EASM) | GuD, KD-GG and BJ-TD | r = 0.438 | r = 0.546 |
| p = 0.003 | p < 0.001 |
EAWM, East Asian winter monsoon; EASM, East Asian summer monsoon.
Regional population dynamics
A marginally significant negative value of Tajima’s D (–1.27, P = 0.070, Table 1) and MDA results (Supplementary Fig. S2) implied that the overall population of R. soongarica underwent a complex expansion of its population structure. Significant negative values of Tajima’s D (−1.55, P = 0.035) and Fu’s Fs (−12.13, P < 0.001) strongly confirmed that the eastern group BJ-TD underwent a population expansion, whereas the western group TaD remained comparatively stable, as only Tajima’s D was significant negative (−1.72, P = 0.021; Table 1). The skyline plots with MIGRATE-N analysis showed that the population size of the overall R. soongarica populations had been increasing significantly (Fig. 4; Supplementary Fig. S3a). In the separate groups, the population size of group BJ-TD increasing more rapidly and reached a maximum at 80 Ka, whereas group TaD exhibited a slower population growth relatively recently (Fig. 4; Supplementary Fig. S4b,d). However, population decreasing was found in the three other groups with genetic admixture/structure: GuD, QB and KD-GG (Fig. 4; Supplementary Fig. S4c,e,f).
Figure 4. Skyline plots with MIGRATE-N estimated for each group of R. soongarica: x-axis, time, years before present (Mya); y-axis, theta (θ = 2Neu).
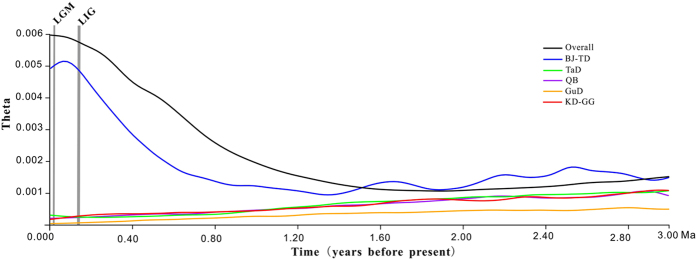
The curves shown in various colors are plotted for the total populations and separated populations distributed in BJ-TD, GuD, TaD, QB and KD-GG regions.
Distribution range shifts in the past
The ENM simulations based on Maxent supported the present distribution range of R. soongarica, which covers nearly all the desert regions across ACA (Fig. 5a). Our modeling data indicated that Bio19 and Bio11, namely, the precipitation and average temperature during the coldest season, together represented more than 60% of the niche contribution and over 80% of the permutation importance (Supplementary Table S4). These results indicated that the EAWM markedly influenced the distribution range of R. soongarica. Compared with the present distribution, the area of suitable habitat determining the range of R. soongarica was extremely contracted and fragmented by the QTP during LGM period, and this tendency further strengthened during LIG period (Fig. 5b,c). However, the local distribution range fluctuated asynchronously: the western region experienced range expansion, and the eastern region underwent more frequent range shifts, following LIG period (Fig. 5b,c). These results indicated that the paleoclimate change could have further driven the habitat fragmentation of R. soongarica and that the EAMS could have markedly affected vegetation distribution in different ways in the two local regions.
Figure 5. Predicted distributions of R. soongarica based on ecological niche modeling using Maxent.
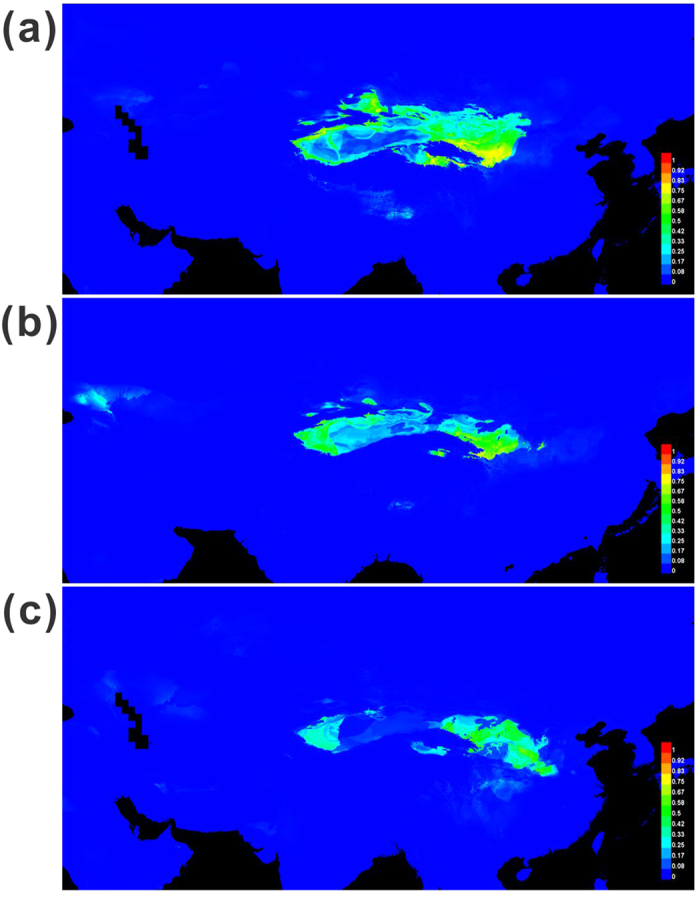
Predicted distributions are shown for (a) the present time, (b) the LGM period, and (c) the LIG period.
Discussion
Based on thorough investigation on genetic structure and distribution range shifts under monsoonal climate scenarios, we intend to understand the mechanism of population divergence and population dynamics of R. soongarica, a Tertiary relic constructive zonal shrub species widely distributed across all the desert ecosystems in ACA. As the formation and evolution of ACA had been affected by the development of EAMS since Pliocene7, the present work supported an idea model to discuss the role of monsoonal climatic changes in shaping the habitat fragmentation which triggered the intraspecific divergence/speciation of the dominant species of ACA.
The cpDNA genealogy network and phyML tree (Fig. 1a; Supplementary Fig. S1a) suggested that R. soongarica diverged into two contrasting lineages. This finding is consistent with our previous study21. The chlorotypes from the western lineage were mainly distributed in the group TaD, whereas those from the eastern lineage were dominant in the group BJ-TD (Fig. 2a). Marked genetic differentiation between these two groups was found in both nrITS and cpDNA (Table 2). Due to the effective population size of the biparentally inherited nuclear genome is twofold larger than that of the uniparentally inherited chloroplast genome, we found incomplete divergence on nuclear ribosome DNA. The most recent common ancestral ribotype, R4, was shared among all groups, excluding GuD (Fig. 1b). However, we detected significant genetic barriers on nrITS between the group TaD and the other groups (Fig. 2b). This marked genetic differentiation between the eastern and western populations in ACA is not specific to R. soongarica. It has also been found in some sympatric plant species such as Nitraria sphaerocarpa18 and N. tangutorum (unpublished data). These findings suggest that geological events common to all populations could have impacted the genetic differentiation of the plants endemic to ACA.
To further clarify the cause of the intraspecific divergence between the two groups, molecular dating was conducted with a relaxed clock method via Bayesian simulation based on substitution rate of 4.87 × 10−10 s/s/y for the chloroplast genome27. The divergence between the two out-group genera could be dated to ca. 43 Mya (95% HPD: 16.63–74.68 Mya, point “b” in Fig. 3b). This result did not conflict with the earliest fossil record of the genus Tamarix29. The crown age of genus Reaumuria and the out-groups could be dated to ca. 66 Mya (95% HPD: 111–27 Mya, point “a” in Fig. 3b). It is consistent with a previous report30, and corresponding to the period of Paleocene-Eocene Thermal Maximum (PETM, Fig. 3a), when a sharp increase in concentration of atmospheric CO2 caused the global surface temperature to increase by approximately 5–6 °C28. The intraspecific divergence of R. soongarica between the eastern and western lineages could be dated to ca. 2.96 Mya (95% HPD: 1.45–5.5 Mya, point “c” in Fig. 3b). This time point is close to the strong tectonic uplift of QTP, especially, a rapid uplift of Qaidam Basin (the northeastern QTP) during the late Pliocene7,14. Such a rapid uplift is consistent with our molecular dating of the intraspecific divergence of R. soongarica to a period prior to the beginning of the dramatic glacial and interglacial oscillations of the global climate (Fig. 6a).
Figure 6. Historical schemes of spatial-temporal evolution historical schemes of paleogeologies and paleoclimatics and the corresponding genetic response of R. soongarica in the ACA region.
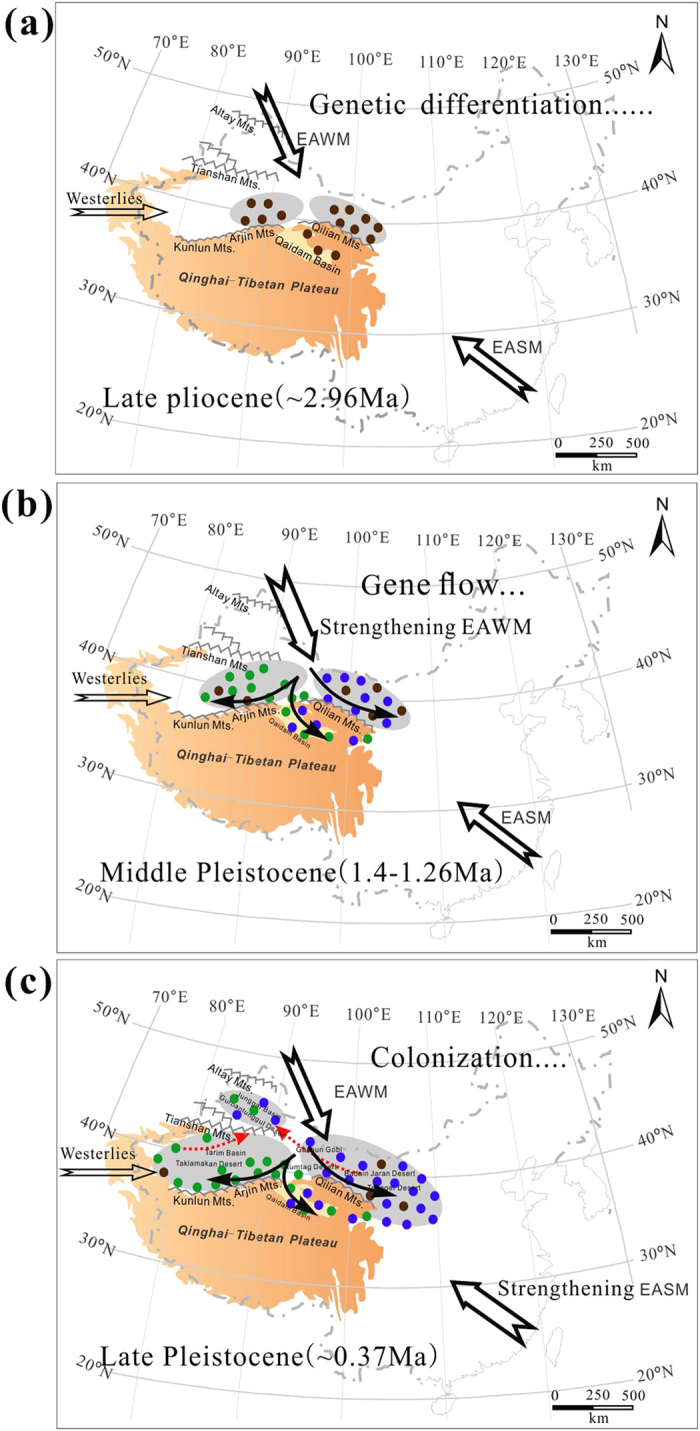
(a) ca. 2.96 Mya: genetic differentiation occurred within R. soongarica triggered by the uplift of the northeastern QTP and EAWM formation; (b) 1.4 ~ 1.26 Mya: maternal gene flow from the KD-GG region (SSG) to the QB and TaD regions under the driving force of the strengthening EAWM; the red line represents the orientation of gene flow; and (c) ca. 0.37 Mya: bi-directional colonization and aridification of the GuD region occurred; the dashed red line represents the directions of colonization. Color coding was consistent with Fig. 2 as follows: the brown spot represents the ancestor of R. soongarica, the blue pie represents the haplotyps from the eastern clade, and the green pie represents ones from the western clade. The shadow represents the expanding arid regions responding to the habitat changes of R. soongarica during different geological periods. These figures were drawn by H.X.Y. using CorelDraw X6 (Corel Corporation, Ottawa, Canada).
However, the abrupt uplift of the QTP had also caused the intensification of EAMS at ca. 3.6 to 2.6 Mya. Actually, based on ENM simulation of the potential distribution range of R. soongarica during the LGM (EAWM dominated) and LIG (EASM dominated), both the glacial and interglacial climates could have greatly contracted the habitat of this species and finally fragmented its distribution range (Fig. 5b,c). On the other hand, the present distribution of R. soongarica was not fragmented at all (Fig. 5a), indicating that the two lineages of R. soongarica should have contacted easily and the two reciprocal habitats could have merged together under the moderate climatic conditions. Therefore, we could not arbitrarily conclude that the geological event was the only condition that triggered the intraspecific divergence of R. soongarica. Based on the considerations above, we proposed that once the isolation occurred as a result of geological cause, the cycles of the monsoonal climatic oscillations was most likely to play a key role in the habitat fragmentation and intraspecific divergence of R. soongarica. After R. soongarica diverged into two regional lineages, further diversification of two lineages began ca. 1.72 Mya (Eastern clade, point “d” in Fig. 3b) and 1.26 Mya (Western clade, point “e” in Fig. 3b), which are fairly concordant with the two main turning points of the EAMS31,32. Stratigraphic data from loess deposits and NW-Pacific ocean deposits suggested that the EAWM steadily strengthened during Pleistocene, whereas the EASM underwent stepwise weakening until ca. 1.8 Mya, when the EASM was increasingly restrengthened. Considering the more fragmented habitat of R. soongarica in the interglacial period such as LIG (Fig. 5c), we speculated that the strengthening EASM of the warming stage ca.1.8 Mya could have fragmented the habitat of R. soongarica and accelerated the diversification of the eastern lineage. However, the western lineage could not start diversification until ca. 1.3 Mya, when the restrengthened EAWM effectively overwhelmed the EASM and induced the further aridification of ACA32. Therefore, we suspected that the EAWM played a crucial role in the diversification of the western lineage of R. soongarica.
Since the asynchronous diversification happened between the two lineages of R. soongarica, could the two regional groups have undergone the similar population dynamics? In fact, from the skyline plots obtained with MIGRATE-N, we found a significant increase of the population size in the eastern groups of R. soongarica (Fig. 4), concordant to significant negative values of the Tajima’s D and Fu’s Fs and star-like shape of the genealogy network (Fig. 1; Table 1). Comparatively, the population size of the western group TaD was much more stable (Fig. 4; Supplementary Fig. S4d). Considering the distribution range of R. soongarica expanding slightly in the western region from LIG to the present based on ENM analysis (Fig. 5b,c), the stable population dynamic of the western group TaD could be related to the geographical isolation caused by the Tianshan-Kunlun-Arjin Mountains and equable climate during the late Pleistocene15,33.
Given that the population size of the western group remained stable while its distribution range expanded, and that the eastern group underwent a remarkable population growth while its distribution range shifted significantly (Figs 4 and 5b,c), the population expansion of the eastern group could be attributed to the frequent range shifts rather than to habitat range expansion. The frequent range shifts during the Pleistocene exerted strong selective forces that facilitated the accumulation of new mutants in the local populations, and consequently induced the effective population to grow more rapidly in the eastern climate-sensitive group than in the western climate-stable group. Previous studies have suggested that during the Pleistocene climate oscillation, population growth was quite common for large-sized species, such as conifers endemic to the climatic-sensitive region, e.g., the southeast QTP4,34 and other plants16 and animals35 endemic to ACA and adjacent regions. Unfortunately, these studies failed to interpret the potential effects of local climate change on the speed and scale of population growth. Here, based on the comparison between the regional groups of the population dynamics and range shifts of R. soongarica, we presented a novel example that the populations in the monsoonal climate zone tended to grow more rapidly than those far from it.
Additionally, an interesting geographical pattern of chlorotypes of R. soongarica showed that frequent genetic admixture of the two lineages occurred in most populations within the QB “corridor” (Fig. 2a). On the other hand, we detected negligible pairwise genetic differentiation (ΦST) among the groups GuD, Ku-GG and QB, which suggested that the latitudinal gene flow had been markedly enhanced (Table 2). To further verify the effects of the EAMS on gene flow among the groups of R. soongarica, we conducted Mantel tests on maternally inherited cpDNA and bi-parentally inherited nrITS along different monsoon directions. Our data strongly supported that the EAWM decisively accelerated maternal gene flow along the latitudinal direction (Table 3). Due to the hairy and light characteristics for the mature seeds of R. soongarica, it is reasonable that the seeds of R. soongarica could be easily dispersed by the EAWM during autumn, accelerating maternal gene flow across the QB “corridor.” Gene flow can minimize intra- and inter-specific genetic differentiation and induce the accumulation of heterozygosity, facilitating the colonization of new habitats and promoting adaptation to climate change36,37. Many well-adapted plant species such as forest trees would utilize wind systems for seeds or pollen dispersal to increase gene flow between diverged populations38,39,40. By dating the historical maternal gene flow occurred among the groups of R. soongarica, we concluded that the EAWM intensified at least after the origin of the chlorotype C9 in the western group (ca. 1.4 Mya, more reasonable after 1.3 Mya, Figs 1a and 6b). In fact, many stratigraphic investigations across the Loess Plateau have recorded that the EAWM intensified after 1.3 Mya7,8,32. Up to date, empirical data are lacking except our new evidence from the desert plant R. soongarica on the scale and direction of the gene flow induced by the EAWM.
More interestingly, phylogenetic data of chlorotypes suggested that R. soongarica has recently (since ca. 0.37 Mya, point “f” in Fig. 3b) colonized into the GuD region, which was concordant to that the Gurbantunggut Desert expanded due to the intensification of the EAWM ca. 0.4 Mya41. This phylogeographic pattern indicates that the aridification of ACA during this time had expanded to the Gurbantunggut Desert region, which had supplied new habitats for the xerophyte R. soongarica (Fig. 6c). The stepwise colonization of plant species has also been found in an alpine conifer species endemic to the QTP42,43. In this study, we provided the first evidence for the stepwise aridification processes of ACA from a molecular phylogeographic perspective and confirmed that the formation time of Gurbantunggut Desert was later than the other deserts, e.g., Badain Jaran and Tengger deserts and Taklimakan Desert in ACA44 The onset of the desertification of ACA has been debated for several decades, with the time estimates ranging from ca. 5.3 Mya to 0.85 Mya45,46,47,48,49. The regional asynchronous diversification of the constructive shrub R. soongarica indicated that the desertification of the eastern Badain Jaran and Tengger deserts regions could have occurred at 1.72 Mya, the western Taklimakan Desert could have formed ca. 1.26 Mya, and the northern Gurbantunggut Desert could have formed ca. 0.4 Mya. Of course, as the corresponding 95% CIs of dating were still wide, more evidence from fossil and molecular records are needed to clarify the accurate formation time of the deserts across ACA.
In summary, our study supported a unique case focused on evaluating the roles of EAMS in impacting the evolutionary history of constructive shrub species endemic to ACA, by triggering the intraspecific divergence between the longitudinal populations and enhancing gene flow among the latitudinal populations. Furthermore, this work supplied new molecular evidence for understanding the spatial and temporal evolution of ACA from a phylogeographic view point.
Methods
Population sampling
A total of 272 individuals were sampled from 34 populations across the ACA geographical ranges (Supplementary Fig. S5; Table S5). Six of these populations were collected in Taklimakan Desert (TaD) region, three in Gurbantunggut Desert (GuD) region, five in Qaidam Basin (QB) region, four in Kumtag Desert and Gashun Gobi (KD-GG) region, and 16 in Hexi corridor and Badain Jaran - Tengger deserts (BJ-TD) regions. Additionally, three individuals of Tamarix amplexicaulis and Myricaria pulcherrima were collected as outgroups to Reaumuria24. In each population, samples of fresh leaves were collected from eight individuals at least 30 meters apart to reduce sampling bias and dried by silica gel. The information on the location of each population, including latitude, longitude and altitude, were recorded with an e-Trex Global Positioning System (Garmin, Taiwan; Supplementary Table S5). All of the sampled voucher specimens were deposited in the Key Laboratory of Stress Physiology and Ecology in Cold and Arid Regions, CAREERI, CAS.
DNA extraction, PCR amplification and sequencing
The total genomic DNA was extracted using DNeasy Plant Kits (Qiagen, Valencia, CA, USA) and then stored at −30 °C in a refrigerator. Six individuals from populations separated by long distances were sequenced to verify the polymorphic sites from 23 pairs of cpDNA fragments (Supplementary Table S6). Due to the maximum polymorphic sites, five cpDNA fragments (petB-petD, trnS2-trnG2, atpH-atpI, ndhA and petN-psbM) and one nrITS fragment were selected for full phylogeographic surveys (Supplementary Table S7), and all six fragments were amplified and sequenced in the 272 individuals of R. soongarica. DNA amplifications were performed in a Gene-Amp PCR system 9700 (Applied Biosystems, USA) with the thermal program referred to Supplementary Method S1. After purification by TIAN quick Midi Purification Kits reagent (TIANGEN, Beijing, China), sequencing with forward and reverse primers in all individuals was performed using an ABI 3130xl Genetic Analyzer (Applied Biosystems, USA) with an ABI Prism BigDye Terminator Cycle version 3.1 Sequencing Kit according to the manufacturer’s instructions. All sequences were double checked by eye in BioEdit version 7.0.5.3 and initially aligned by ClustalX version 1.8150. All newly obtained sequences for each variety have been deposited in GenBank (accessions: KM242133-KM242261).
Nucleotide diversity and genealogy
Sequences from each cpDNA fragment were concatenated into a supergene matrix by the program DnaSP Version 5.051. For nrITS sequences, if double peaks occurred in the same position in both directions and the weakest signal reached at least 25% of the strongest signal, we considered the site to be heterozygous52. Each two ribotypes of all nrITS in heterozygous individuals were separated under the default parameters with PHASE implemented in the program DnaSP Version 5.0. The molecular diversity for the concatenated sequences of five cpDNA fragments with the numbers of haplotypes (n), segregating sites (s), total haplotype diversity (hd) and nucleotide diversity (pi) for each population were also estimated with DnaSP Version 5.0. The genealogical topologies of chlorotypes and ribotypes were constructed using the program NETWORK Version 4.2.0.1 with a median-joining model (available at http://www.fluxus-engineering.com/)53.
Population genetic structure
Population gene diversity (HS, HT) and between-population divergence (GST, NST) were estimated for each region based on the cpDNA using the program PERMUT with 1,000 permutation tests (available at http://www.pierroton.inra. fr/genetics/labo/Software/PermutCpSSR)54. In addition, spatial genetic structures of chlorotypes and ribotypes were analyzed with SAMOVA Version 1.055, using a simulated annealing procedure to identify the number of population groups (K) that were geographically homogeneous and maximally differentiated from each other. According to the various geographical locations and SAMOVA results, all populations of R. soongarica were divided into five groups as follows: QB, BJ-TD, TaD, GuD and KD-GG. Notably, the populations from the KD-GG group were considered one group due to the geological “junction” location of the GuD, TaD, QB and BJ-TD groups. The populations collected from the Hexi Corridor and from the Badain Jaran-Tengger deserts were combined as the BJ-TD group according to the analysis of the initial polymorphism survey and geographical locations (Supplementary Fig. S5). Further genetic differentiation among groups (FCT), among populations within groups (FSC), and within populations (FST) was calculated with a hierarchical analyses of molecular variance (AMOVA) using Arlequin version 3.1156, with significance tests of variance components and differentiation indices with 1,000 permutations. Pairwise differentiations (ΦST) between all pairs of groups were also estimated with 1,000 permutations.
We used the software BARRIER version 2.257 to identify the areas of genetic discontinuity or barriers to gene flow across the sampled species’ ranges. One hundred bootstrapped datasets were generated by re-sampling individuals within populations with replacement using PopTools version 3.2.5 (available at http://www.poptools.org/). Under ideal circumstance, the pairwise genetic differentiation between the populations could not reject the Mantel test supporting isolation by distance58. To test the significance of the impact of wind systems on maternal gene flow limited by physical distance, Mantel tests along different East Asian monsoon directions were performed in IBDWS (available at http://ibdws.sdsu.edu/~ibdws/) based on the pairwise FST between populations using 1,000 randomizations59.
Phylogenetic analyses and estimates of divergence time
The phylogenetic relationships among chlorotypes and ribotypes were reconstructed by PhyML version 3.0 with maximum likelihood (ML)60 and by MrBayes Version 3.2.1 with mixed models using Bayesian inference (BI) and MCMC sampling61,62, respectively. The best-fit nucleotide substitution model for the codon position was evaluated by the Akaike Information Criterion63 with the program jModeltest Version 0.1.1 under the ML criterion64 and then by GTR+G and TrN+I+G models for cpDNA and nrITS, respectively. The ML analyses were conducted with the heuristic search option over 100 random-taxon-addition replicates with tree bisection–reconnection (TBR) branch swapping. Trees were sampled for every 100 generations, resulting in 100,000 trees, and the first 25% were discarded as burn-in. Bayesian analyses were performed over 50 million generations with one cold and three incrementally heated Markov Chain Monte Carlo (MCMC) analyses, starting from a random tree and sampling every 1,000 generations. The first 10% of trees were treated as burn-in and discarded, and all remaining trees were used to construct a Bayesian consensus tree. All trees were then modified using FigTree version 1.3.165.
To estimate the divergence time between the genealogical lineages, we carefully chose a mutation rate for cpDNA of 4.87 × 10−10 s/s/y, which is suitable for shrub species27. Since neither any pollen nor fossil records of Reaumuria could be found from the deposit strata published, we chose two closely related genera (Tamarix and Myricaria) within Tamaraceae to calibrate the molecular clock. Based on this mutation rate and its assumed standard deviation (10%), the divergence time and coalescence time of lineages were estimated using a Bayesian approach implemented in BEAST Version 1.7.5 with the criteria set by BEAUti (available at http://BEAST.bio.ed.ac.uk)65. All simulations were based on an uncorrected log-normal relaxed clock distribution of branch lengths, started with a prior constant population size and a randomly generated starting tree. The posterior distributions of the parameters in the MCMC analyses were approximated with 80 million steps in each analysis and sampled every 8,000 generations. Convergence of the parameters sampled was checked with the program TRACER version 1.5 to examine the highest effective sampling size values (ESSs > 200) for all parameters66, and the burn-in steps were discarded to obtain an estimation of the posterior probability distribution of divergence time at the relevant node. The maximum lineage credibility tree was then summarized by TreeAnnotator version 1.7.5 (available at http://BEAST.bio.ed.ac.uk/TreeAnnotator/).
Inference of demographic history
Possible population expansions were assessed by neutrality tests with Tajima’s D and Fu’s Fs and mismatch distribution analysis (Supplementary Method S2). MIGRATE-N version 3.6.425 was used to estimate the variation in the historical effective population size. The default settings were conducted in the MIGRATE-N program with some modified options as follows: the Bayesian inference module; one single long run using heating with temperatures of 1.0, 1.5, 3.0 and 10,000, totaling 50,100,000 visited parameter and genealogy changes in the cold (1.0) chain; sampling 10 replicates of each 50,000 in intervals of 100; after discarding the first 100,000 visits. The mutation-scaled population size theta (θ) was displayed by the uniform prior distributions with ranges from 0 to 0.1, namely the effective population size Ne times the mutation rate μ per site and generation. Moreover, Bayesian skyline plots (BSPs) for population dynamics were also conducted on cpDNA (Supplementary Method S3).
Ecological niche modeling
To predict the distribution of R. soongarica based on habitat suitability, ecological niche modeling was conducted with Maxent26 with the WorldClim database from 1955 to 2000 (http://www.worldclim.org/). The pairwise correlation for all 19 biological factors were tested, and 12 independent biological factors were used for further predictions. The distribution sites consisted of those examined in the field investigation together with the specimen records from the Chinese Virtual Herbarium and with coordination from the Global Positioning System (http://www.cvh.org.cn/); 75% of the sites were randomly selected to design a prediction scheme on consideration of the commission error, and the remaining 25% of the sites were used for evaluations of the goodness of the prediction.
The contemporary species-climate relationships were then projected to past climate layers to predict the potential range of the species during LGM (ca. 21,000 years BP) and LIG (ca. 120,000 – 140,000 years BP) periods. The project bioclimatic layers for LIG (resolution of 30 arc-second) and LGM (resolution of 2.5 arc-minute) were obtained from the WorldClim database version 1.467. We employed a “maximum training sensitivity plus specificity” threshold to determine suitable/unsuitable habitat because sensitivity–specificity combined approaches have been suggested as good thresholds68. An AUC value estimation greater than 0.9 for each estimate was used as the criterion for a good model. Additionally, 100 permutations were conducted in the modeling process for LGM and LIG periods based on the MIROC model. The relative contribution of each variable was evaluated with the covariance in terms of increased predictive performance resulting from a given environmental variables. Only the ecological variables that explained greater than 10% of the prediction variance were viewed as the principal habitat suitability factors limiting the distribution range of R. soongarica.
Additional Information
How to cite this article: Yin, H. et al. The role of East Asian monsoon system in shaping population divergence and dynamics of a constructive desert shrub Reaumuria soongarica. Sci. Rep. 5, 15823; doi: 10.1038/srep15823 (2015).
Supplementary Material
Acknowledgments
We sincerely appreciate the valuable comments and suggestions on this manuscript from Xiao-Ru Wang and Martin Lascoux. We are in debt to the heartful help on the sampling works from Gang Wang and Haikui Chen, laboratory assistant from Pengshan Zhao and Xin Zhao, data analysis from Jiabing Zou, Faqi Zhang, Pengcheng Fu and Dongrui Jia. Thanks for all the inspiring comments from Xiaoguang Qin, Tao Wang, Yunfa Miao and Xueyong Zhao. This work was supported by National Natural Science Foundation of China (NSFC, Grant No. 31370395), the “One Hundred Talents” projects of Chinese Academy of Sciences (Grant No. 29Y127E71) and International S & T Cooperation Program of China (Grant No. 2012DFR).
Footnotes
Author Contributions X.-F.M. conceived the ideas and designed the investigations; H.X.Y. and X.Y. collected the data; H.X.Y., X.Y. and X.-F.M. analyzed the data and led the writing; Y.S., C.J.Q., L.R.W. and Y.L. collected samples; W.Z. and Y.S. assisted the experiments and data analysis; Z.H.L. and X.Z.L. participated in project discussion; G.X.C. and X.R.L supported experiment facilities; E.N. provided improvements to the manuscript.
References
- Hewitt G. M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 183–195 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J. C. Molecular markers, natural history, and evolution. (Sunderland Sinauer Associates, 2004). [Google Scholar]
- Liu J. Q., Duan Y. W., Hao G., Ge X. J. & Sun H. Evolutionary history and underlying adaptation of alpine plants on the Qinghai–Tibet Plateau. J. Syst. Evol. 52, 241–249 (2014). [Google Scholar]
- Qiu Y. X., Fu C. X. & Comes H. P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244 (2011). [DOI] [PubMed] [Google Scholar]
- Favre A. et al. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 90, 236–253 (2015). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. Holocene moisture evolution in arid central Asia and its out-of-phase relationship with Asian monsoon history. Quat. Sci. Rev. 27, 351–364 (2008). [Google Scholar]
- An Z. S., John E. K., Warren L. P. & Stephen C. P. Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature 411, 62–66 (2001). [DOI] [PubMed] [Google Scholar]
- Guo Z. T. et al. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 416, 159–163 (2002). [DOI] [PubMed] [Google Scholar]
- Harrison T. M., Copeland P., Kidd W. S. & Yin A. Raising tibet. Science 255, 1663–1670 (1992). [DOI] [PubMed] [Google Scholar]
- Li J. J., Shi Y. F. & Li B. Y. Uplift of the Qinghai-Xizang (Tibet) plateau and global change (Lanzhou University Press, 1995). [Google Scholar]
- Shi Y. F., Li J. J. & Li B. Y. Uplift and environmental changes of Qinghai-Tibetan plateau in the late Cenozoic. (Guangdong Science & Technology Press, 1998). [Google Scholar]
- Spicer R. A. et al. Constant elevation of southern Tibet over the past 15 million years. Nature 421, 622–624 (2003). [DOI] [PubMed] [Google Scholar]
- Kutzbach J. E., Prell W. L. & Ruddiman W. F. Sensitivity of Eurasian Climate to Surface Uplift of the Tibetan Plateau. J. Geol. 101, 177–190 (1993). [Google Scholar]
- Miao Y., Herrmann M., Wu F., Yan X. & Yang S. What controlled Mid–Late Miocene long-term aridification in Central Asia? — Global cooling or Tibetan Plateau uplift: A review. Earth-Sci. Rev. 112, 155–172 (2012). [Google Scholar]
- Lu H. et al. Variation of East Asian monsoon precipitation during the past 21 kya and potential CO2 forcing. Geology 41, 1023–1026 (2013). [Google Scholar]
- Meng H. H. & Zhang M. L. Diversification of plant species in arid Northwest China: species-level phylogeographical history of Lagochilus Bunge ex Bentham (Lamiaceae). Mol. Phylogenet. Evol. 68, 398–409 (2013). [DOI] [PubMed] [Google Scholar]
- Meng H. H., Gao X. Y., Huang J. F. & Zhang M. L. Plant phylogeography in arid Northwest China: Retrospectives and perspectives. J. Syst. Evol. 53, 33–46 (2015). [Google Scholar]
- Su Z. H & Zhang M. L Evolutionary response to Quaternary climate aridification and oscillations in north-western China revealed by chloroplast phylogeography of the desert shrub Nitraria sphaerocarpa (Nitrariaceae). Biol. J. Linn. Soc. 109, 757–770 (2013). [Google Scholar]
- Hou X. Geographical distribution of vegetation in arid desert area of Chinese temperate zones. Botany Bulletin 2, 37–66 (1987). [Google Scholar]
- Shi Y. et al. Transcriptomic Analysis of a Tertiary Relict Plant, Extreme Xerophyte Reaumuria soongorica to Identify Genes Related to Drought Adaptation. PLoS ONE 8, e63993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. H. et al. Response of a desert shrub to past geological and climatic change: A phylogeographic study of Reaumuria soongarica (Tamaricaceae) in western China. J. Syst. Evol. 50, 351–361 (2012). [Google Scholar]
- Gaskin J. F., Ghahremani-nejad F., Zhang D. Y. & Jason P. L. A Systematic Overview of Frankeniaceae and Tamaricaceae from Nuclear rDNA and Plastid Sequence Data. Ann. Mo. Bot. Gard. 91, 401- 409 (2004). [Google Scholar]
- Gaskin J. F. & Schaal B. A. Molecular Phylogenetic Investigation of U.S. Invasive Tamarix. Syst. Bot. 28, 86–95 (2003). [Google Scholar]
- Liu Y. F, Wang Y. & Huang H. W. Species-level phylogeographical history of Myricaria plants in the mountain ranges of western China and the origin of M. laxiflora in the Three Gorges mountain region. Mol. Ecol. 18, 2700–2712 (2009). [DOI] [PubMed] [Google Scholar]
- Beerli P. Effect of unsampled populations on the estimation of population sizes and migration rates between sampled populations. Mol. Ecol. 13, 827–836 (2004). [DOI] [PubMed] [Google Scholar]
- Phillips S. J., Anderson R. P. & Schapire R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 (2006). [Google Scholar]
- Wang H. et al. Phylogeographic structure of Hippophae tibetana (Elaeagnaceae) highlights the highest microrefugia and the rapid uplift of the Qinghai-Tibetan Plateau. Mol. Ecol. 19, 2964–2979 (2010). [DOI] [PubMed] [Google Scholar]
- Zachos J., Pagani M., Sloan L., Thomas E. & Billups K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 292, 686–693 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang D., Pan B. & Yin L. The photogeographical studies of Tamarix (Tamaricaceae). Acta Botanica Yunnanica 25, 415–427 (2003). [Google Scholar]
- Zhang M. L. et al. Spatiotemporal evolution of Reaumuria (Tamaricaceae) in Central Asia: insights from molecular biogeography. Phytotaxa 167, 15 (2014). [Google Scholar]
- Kukla G. & An Z. S. Loess stratigraphy in Central China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 72, 203–225 (1989). [Google Scholar]
- Meng X., Xia P., Zheng J. & Wang X. Evolution of the East Asian monsoon and its response to uplift of the Tibetan Plateau since 1.8 Ma recorded by major elements in sediments of the South China Sea. Chin. Sci. Bull. 56, 547–551 (2011). [Google Scholar]
- Liu W. et al. Late Miocene episodic lakes in the arid Tarim Basin, western China. Proc. Natl Acad. Sci. USA 111, 16292–16296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Hou Z. H., Ai L. & Tan L. C. Precipitation at Lake Qinghai, NE Qinghai–Tibet Plateau, and its relation to Asian summer monsoons on decadal/ interdecadal scales during the past 500 years. Palaeogeogr. Palaeoclimatol. Palaeoecol. 254, 541–549 (2007). [Google Scholar]
- Shi C. M., Ji Y. J., Liu L., Wang L. & Zhang D. X. Impact of climate changes from Middle Miocene onwards on evolutionary diversification in Eurasia: insights from the mesobuthid scorpions. Mol. Ecol. 22, 1700–1716 (2013). [DOI] [PubMed] [Google Scholar]
- Clegg S. M. & Phillimore A. B. The influence of gene flow and drift on genetic and phenotypic divergence in two species of Zosterops in Vanuatu. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1077–1092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho C. & Hey J. Divergence with Gene Flow: Models and Data. Annu. Rev. Ecol. Evol. Syst. 41, 215–230 (2010). [Google Scholar]
- Calviño-Cancela M. et al. The role of seed dispersal, pollination and historical effects on genetic patterns of an insular plant that has lost its only seed disperser. J. Biogeogr. 39, 1996–2006 (2012). [Google Scholar]
- Gerber S. et al. High Rates of Gene Flow by Pollen and Seed in Oak Populations across Europe. PLoS ONE 9, e85130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X.-S. & Ennos R. A. On estimation of the ratio of pollen to seed flow among plant populations. Heredity 79, 541–552 (1997). [Google Scholar]
- Shi Z. T., Song Y. G. & An Z. S. Evolution of Gurbantunggut Desert recorded by Tianshan Loess. Journal of Desert Research 26, 675–679 (2006). [Google Scholar]
- Gao J. et al. Demography and speciation history of the homoploid hybrid pine Pinus densata on the Tibetan Plateau. Mol. Ecol. 21, 4811–4827 (2012). [DOI] [PubMed] [Google Scholar]
- Ge X. J. et al. Conservation genetics and phylogeography of endangered and endemic shrub Tetraena mongolica (Zygophyllaceae) in Inner Mongolia, China. BMC Genet. 12, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. X. A Study on Origin and Formation of the Chinese Desert Floras. Acta Phytotaxonomica Sinica 33, 131–143 (1995). [Google Scholar]
- Fang X. M. et al. Loess in the Tian Shan and its implications for the development of the Gurbantunggut Desert and drying of northern Xinjiang. Chin. Sci. Bull. 47, 1381–1387 (2002). [Google Scholar]
- Guan Q., Pan B., Li N., Zhang J. & Xue L. Timing and significance of the initiation of present day deserts in the northeastern Hexi Corridor, China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 306, 70–74 (2011). [Google Scholar]
- Sun J. M & Liu T. S. The age of the Taklimakan Desert. Science 312, 1621 (2006). [DOI] [PubMed] [Google Scholar]
- Sun J. M, Zhang Z. Q & Zhang L. Y. New evidence on the age of the Taklimakan Desert. Geology 37, 159–162 (2009). [Google Scholar]
- Zhu Z. D., Chen Y. P. & Wu Z. Study on the Aeolian geomorphology of the Taklamakan Desert (Science Press, 1981). [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Fuertes Aguilar J. & Nieto Feliner G. Additive polymorphisms and reticulation in an ITS phylogeny of thrifts (Armeria, Plumbaginaceae). Mol. Phylogenet. Evol. 28, 430–447 (2003). [DOI] [PubMed] [Google Scholar]
- Bandelt H. J., Forster P. & Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- Pons O. & Petit R. J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupanloup I., Schneider S. & Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581 (2002). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 (2005). [PMC free article] [PubMed] [Google Scholar]
- Manni F., Guerard E. & Heyer E. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier's algorithm. Hum. Biol. 76, 173–190 (2004). [DOI] [PubMed] [Google Scholar]
- Hardy O. J. & Vekemans X. Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity 83, 145–154 (1999). [DOI] [PubMed] [Google Scholar]
- Jensen J., Bohonak A. & Kelley S. Isolation by distance, web service. BMC Genet. 6, 13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Nylander J. A., Olsson U., Alstrom P. & Sanmartin I. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst. Biol. 57, 257–268 (2008). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. Automatic Control, IEEE Transactions on 19, 716–723 (1974). [Google Scholar]
- Kelchner S. A. & Thomas M. A. Model use in phylogenetics: nine key questions. Trends Ecol. Evol. 22, 87–94 (2007). [DOI] [PubMed] [Google Scholar]
- Drummond A. J. & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. & Drummond A. J. Tracer v1.5. (2009). Available at: http://beast.bio.ed.ac.uk/Tracer. (Accessed: 18th January 2015).
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G. & Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
- Liu C., Berry P. M., Dawson T. P. & Pearson R. G. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28, 385–393 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.