Abstract
Multivariate analysis (MVA) is a class of statistical and pattern recognition methods that involve the processing of data that contains multiple measurements per sample. MVA can be used to address a wide variety of medical neuroimaging-related challenges including identifying variables associated with a measure of clinical importance (i.e. patient outcome), creating diagnostic tests, assisting in characterizing developmental disorders, understanding disease etiology, development and progression, assisting in treatment monitoring and much more. Compared to adults, imaging of developing immature brains has attracted less attention from MVA researchers. However, remarkable MVA research growth has occurred in recent years. This paper presents the results of a systematic review of the literature focusing on MVA technologies applied to neurodevelopmental disorders in fetal, neonatal and pediatric magnetic resonance imaging (MRI) of the brain. The goal of this manuscript is to provide a concise review of the state of the scientific literature on studies employing brain MRI and MVA in a pre-adult population. Neurological developmental disorders addressed in the MVA research contained in this review include autism spectrum disorder, attention deficit hyperactivity disorder, epilepsy, schizophrenia and more. While the results of this review demonstrate considerable interest from the scientific community in applications of MVA technologies in pediatric/neonatal/fetal brain MRI, the field is still young and considerable research growth remains ahead of us.
Keywords: Multivariate analysis, Machine learning, Fetal, Neonatal, Pediatric, Brain MRI
Highlights
-
•
Literature review of multivariate brain MRI neurodevelopmental disorders research
-
•
Focused on fetal, neonatal and pediatric populations
-
•
Neurodevelopmental disorders: autism, ADHD, epilepsy, neuropsychiatric disorders, etc.
-
•
Enormous growth in multivariate analyses in recent years, more to come
-
•
Multivariate analysis has tremendous potential in developing population applications.
1. Introduction
The developing brain is a distributed processing machine undergoing rapid change. Basic functional tasks require the coordination and cooperation of neurons in multiple brain regions all of which are in a state of rapid growth. Differences in brain activity between children and adults (Casey et al., 1997, Thomas et al., 2001, Bunge et al., 2002), the structural changes that many regions undergo during development (Reiss et al., 1996, Gogtay et al., 2004, Fair et al., 2009, Supekar et al., 2009) and the recruitment of large cohorts of age matched subjects are major challenges facing researchers in fetal, neonatal and pediatric neuroimaging. Important information regarding brain function is encoded in distributed patterns of brain activity and structure (Mesulam, 1981, McIntosh et al., 1996, Fox et al., 2005, Vaadia et al., 1995) and assessing/identifying these distributed patterns is particularly challenging in a pediatric/neonatal/fetal population due to small brain sizes, a rapidly changing physiology, a high degree of brain plasticity, patient motion, increased metabolism and an incomplete understanding of brain development.
Multivariate analysis (MVA) techniques (i.e. multivariate regression, multivariate analysis of variance, machine learning, etc.) are advanced statistical, computational and pattern recognition technologies that evaluate multiple variables/measurements simultaneously. MVA technologies provide a theoretical improvement over traditional univariate techniques which examine each acquired measurement individually. MVA has particularly large potential in magnetic resonance imaging (MRI) as many physiological and anatomical parameters can be measured, new measurements are constantly under development and distributed measurements across the entire brain are acquired. The ideal way to combine a distributed set of a variety of different physiological and anatomical measurements for any particular disease population is not known a priori, making MVA research applied to the MRI-assessed developing brain a fertile field of ongoing investigation.
MVA techniques can be employed to discover what brain regions and what physiological/anatomical measurements best help characterize different types of neurodevelopmental disorders. Furthermore, reliably predicting disease onset, detecting and diagnosing disease and monitoring treatment response are extremely important challenges in clinical pediatric neuroimaging, an area in which MVA technologies are capable of assisting. MVA techniques can also be used to identify clinical factors and imaging parameters that are associated with important issues such as patient outcomes, disease progression and more.
MRI provides a wide variety of different physiological/anatomical measurements distributed across a subject's brain, thus providing a wealth of information that may assist in an array of research problems in both clinical applications and basic research. The most common MRI method produces basic structural information related to the concentration of hydrogen protons. Water is the most abundant molecule in the human body and two hydrogen protons are found in each molecule. Since the body regulates many tissues and organs by controlling the concentration of water across membranes, structural MRI provides excellent tissue contrast. Perfusion MRI measures blood perfusion by tagging fast moving hydrogen protons in the blood stream and monitoring the tissues to which they are deposited. Functional MRI (fMRI) measures a blood oxygen level-dependent signal which is associated with brain activity and is an important method for monitoring brain function during an assigned task. Functional MRI is also used to monitor normal blood oxygen levels in the brain while the subject is at rest (rs-fMRI). Functional MRI is also extended to functional connectivity analyses which often employ MVA technology to identify regions of the brain with similar activation patterns. It should be noted that MRI based functional connectivity analyses merely infer connectivity based on similar functional behavior; connectivity is not measured directly. Diffusion weighted imaging (DWI) is focused on acquiring measurements of water diffusion which can help identify a wide variety of tissue conditions. Diffusion tensor imaging (DTI) is a directional extension of DWI which measures water diffusion in many different spatial directions at each pixel/voxel location in the brain. DTI allows the tracking of tissue coherence that is often associated with neural fiber tracts whose presence is inferred based on voxels exhibiting water diffusion directed towards adjacent voxels. Thus DTI provides a direct measurement from which connectivity is inferred. MRI can acquire considerably more types of images as well that at present have not had a major impact in studies focused on imaging-based MVA of pediatric, neonatal and fetal neurodevelopmental disorders such as chemical exchange saturation transfer imaging (which includes pH sensitive amide proton transfer imaging) and MR spectroscopy (which is often not spatially resolved but a single measurement that is acquired across the entire brain).
Variations in MRI modalities allow us to acquire a wide variety of measurements distributed across the pediatric, neonatal and fetal brain in vivo. Furthermore, neurodevelopmental disorders can be associated with abnormal physiological/anatomical measurements of various types at a wide variety of different locations within the brain. Thus powerful MVA techniques which combine multiple measurements have the potential to assist in identifying the physiological/anatomical conditions associated with the formation and presence of neurodevelopmental disorders. MVA techniques also have the potential to inform prognosis prediction and to monitor disease progression and therapeutic response.
MVA techniques have seen remarkable growth in interest from the pediatric, neonatal and fetal imaging scientific research community in recent years. An excellent review article on the use of MVA classification technologies in developmental brain imaging was published in 2009 (Bray et al., 2009), however, at the time of publication the number of research studies using MVA in a pediatric, neonatal and fetal population was limited. In the years since 2009, pediatric brain MRI studies employing MVA technologies have exhibited such remarkable growth that the topic warrants a systematic review. The goal of this manuscript is to provide a concise review of the state of the scientific literature's studies employing brain MRI and MVA in a pre-adult population and focuses on neurodevelopmental disorders.
2. Materials and methods
2.1. Multivariate analysis techniques
MVA techniques can be divided into several classes of technology. Multivariate statistical techniques are quite varied in their potential applications, with a prominent example being techniques focused on the identification of measurements that are correlated with an important clinical variable. With a large set of measurements available, MVA techniques such as multivariable linear regression (Rencher and Christensen, 2012) can be used to identify a subset of variables that are associated with an important clinical outcome. MVA techniques such as multivariate analysis of variance (MANOVA) (Warne, 2014) can help assess the effect that changes in one variable have on dependent variables and can generally help elucidate the existing relationships between dependent and independent variables. Multivariate analysis of covariance (MANCOVA) (Smith, 1958) is a technique similar to MANOVA but allows the factoring out of noise or error introduced by a covariant variable. This review will discuss many applications of multivariate statistics in a pediatric, neonatal and fetal population which will help to further illustrate the wide variety of potential uses of these techniques in a medical research context.
Multivariate statistical techniques can also create new measurements that are a combination of existing measures creating customized factors/components that can represent underlying physiological conditions. This is typically accomplished by transforming a large matrix of example measurements in an effort to isolate underlying patterns represented in the dataset. One example technique is Factor Analysis (Harman, 1960), a statistical method used to describe variability among observed, correlated variables in terms of a potentially lower number of unobserved variables called factors. Another example of this type of technique is Principal Components Analysis (PCA) (Dunteman, 1989), which uses an orthogonal transformation to convert a set of measurements of possibly correlated variables into a set of linearly uncorrelated variables called principal components obtained by maximizing the variance in the initial dataset represented by the computed components. Independent Component Analysis (ICA) (Hyvarinen et al., 2004) represents an alternative technique capable of revealing hidden factors that underlie sets of random measurements by assuming that these underlying components are non-Gaussian signals. These data reduction methods involve summarizing many measurements with few measurements and bridge the gap between statistical analysis techniques (which are typically employed by researchers as a step in their study's data analysis) and related computational technologies (which can be integrated into automated and semi-automated applications that can be used by physicians in a clinical capacity).
A related class of analysis techniques have considerable overlap with multivariate statistical approaches in that many also involve the selection of measurements and employ data reduction, however, they are often considered technologies rather than statistical analysis techniques with a prime example being machine learning (Carbonell et al., 1983). Machine learning is further divided into two main approaches: supervised and unsupervised learning. Supervised learning is a class of technologies that use training data which is a collection of measurements associated with multiple groups such as healthy controls and subjects with a known neurodevelopmental disorder. The training data is used to inform future predictions, allowing the computer algorithm to assign new unknown samples to one of the training groups for which it was provided example sets of measurements. Some supervised learning algorithms include feature selection as part of the overall technology (the process of selecting which measurements to rely upon for prediction), however, many do not and feature selection is often addressed as a separate topic in the scientific literature. Example supervised learning technologies include the support vector machine (SVM) (Vapnik, 1995), artificial neural networks (ANN) (Yegnanarayana, 2009), linear discriminant analysis (LDA) (McLachlan, 2004), random forests (RF) (Breiman, 2001), k nearest neighbor (kNN) (Altman, 1992), Bayesian classification (Devroye et al., 1996) and the generalized linear model (GLM) (Nelder and Wedderburn, 1972) which is also sometimes referred to as a multivariate statistical analysis technique, highlighting the considerable overlap between machine learning technologies and traditional statistical MVA techniques.
Unsupervised learning differs from supervised learning in that these MVA technologies are not provided with a set of example training data on which to base predictions from new unknown samples. Instead, unsupervised learning technologies are tasked with performing a basic level of pattern recognition on a medical imaging examination based on the examination itself and the analysis strategies employed by the unsupervised learning technology. This typically involves dividing a medical examination into multiple regions-of-interest which can facilitate a variety of in depth analyses (this process is also referred to as image segmentation). These technologies can be applied to isolating a particular structure in the brain and can be used to monitor changes due to healthy brain development or malformations due to the advent of neurodevelopmental disorders. Unsupervised learning technology can assist in the extraction of regional physiological statistics and in turn can play a critical role in computer-aided diagnosis systems, supporting high level patient wide diagnoses. Unsupervised learning technologies can also play a useful role in tissue outcome prediction studies by delineating the extent of tissue damage at follow-up imaging. Example unsupervised learning technologies include the ISODATA algorithm (Ball and Hall, 1965) and cluster analysis (Manton et al., 2014), a family of techniques that include k-means, hierarchical clustering and so on.
2.2. Validation overview
Validating the performance of the techniques and technologies used in the analyses covered in this review is challenging as many MVA approaches rely on provided training data (samples indicative of known conditions). The training data is used to guide the MVA technique (for example: supervised machine learning) allowing it to process new samples and classify them as belonging to one of the groups of training samples it was provided with. Ideally, training samples would be extracted from one dataset and testing would be performed on an independent collection of patient data in order to ensure there is no overlap between the training and testing samples employed as such an overlap could result in unrealistically promising test performance metrics. In practice it is very common for a researcher to only have access to a single dataset collected at a research center with which they (or their collaborators) are affiliated due to laws protecting patient data which prevents their medical examinations from being shared publicly. As such validation techniques are required in order to accurately assess the performance of multivariate analysis technologies when only a single dataset is available to the researchers.
A variety of validation strategies are available for the evaluation of MVA technologies on a single dataset. In leave-one-out (LOO) validation the MVA technology is provided with all but one of the samples in the dataset for training (Kearns and Ron, 1999). Testing is performed on the single sample that was excluded. This process is repeated exhaustively such that each sample in the dataset is excluded from training and assigned as the testing sample during one of the validation iterations. A variety of possible performance metrics are computed in each validation iteration and after completion of all validation runs, the performance metrics are combined using standard group based statistical analyses (average values, standard deviations, etc.). K-fold cross validation (CV) randomly assigns the dataset into a user defined number of groups at which point a group based leave-one-out procedure is repeated such that each group is left out of the training samples and reserved for testing (Kohavi, 1995).
Another validation procedure involves performing a basic randomized validation trial which typically involves randomly selecting a predefined percentage of samples for training and the remaining samples are reserved for testing (Levman et al., 2008). This randomization is repeated for a user specified number of iterations and summary statistics are collected across all of the validation runs. Efron's bootstrap (Efron and Tibshirani, 1994) is a variation of the basic randomized trial involving the selection of n samples from a dataset with n total samples for training (note that this allows multiple selection of a single sample into the training group yielding approximately 63.8% of the total samples for the training group). The testing group is made of the remaining samples and the trial is repeated for a user specified number of iterations. Note that although Efron's bootstrap allows a single sample to be selected multiple times for the training data in a single iteration of the validation, it ensures that there is no overlap between the training and testing groups in each randomized validation iteration.
In another validation strategy, a single dataset is partitioned into two temporally contiguous datasets. In this situation, the first dataset comprises the examinations from the first half of the retrospective review period and the second dataset comprises the examinations from the second half of the review period. This allows a pseudo-prospective validation structure within a strictly retrospective analysis, whereby the machine learning technology is trained on the samples during the first time period and is tested on those samples acquired after those that were used for training. This testing strategy allows a simulation of what one might expect from a prospective assessment of the technology being evaluating.
Each time a validation strategy is employed, evaluative metrics are calculated which help assess the quality of a given supervised learning based technology. Sensitivity is a metric that evaluates the percentage of patients with a given neurological developmental disorder correctly diagnosed by the test as having that neurodevelopmental disorder (Youngstrom, 2014). Specificity evaluates the percentage of patients without a given neurodevelopmental disorder correctly diagnosed as not having that disorder (Youngstrom, 2014). Overall accuracy is a combination of what Sensitivity and Specificity assess in a single evaluative metric that measures the percentage of all subjects (with and without the neurological developmental disorder being tested for) that are correctly diagnosed by the supervised learning technique. Receiver operating characteristic curve analysis is a visual plot of the trade-off between a test's Sensitivity and its Specificity (Youngstrom, 2014) as the test bias is varied (the test bias is an adjustable parameter that inclines the test to be more or less likely to assign a given sample to the pathological or normal groups). The area under the receiver operating characteristic curve (AUC: Area Under the Curve) is a common metric for evaluating the robustness of a test independent of a fixed test bias setting, with a value of 1 indicating a perfect test and a value of 0.5 representing a test that randomly guesses. Receiver operating characteristic curve analyses represent one of the most robust testing methodologies available for the evaluation of MVA technologies.
After assessment of the performance of a supervised learning technology with the aforementioned validation techniques, performance metrics are interpreted and if deemed to be of particularly high quality, the technology under consideration can be promoted to the gold standard method for validation, a prospective trial. In a prospective trial, the technology under consideration is incorporated into the clinical care of patients as they are being evaluated, assessed and treated in an ongoing clinical setting.
2.3. Review Parameters
The search engine MEDLINE/PubMed was used for this systematic review on May 4th, 2015. The search terms included were <Multivariate pediatric brain MRI> or <machine learning pediatric brain MRI> or <Multivariate neonatal brain MRI> or <machine learning neonatal brain MRI> or <Multivariate fetal brain MRI> or <machine learning fetal brain MRI>. This yielded 166 articles whose titles and abstracts were reviewed for appropriateness. Articles were excluded if brain MRI was not performed in a fetal, neonatal or child/adolescent population. Articles were excluded if they did not involve an MVA that included the brain MRI data acquired as an important component of the analysis (this includes measurements from MRI examinations being incorporated into the MVA, MRI informed clinical diagnoses allowing group stratification prior to MVA, etc.). Articles were excluded if they were not authored in English. Articles were excluded if they were focused on healthy brains, normal neurological development, brain cancer, stroke, hemorrhage, brain injury, brain damage due to a medical condition located outside the nervous system and related surgeries, thus allowing this article to focus on a wide variety of neurological developmental disorders. Articles from this search process that were not excluded were analyzed for this systematic review and noteworthy citations within these articles were considered for inclusion in this review (subject to the same exclusion criteria).
3. Results
The results of this systematic review are divided into subsections focusing on the main neurological developmental disorders addressed in the literature, including autism, attention-deficit hyperactivity disorder, epilepsy and neuropsychiatric disorders followed by a section addressing the many neurodevelopmental disorders for which there were very few studies that met our inclusion criteria.
3.1. Autism
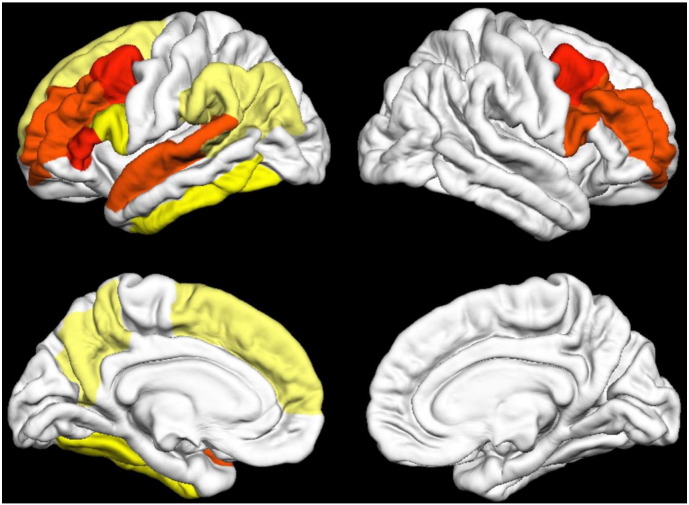
Autism is a neurodevelopmental disorder defined in the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) as characterized by persistent deficits in social communication and social interaction that manifests as deficits in social–emotional reciprocity, deficits in nonverbal communicative behaviors used for social interaction, deficits in developing, maintaining and understanding relationships and restricted, repetitive patterns of behavior (American Psychiatric Association, 2013). The use of structural MRI in autism spectrum disorder (ASD) is summarized in a previously published review article (Chen et al., 2011), whereas the review presented here includes all types of MRI acquisition (structural, diffusion, diffusion tractography, functional, etc.). While some relevant studies have been published in conferences (Ghiassian et al., 2013, Cates et al., 2008, An et al., 2010), Table 1 is provided to summarize the multivariate analyses applied to ASD presented in the literature in journal form. Due to the inherent heterogeneity in these datasets it is challenging to accurately compare the results between studies. However, the most commonly presented performance metric (Overall Accuracy — referred to simply as Accuracy in the Tables in this review) is presented and alternative metrics such as Sensitivity, Specificity and the area under the receiver operating characteristic curve (AUC) are provided as well under the Results column. These results are performance metrics typically assessing the developed technique's ability to discriminate between pathological patients and healthy controls. The number of subjects with ASD is provided under the column labeled n. The feature measurements used in the study are provided under the heading Features relied upon, the MVA methodology used is provided under the heading MVA and the validation method employed is provided under the heading Validation. If an entry is not available for a given study “N/A” is entered into the table. Fig. 1 provides a three-dimensional rendering of a structural MRI examination of a child with autism with an overlaid color map representing cortical thickness as a demonstration of some of the creative methods available for the analysis of the large amounts of data acquired in modern MRI examinations.
Table 1.
Summary of multivariate analyses applied to autism spectrum disorder.
| Author | Year | n | Results | Features relied upon | MVA | Validation |
|---|---|---|---|---|---|---|
| Jiao et al. (2010), also Jiao et al. (2011) | 2010 | 22 | AUC = 0.93 Accuracy = 87% | Regional cortical thickness (left & right pars triangularis, left medial orbitofrontal gyrus, left parahippocampal gyrus, left frontal pole, left precuneus, left caudal anterior cingulate) | SVM, ANN, Functional Trees, Logistic Model Trees | 10 fold CV |
| Zhou et al. (2014) | 2014 | 127 | Accuracy = 70% | Caudate volume, rs-fMRI: caudate-cortical & inferior frontal gyrus connectivity | RF | 10 fold CV |
| Wee et al. (2014) | 2014 | 58 | AUC = 0.995 Accuracy = 96% | Regional measurements combined with interregional measurements (cortical thickness, etc.) | SVM | 2 fold CV |
| Crippa et al. (2015) | 2015 | 15 | Accuracy = 97% | Upper limb motor movement feature measurements | SVM | LOO |
| Deshpande et al. (2013) | 2013 | 15 | Accuracy = 96% | DTI: measurements based on effective connectivity paths | SVM | 10 fold CV |
| Ecker et al. (2010) | 2010 | 22 | Sensitivity = 88% Specificity = 86% |
Limbic fronto-striatal, fronto-temporal, fronto-parietal & cerebellar measures | SVM | LOO |
| Ingalhalikar et al. (2011) | 2011 | 45 | Sensitivity = 74% Specificity = 84% |
Atlas based regional feature measurements extracted from DTI | SVM | LOO |
| Calderoni et al. (2012) | 2012 | 38 | AUC = 0.80 | left superior frontal gyrus (female autism patients exhibited more gray matter) | SVM | LOO (paired) |
| Lange et al. (2010) | 2010 | 30 + 12 | Accuracy = 92% | Hemispheric asymmetry measurements with DTI | SVM | LOO |
| Plitt et al. (2015) | 2015 | 59 | Accuracy = 95% | Behavioral measures and resting state fMRI measurements. Behavioral measurements outperformed. | RF, kNN, SVM, Bayes, LDA, logistic regression | LOO |
| Adluru et al. (2012) | 2012 | 11 | Accuracy = 81% | Feature measurements derived from Epsilon Radial Networks | SVM | LOO |
| Lombardo et al. (2015) | 2015 | 60 | AUC = 0.81 | fMRI, language measures, autism diagnostic observation scale for predicting language outcome in children with autism | LDA | 5 fold CV |
| Lim et al. (2013) | 2013 | 19 | Accuracy = 85% | Gray matter volumetric data | Gaussian process | LOO |
| Uddin et al. (2011) | 2011 | 24 | Accuracy = 90% | Gray matter in the posterior cingulate cortex, medial prefrontal cortex and bilateral medial temporal lobes | SVM | 10 fold CV |
| Nielsen et al. (2013) | 2013 | 447 | Accuracy = 60% | rs-fMRI: default mode network, parahippocampal and fusiform gyri, insula, Wernicki Area, intraparietal sulcus | GLM | LOO |
| Chen et al. (2015) | 2015 | 126 | Accuracy = 91% | rs-fMRI: somatosensory, default mode, visual and subcortical regions | RF | Native to RF |
| Chen et al. (2016) | 2016 | 112 | Accuracy = 79% | rs-fMRI: atypical connections between the default mode, fronto-parietal and cingulo-opercular networks | SVM | LOO |
| Juranek et al. (2006) | 2006 | 42 | Anxiety/depression correlated with right amygdala volume | Amygdala volume and anxiety/depression levels measured in autistic children. | Linear regression with multivariate adjustment of covariates | N/A |
Fig. 1.
A 3D surface rendering of a structural T1 examination of a child with autism with overlaid cortical thickness measurements (see color bar) (Jiao et al., 2010). Figure is reproduced with permission.
The MVA autism literature demonstrates the feasibility of creating technology that can contribute to the differential diagnosis of autism relative to normal controls, ADHD and Asperger's syndrome. Furthermore, the autism literature demonstrates the feasibility of identifying areas of the brain with abnormal structural and functional organization among patients with autism.
3.2. Attention deficit hyperactivity disorder (ADHD)
Attention deficit hyperactivity disorder (ADHD), defined in DSM-5 is a developmental disorder with symptoms arising prior to the age of 12 characterized by a pattern of behavior that can result in performance issues in social, educational or work settings (American Psychiatric Association, 2013). ADHD is further subdivided into its Inattentive form (children who exhibit distraction, forgetfulness, lack of attention to detail, carelessness, etc.), its Hyperactive–Impulsive form (children who exhibit fidgeting, squirming, interrupting, excessive talking, etc.) and the Combined form (children with both Inattentive and Hyperactive–Impulsive symptoms). While some relevant studies have been published in conferences (Ghiassian et al., 2013, Mahanand et al., 2013, Liang et al., 2012, Zhu et al., 2005, Wang et al., 2011), Table 2 is provided to summarize the multivariate analyses applied to ADHD presented in the literature in journal form. Due to the inherent heterogeneity in these datasets it is challenging to compare the results between studies. Table 2 is provided to summarize the results and facilitate comparisons and is structured similarly to Table 1. The number of subjects with ADHD is provided under the column labeled n, where entries marked ADHD-200 indicate that the authors used the dataset provided by the ADHD-200 Consortium's machine learning competition which included approximately 285 examinations of patients with ADHD as well as controls, however the studies involved in the competition varied considerably in the exclusion of samples from their analyses. Fig. 2 provides example structural MRI examinations of a patient with ADHD as well as gray matter membership masks which allow for cortical assessment independent of white matter development.
Table 2.
Summary of multivariate analyses applied to attention deficit hyperactivity disorder.
| Author | Year | n | Results | Features relied upon | MVA | Validation |
|---|---|---|---|---|---|---|
| Fair et al. (2012) | 2012 | ADHD-200 | Accuracy = 81% Inattentive = 81% Combined = 77% |
rs-fMRI: cerebellum and dorsolateral prefrontal cortex. | SVM | LOO |
| Anderson et al. (2014) | 2014 | ADHD-200 | Accuracy = 67% | fMRI: posterior cingulate, precuneus, parahippocampal regions (Inattentive ADHD) | Decision Tree | LOO |
| Subramanian et al. (2014) | 2014 | ADHD-200 | Sensitivity = 62% Specificity = 74% |
Amygdala regional anatomy | McIT2FIS | 10 fold CV |
| Chang et al. (2012) | 2012 | ADHD-200 | Accuracy = 70% | Texture analysis of anatomical brain MRI. Gray matter is most important |
SVM | 10 fold CV |
| Eloyan et al. (2012) | 2012 | ADHD-200 | Sensitivity = 21% Specificity = 94% |
rs-fMRI and structural MRI | RF, SVM | Data Splitting |
| Dai et al. (2012) | 2012 | ADHD-200 | Accuracy = 68% | rs-fMRI, cortical thickness | SVM | 10 fold CV |
| Sato et al. (2012) | 2012 | ADHD-200 | Accuracy = 67% inattentive vs. combined ADHD | ICA maps, regional homogeneity, amplitude of low frequency fluctuations | Logistic regression | Monte Carlo Subsampling |
| Colby et al. (2012) | 2012 | ADHD-200 | Accuracy = 55% | Structural cortical measures: frontal, temporal and cingulate regions | SVM | 10 fold CV |
| Cheng et al. (2012) | 2012 | ADHD-200 | Accuracy = 76% | Frontal and cerebellar regions | SVM | LOO |
| Peng et al. (2013) | 2013 | 55 | Accuracy = 90% | Right pars opercularis, left paracentral lobule, left & right transverse temporal, left middle temporal, left & right cuneus, left lingual, left & right insular regions | Extreme learning | LOO |
| Igual et al. (2012a), also Igual et al., 2011, Igual et al., 2012b, Igual et al., 2012c | 2012 | 39 | Accuracy = 72% | Caudate nuclei, dissociated dipoles | SVM, Adaboost | 5 fold CV |
| Zhu et al. (2008) | 2008 | 9 | Accuracy = 85% | Regional homogeneity of resting state fMRI investigated | Fisher Discriminant Analysis | LOO |
| Iannaccone et al. (2015) | 2015 | 20 | Accuracy = 78% | Posterior cingulate, temporal and occipital cortex | SVM | LOO |
| Lim et al. (2013) | 2013 | 29 | Accuracy = 85% ADHD vs. autism | Gray matter volumetric data | Gaussian process | LOO |
| Hart et al. (2014a), also Hart et al. (2014b) | 2014 | 30 | Accuracy = 77% | Earlier developing ventromedial fronto-limbic regions | Gaussian process | LOO |
| Shehzad et al. (2014) | 2014 | ADHD-200 | Associations within superior parietal cortex | rs-fMRI distributed across subregions of the brain | Multivariate distance matrix regression | Randomized bootstrap |
| Shi et al. (2013) | 2013 | 17 | Abnormalities in anterior/inferior putamen | Morphology of ventral aspect of striatum for analysis of preterm neonates | Multivariate tensor morphometry | N/A |
Fig. 2.
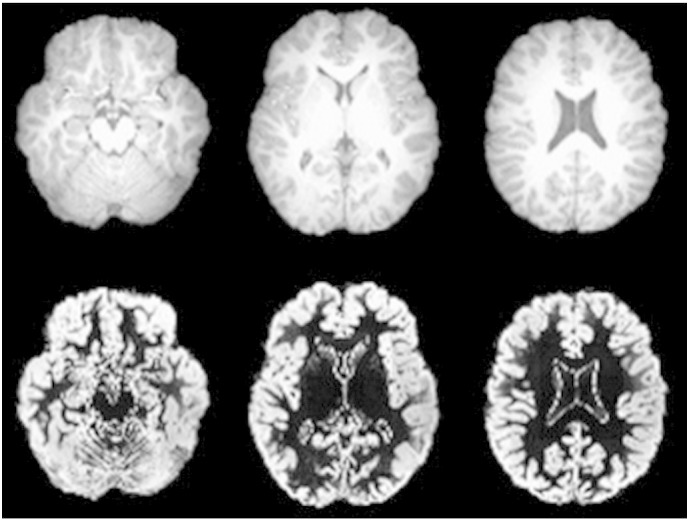
Structural MRI examinations (top row) and gray matter classified membership maps (bottom row) of a subject from an ADHD research study (Zhu et al., 2008). Figure is reproduced with permission.
As a related aside, prenatal exposure to cocaine is associated with ADHD as well as premature birth and birth defects. Deshpande et al. (2010) demonstrated that multivariate analysis technologies can predict with 90% accuracy whether a subject was exposed to cocaine prenatally based on functional MRI data.
The ADHD-200 competition provides an unusual opportunity to compare a variety of different MVA approaches to an identical dataset (see Table 2). Fair et al. (Fair et al., 2012) produced the most accurate system for the identification of ADHD from MRI examinations based on this competition with 81% overall accuracy using the support vector machine (Vapnik, 1995). The widely varying reported accuracies from those studies that participated in the ADHD-200 competition (see Table 2) helps illustrate the remarkable challenge associated with producing high quality MVA research applied to brain MRI in pre-adult populations.
The MVA ADHD literature demonstrates the feasibility of creating technology that can contribute to the diagnosis of ADHD. Furthermore, the literature demonstrates the feasibility of identifying areas of the brain with abnormal structural and functional organization among patients with ADHD.
3.3. Epilepsy and focal cortical dysplasia
Epilepsy, defined by the International League Against Epilepsy (ILAE), is characterized as any one of the following conditions: at least two unprovoked seizures occurring > 24 h apart, one unprovoked seizure and a probability of further seizures (at least 60%) over the next 10 years after two unprovoked seizures or diagnosis of an epilepsy syndrome (Fisher et al., 2014). Epilepsy has a clear cause in only a minority of patients diagnosed with the condition. Focal cortical dysplasia (FCD) is a congenital abnormality characterized by neurons failing to migrate in the proper formation in utero leading to misfiring neurons that can cause seizures and is one potential cause of intractable epilepsy. Due to the inherent heterogeneity in these datasets it is challenging to accurately compare the results between studies. Table 3 is provided to summarize the results and facilitate comparisons and is structured similarly to Table 1, Table 2. When randomized validation is performed it is entered in the final column as “Random” followed by the percentage of samples devoted to training and testing respectively.
Table 3.
Summary of multivariate analyses applied to epilepsy and focal cortical dysplasia.
| Author | Year | n | Results | Features relied upon | MVA | Validation |
|---|---|---|---|---|---|---|
| Paldino et al. (2014) | 2014 | 33 | Sensitivity = 100% Specificity = 95% |
DTI: tracts of corpus callosum, corticospinal tracts. Inferior longitudinal, inferior fronto-occipital, uncinate and arcuate fasciculi | RF | Random 67/33 |
| You et al. (2009) | 2009 | 122 | Accuracy = 96% | PCA derived measurements of fMRI data | SVM | Random 50/50 |
| Amarreh et al. (2014) | 2014 | 20 | Sensitivity = 90 to 100% Specificity = 100 to 97% |
DTI: mean, radial & axial diffusivity as well as fractional anisotropy | SVM | 10 fold CV |
| Wang et al. (2014) | 2014 | 58 | Accuracy = 95% | fMRI measurements for separation of children between left dominant, right dominant, bilateral, other | Nearest neighbor & distance based fuzzy classifier | Random 50/50 |
| Krsek et al. (2013) | 2013 | 33 | Complete removal of epileptogenic tissue detected on MRI predicts seizure free outcome | Resection of epileptogenic tissue, EEG measurements | Multivariate statistical Cox's model | N/A |
| Siniatchkin et al. (2007) | 2007 | 7 | 4.75 times more BOLD signal compared to traditional method | EEG and fMRI based measurements | PCA | Custom |
| Loyek et al. (2008) | 2008 | 5 | Sensitivity = 85 to 98% Specificity = 73 to 92% FCD detection |
Statistical, gray level co-occurrence, gray level run length measurements | SVM | N/A |
| Kobashi et al. (2011) | 2011 | 3 | Sensitivity = 94% Specificity = 85% |
MR texture features, fractal dimension for FCD detection | SVM | LOO |
| Kawakami et al. (2010) | 2010 | 3 | Sensitivity = 91% Specificity = 93% |
T1 structural measurements for FCD detection | SVM | LOO |
| Daley et al. (2006) | 2006 | 19 | Epileptic patients have smaller anterior hippocampal volumes | MRI based hippocampal volumes | Maximum likelihood classification | N/A |
Additionally, Galka et al. (2011) presented a technique that acts as an alternative to Independent Components Analysis and their study included a demonstration of their approach on a pediatric patient with epilepsy.
As a related aside, seizure disorders (epilepsy included) are rarely treated with hemispherectomy, a surgical procedure involving the removal or disabling of one of the hemispheres of the brain. Moosa et al. (2013) used MVA to demonstrate that multilobar MRI abnormalities in the contralateral hemisphere were correlated with poor language outcome in pediatric patients with seizure disorders who had underwent hemispherectomy.
The MVA literature focused on epilepsy and focal cortical dysplasia demonstrates the feasibility of creating technology that can contribute to the detection and diagnosis of these conditions. Furthermore, the literature demonstrates the feasibility of identifying areas of the brain with abnormal structural and functional organization with the help of MVA techniques and technologies.
3.4. Neuropsychiatric disorders
Schizophrenia (also known as schizophrenic spectrum disorders — SSD), defined in DSM-5 as being characterized by two or more of the following: delusions, hallucinations, disorganized speech, grossly disorganized or catatonic behavior, alogia or avolition (American Psychiatric Association, 2013). Due to the inherent heterogeneity in these datasets it is challenging to accurately compare the results between studies. Table 4 is provided to summarize the results and facilitate comparisons and is structured similarly to Table 1, Table 2, Table 3. Fig. 3 provides a three-dimensional rendering of structural T1 MRI examinations demonstrating the variability in cortical differences between healthy controls and children with schizophrenia.
Table 4.
Summary of multivariate analyses applied to schizophrenia.
| Author | Year | n | Results | Features relied upon | MVA | Validation |
|---|---|---|---|---|---|---|
| de Arruda et al. (2014) | 2014 | 19 | Sensitivity = 90% Specificity = 74% |
fMRI across 140 cortical regions | Bayes, Decision Trees | LOO |
| Pina-Camacho et al. (2015) | 2015 | 81 | Accuracy = 99% Accuracy = 81% (SSD & other psychosis) |
Clinical, neuropsychological & neuroimaging measurements | SVM | LOO and 5 fold CV |
| Greenstein et al. (2012) | 2012 | 98 | Accuracy = 74% | left temporal lobes, bilateral dorsolateral prefrontal regions, left medial parietal lobes | RF | Native to RF |
| Rapoport et al. (1999) | 1999 | 15 | 11% decrease in cortical gray matter volume | Gray matter volumes and white matter volumes | MANOVA | N/A |
| Tang et al. (2013) | 2013 | 32 | Increased connectivity: medial frontal gyrus and default mode network | fMRI default mode network measurements for assessment of early onset schizophrenia | ICA | N/A |
Fig. 3.
Colored cortical regions demonstrating differences between patients with childhood onset schizophrenia and healthy controls based on structural T1 MRI (Greenstein et al., 2012). Red regions demonstrate larger cortical differences between schizophrenic patients and controls, yellow regions demonstrate smaller differences. White/gray regions represent no difference. Figure reproduced with permission.
Bipolar disorder, defined in DSM-5, is characterized by extreme changes in mood that range from depression to mania (American Psychiatric Association, 2013). Frazier et al. (2005) demonstrated that pediatric bipolar subjects exhibited smaller cerebral volumes and that female pediatric bipolar patients exhibited smaller hippocampal volumes. Their study was supported by a multivariate analysis of variance (MANOVA) (Frazier et al., 2005). Mwangi et al. (2014) focused on differential diagnosis between healthy controls and subjects with pediatric bipolar disorder based on neuroanatomical abnormalities of the amygdala. Their methodology yielded a diagnostic accuracy of 78%. Ahn et al. (2007) performed a MANOVA analysis and concluded that subjects with bipolar disorder had larger right nucleus accumbens volumes, a finding that was most pronounced in the prepubescent group.
Depression is defined in DSM-5 as being characterized by five or more of the following symptoms which cause clinically significant distress or impairment in social or occupational functioning not caused by the direct physiological effect of a substance or other medical condition: depressed mood most of the day nearly every day, diminished interest in almost all activities, weight loss, insomnia or hypersomnia nearly every day, psychomotor agitation, fatigue, feelings of worthlessness or guilt, diminished ability to think or concentrate and recurrent thoughts of death (American Psychiatric Association, 2013). Wu et al. (2015) demonstrated that multivariate analysis techniques can be applied to the diagnosis of depression with 78% accuracy. Juranek et al. (2006) investigated the association between amygdala volume and depression/anxiety levels in autistic children with MRI and multivariate analyses. Their results indicated that symptoms of anxiety and depression were highly correlated with right amygdala volume, but not with left amygdala volume.
Obsessive compulsive disorder (OCD) is defined in DSM-5 as being characterized by obsessions and compulsions that are time consuming, not attributable to the physiological effects of a substance or other medical condition and not better explained by the symptoms of another mental disorder (American Psychiatric Association, 2013). Obsessions are defined by recurrent or persistent thoughts, urges or impulses experienced as intrusive or unwanted and causing anxiety or distress with the individual attempting to ignore or suppress such thoughts. Compulsions are defined by repetitive behaviors performed in response to an obsession as well as behaviors aimed at preventing or reducing anxiety or distress but are clearly excessive or are not connected with a realistic way to neutralize or prevent the subject's anxiety. Li et al. (2014) demonstrated that multivariate analysis technology can be applied to the diagnosis of OCD from DTI images yielding an overall test accuracy of 84%. The study's results revealed white matter abnormalities based on measurements from the bilateral prefrontal and temporal regions, inferior frontal–occipital fasciculus, superior fronto-parietal fasciculus, splenium of the corpus callosum and left middle cingulum bundle. Szeszko et al. (2004) performed a MANCOVA multivariate analysis and determined that pediatric patients with obsessive compulsive disorder had smaller globus pallidus volumes than healthy controls and more total gray matter in the anterior cingulate gyrus but not the superior frontal gyrus.
Gender dysphoria is defined in DSM-5 as characterized by individuals whose gender at birth is contrary to the one they identify with (American Psychiatric Association, 2013). Hoekzema et al. (2015) demonstrated with MVA techniques that subjects with gender dysphoria could be more accurately automatically discriminated from individuals sharing their gender identity than from those sharing their inherited gender based on spatially distributed gray matter patterns. The authors studied gray matter distributions and regional volumes and reported subtle deviations from natal sex in sexually dimorphic structures which could represent signs of partial sex-atypical differentiation of the brain in subjects with gender dysphoria.
The MVA literature focused on neuropsychiatric disorders demonstrates the feasibility of creating technology that can contribute to the diagnosis of schizophrenia, bipolar disorder and more. Furthermore, the literature demonstrates the feasibility of identifying areas of the brain with abnormal structural and functional organization among patients with neuropsychiatric disorders.
3.5. Other developmental disorders
Agenesis refers to the failure of a particular organ or tissue to develop during embryonic growth. Agenesis of the corpus callosum is a congenital abnormality characterized by the complete or partial absence of the corpus callosum. Jakab et al. (2015) investigated agenesis of the corpus callosum with diffusion tensor imaging and the general linear model multivariate analysis technique. Their results demonstrated that they were able to reveal pathological fiber tracts in 92% of third trimester patients. An example of diffusion tensor imaging results of an in utero fetus with agenesis of the corpus callosum is provided on the left of Fig. 4 and a DTI of a normally developing fetus on the right. Note the comparative lack of fiber tracts connecting the left and right hemispheres of the brain in the subject exhibiting agenesis of the corpus callosum.
Fig. 4.
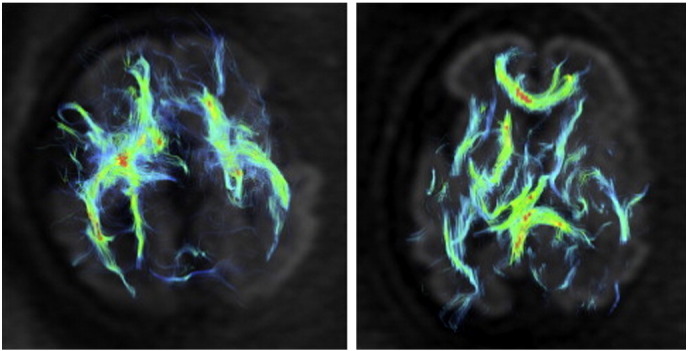
Diffusion tensor imaging of an in utero fetus with agenesis of the corpus callosum (left) and a normally developing fetus (right) at the 30th week of gestation (Jakab et al., 2015). Figure reproduced with permission.
Holoprosencephaly is a neurodevelopmental disorder characterized by malformation of the brain induced by incomplete cleavage of the cerebral hemispheres and deep brain structures. Hahn et al. (Hahn et al., 2006) investigated the neuroanatomical and clinical characteristics of holoprosencephaly using the multivariate technique Factor Analysis. Their analysis identified neuroimaging characteristics as a significant factor towards characterization of the variance exhibited in their data. Neuroimaging combined with mobility, upper extremity/hand function and expressive language accounted for 31% of the variance in their data.
Malformations of the cerebral cortex are a common cause of developmental delay and neurological deficits. The multivariate diagnostic system proposed by Alayon et al. (2007) was reported to yield a sensitivity of 100% and a specificity of 100% in the diagnosis of cortical malformations. The corpus callosum, which is located beneath the cortex is the subject of an additional study (Peruzzo et al., 2014) which indicated that multivariate technology can assist in the radiological diagnostic process.
Cerebral palsy is a group of permanent movement disorders that develop in early childhood and is characterized by poor coordination, stiff & weak muscles, trouble swallowing or speaking and tremors. Griffiths et al. (2010) used multivariate analyses to identify predictors in subjects with acute hypoxic–ischemic injury that can assist in differentiating dyskinetic cerebral palsy (subjects exhibit involuntary movement especially when attempting to move) from spastic cerebral palsy (subjects exhibit muscle engagement in muscle groups that are supposed to only be engaged in a mutually exclusive manner). Children with dyskinetic cerebral palsy were shown to have more frequent injury to the subthalamic nucleus. Children with spastic cerebral palsy were shown to have more severe damage to the white matter in the vicinity of the paracentral lobule. Huang et al. (2006) presented an analysis of DTI MRI findings of fetal, neonatal and pediatric populations with cerebral palsy using MVA based analysis of the diffusion tensor. Their results demonstrated that limbic white matter fibers are first to develop and association fibers develop last.
Dyslexia is a condition characterized by difficulties with accurate word recognition as well as by poor spelling and decoding abilities. Hoeft et al. (2011) have demonstrated that MVA techniques could produce a test to indicate which children's reading skills would improve with 72% accuracy based on fMRI and DTI. This prediction was based on greater right prefrontal activation and right superior longitudinal fasciculus white-matter organization. Froehlich et al. (2014) investigated the ability of multivariate technologies applied to functional MRI data to distinguish between subjects resting and subjects engaged in a reading task in an effort to better understand the neural underpinnings of dyslexia. They reported 90% accuracy for discerning between subjects reading and subjects resting.
Fragile X syndrome is a disorder caused by a single gene mutation resulting in abnormal dendritic and synaptic pruning. Hoeft et al. (2008) demonstrated that the application of multivariate pattern analysis technologies can yield a test that differentially diagnoses subjects with fragile X syndrome from developmentally delayed subjects and healthy controls with 90% accuracy. The approach presented relies upon increased and decreased brain volumes in the posterior vermis, amygdala and hippocampus. Turner syndrome is a genetic condition caused by complete or partial absence of one X-chromosome in a female. The condition is characterized by prominent cognitive weaknesses in executive and visuospatial functions. Marzelli et al. (2011) used multivariate machine learning technology to discriminate between patients with Turner syndrome and healthy controls. Machine learning results identified neuroanatomical variations in individuals with Turner syndrome localized in the hippocampus, orbitofrontal cortex, insula, caudate and cuneus.
Sanz-Cortes et al. (2013) demonstrated that multivariate technologies based on texture analysis could predict abnormal neurobehavior from fetal MR imaging with an accuracy of 95% when based on the frontal lobe, 96% when based on the basal ganglia, 93% when based on the mesencephalon and 83% when based on the cerebellum. Abnormal neurobehaviour was assessed by the Neonatal Behavioral Assessment Scale applied at an average of 42 weeks post birth. Cho and Shin (2001) reported that MVA technologies could be applied to assist in the diagnosis of brain dysfunction.
Ventriculomegaly is a disorder of the brain's ventricle system characterized by dilated lateral ventricles induced by loss of brain volume, infection, infarction, impaired outflow of cerebrospinal fluid from the ventricle system, impaired absorption of cerebrospinal fluid or disorders of the interventricular foramen (congenital malformation, infection, hemorrhage or tumor). Baffero et al. (Baffero et al., 2015) employed MVA which indicated that progression of ventricular enlargement on serial ultrasound examinations was a significant predictor of major cerebral abnormalities. Their analysis supports the use of fetal brain MRI in cases of persistent or progressive ventriculomegaly.
Hydrocephalus is a condition in which there is an abnormal accumulation of cerebrospinal fluid in the brain. Mandell et al. (2015) employed multivariate supervised learning technology to determine that large fluid volume was associated with a decrement in subject cognition. Fukuhara and Luciano (2001) investigated patients with late-onset idiopathic aqueductal stenosis, a narrowing of the aqueduct of Sylvius which blocks the flow of cerebrospinal fluid in the ventricular system. Their subjects were divided into those with symptomatic headaches and those with hydrocephalus. Their study included MRI examinations and MVA which indicated that age independently affected the type of chronic symptoms.
Multiple sclerosis is a disease in which the subject's immune system attacks the protective sheath known as myelin that covers nerves. Peche et al. (2013) employed MVA and reported that the Callen MRI criteria (subject exhibits at least two of the following three criteria: five or more lesions, two or more periventricular lesions, or one brainstem lesion (Callen et al., 2009)) predicts the diagnosis of multiple sclerosis in a pediatric population.
Infants with extremely low birth weight are at increased risk of inhibited growth, cognitive development and chronic diseases later in life. Parikh et al. (2013) employed MVA techniques and showed that surgery for retinopathy (damage to the retina of the eye) and surgery for necrotizing enterocolitis (intestinal tissue dies off) or spontaneous intestinal perforation (a lesion or hole in the intestine) were significantly associated with increasing volume of white matter hyperintensities on MRI in infants with extremely low birth weight.
Sturge–Weber syndrome is a congenital neurological and skin disorder often associated with glaucoma, seizures, intellectual disability, cerebral malformations and benign tumors (angiomas). Juhasz et al. (2007) investigated the association between white matter volume and cognitive function in children with Sturge–Weber syndrome using volumetric MRI and MVA techniques which showed that hemispheric white matter volume ipsilateral to the angioma was an independent predictor of a subject's Intelligence Quotient (IQ) and negatively correlated with age. No correlation was found with gray matter volume.
Several studies exist using MVA and brain MRI applied to a variety of different benign tumors. Hausler et al. (2003) investigated neurological inflammatory pseudotumors which are mostly benign growths characterized by fibrotic ground tissue and polyclonal mononuclear infiltrate. Their study included MR imaging and MVA. Their multivariate model indicated that incomplete resection and female gender were associated with a higher risk of recurrence after surgical removal. Arachnoid cysts are a congenital disorder characterized by cerebrospinal fluid covered by collagen and arachnoidal cells. Al-Holou et al. (2010) employed multivariate analyses which demonstrated that initial cyst size and intracranial location correlated with the presence or development of symptoms and surgical treatment. Neurofibromatosis is a genetic condition characterized by cognitive dysfunction and carries a high risk of tumor formation, particularly in the brain. Duarte et al. (2014) demonstrated that the application of MVA technologies can discriminate individuals with neurofibromatosis from controls with 94% accuracy. Huang et al. (2005) created an image matching system for retrieving similar exams to a provided case of clinical interest. Their testing of the matching algorithm included an examination of a patient with neurofibromatosis, an examination of a patient with an arachnoid cyst and a subject with an arterial venous malformation. Their image matching results were compared to the opinion of a pediatric neuroradiologist which demonstrated that the proposed approach exhibited highly varying performance with 0 to 60% of the matches yielding a correct diagnosis.
Batchelor et al. (2002) presented a set of measurements meant to help quantify folding patterns in the brain from fetal MRI examinations. They propose a multivariate approach to studying the shape of the cerebral cortex to assist in quantifying brain folding. Their study focused on the imaging of ex vivo brain specimens which included a wide variety of pathological findings.
Auditory processing disorders affect the way the brain processes information from the ears. These disorders are characterized by difficulties in recognizing and interpreting sounds despite normal ear structure and function. Schmithorst et al. (2013) applied MVA technology to fMRI and DTI data of subjects with left ear advantage, which is often regarded by clinical audiologists as an indicator for auditory processing disorders. The results indicated that left ear advantage was predicted by increased axial diffusivity in the left internal capsule and decreased functional activation in the left frontal eye fields. Farah et al. (2014) used a generalized linear model based multivariate analysis in their research which concluded that white matter abnormalities underlie listening difficulties in children suspected of auditory processing disorders.
4. Discussion
The results of this systematic review demonstrate a remarkably wide variety of applications of multivariate analysis (MVA) techniques in neurological developmental disorders in pediatric, neonatal and fetal populations. Since the ideal combination of MVA technique and medical imaging derived clinical information is unknown a priori for applications focused on any given neurodevelopmental disorder, an enormous amount of research is required to fully optimize MVA's potential in this domain. Recent years have exhibited ample growth in MVA applied to neurodevelopmental disorders with development in this field ongoing. However, future research growth will be accompanied by substantial challenges. There are many MVA techniques and technologies available to the medical researcher and there is a wide array of associated challenges that will contribute to making this an exciting and difficult research endeavor.
One major obstacle in this application domain is caused by patient motion which is particularly challenging in children who tend to have a harder time remaining still during imaging. Children instructed to remain still in the scanner may forget over the course of the examination. The problems with patient motion are even more poignant for many pediatric neurodevelopmental disorders such as ADHD or epilepsy which make remaining motionless during MRI all the more challenging for the subject. Image registration is a class of technology used to compensate for patient motion but this is a challenging problem for which no gold standard accepted solution is available. Spatial registration to standard templates (or brain atlases) are typically based on the adult brain (Talairach and Tournoux, 1988) and it has been shown that normalization procedures used can cause distortions in the brain examinations of children 6 years old and under (Muzik et al., 2000). Distortional effects may negatively impact MVA results and so care should be taken to avoid providing data exhibiting distortional artifacts to MVA technology. As can be seen from this review, many studies exist that apply MVA to patient populations prone to movement during image acquisition such as ADHD and epilepsy. Typically such studies employ image registration technology to compensate for patient motion, however, it should be noted that many techniques are available and the methods available are not perfect and so the use of such technologies are associated with the introduction of an unknown quantity of experimental error. This unknown experimental error applies regardless of whether registration is performed for within-subject motion correction or if registration is employed to align patient data in a group-wise fashion which may be accomplished with atlases.
Overfitting is an unwanted effect characterized by supervised learning algorithms that are overly tuned to the training data provided. Overfitting can yield extremely impressive test evaluation metrics while simultaneously not representing a robust test that will perform well on an independent dataset. Thus reported impressive test evaluation metrics in the literature may be unrealistic if the authors are reporting results based on an overfitted technology. The earliest studies targeting any given developmental condition typically have to rely on only a single dataset due to the availability of appropriate data. Further testing on independent datasets is a requisite for advancing an MVA technology beyond preliminary analyses. Additionally, it is known that the number of measurements an MVA technology relies upon needs to be much smaller than the total number of samples provided in order to produce robust and reliable results (Raudys and Jain, 1991). When MVA technologies are provided with an inappropriately large number of measurements, the input data space is so large that many MVA techniques might fully or nearly fully separate the data merely due to the large amount of separation between neighboring samples of any of the classes/groups provided. Thus MVA technologies are prone to overfitting unless the number of measurements is much lower than the number of samples. Keeping the number of measurements at one tenth of the total number of samples is a reasonable way to avoid overfitted solutions of this type, which is supported by Raudys and Jain's seminal analysis on this subject (Raudys and Jain, 1991). Whole brain analyses performed on a voxel-wise basis are capable of generating thousands of sample measurements per subject. If those distributed measurements from a single subject were truly independent of each other then the total number of samples will always be very large even for studies with very few subjects therein and thus might be able to help overcome creating overfitted solutions based on too few samples relative to the number of measurements. However, caution is warranted in this situation, as a neurodevelopmental disorder can have a similar effect on multiple voxels, thus providing multiple measurements of the same underlying physiological condition from the same subject. It cannot be guaranteed that such distributed measures actually provide an appropriate distribution of example measurements to facilitate avoiding producing overfitted solutions. Furthermore, ground truth as to the presence of a neurodevelopmental disorder is typically known at a subject level (i.e. patient wide diagnoses) and often not known on a voxel-by-voxel level (which often requires complicated experiments combining histology and coregistration to MRI). Without voxel-by-voxel ground truth data, a validation procedure will be unable to separate distributed samples from a subject's brain MRI into tissue representative of neurodevelopmental abnormalities and tissue that is progressing normally. As such the most reliable approach to validation involves the acquisition of data from a large cohort of independent subjects.
Additionally, it should be noted that one of the main shortcomings of the studies presented in this review is that they are evaluated on a single dataset. Ideally, a system for detecting/diagnosing a given condition validated on a single dataset would be further validated on an independent dataset so as to avoid the potential for producing unrealistically high performance metrics due to the computational learning approach being overly tuned to the nature of the single dataset from which it was provided training data. This would represent an additional type of overfitted solution. It is unknown how substantial this effect is on any given study, providing a substantial amount of uncertainty in the interpretation of the findings presented in this review.
The validation techniques outlined in this study and tabulated in Table 1, Table 2, Table 3, Table 4 are designed to avoid the problem of overfitting in the situation where the researchers only have a single dataset on which to conduct their research. The most common validation technique employed in this paper's reviewed research studies is leave-one-out (LOO) validation. This technique's main strength lies in its ease of applicability, particularly when the dataset being evaluated is relatively small. LOO reserves the largest proportion of data samples for training which is advantageous when very few samples are available. LOO's shortcomings are linked to the same issue; by providing the largest percentage of samples for training, LOO will be more prone than alternative validation techniques to yielding an overfitted solution excessively tuned to the large amount of data the machine learning technique was provided for training. Randomized trials devoting 50% of the dataset to training and 50% to testing would be an ideal validation process for a single dataset study that contains many samples, however, on small datasets this validation approach suffers from widely varying performance metrics in each validation iteration, especially when dealing with a heterogeneous dataset which is common in studies of neurodevelopmental disorders.
While feature selection can be addressed as a class of technology independent of supervised learning, some supervised learning techniques incorporate feature selection into their learning procedure while others do not. Feature selection is particularly important in the characterization, detection and diagnosis of developmental brain disorders because we acquire a multitude of measurements distributed across the brain and we do not know a priori all the regions that are involved in the developmental disorder being investigated. Feature selection can theoretically improve our understanding of brain development and could lead to new technologies to assist in the characterization, detection and diagnosis of a variety of medical conditions as well as to assist in clinical therapeutic assessment.
While predicting a sample as belonging to a particular group based on training data is the most common approach to the use of supervised machine learning algorithms, they can also be adapted to produce a unidimensional measurement of the severity of a given condition. This is a very powerful and underdeveloped area of research that allows the MVA technology to create composite medical images that combine multiple pre-existing images to create useful customized medical images (based on the use of the machine learning technology in regression mode, which is in turn closely related to some multivariate statistical analysis techniques). Examples of customized composite medical images that can be created with this technology include images of tissue stress, images of the likelihood of the presence of disease or images of the likelihood of a given region of tissue exhibiting a physiological condition that could lead to the development of a neurological disorder. An extension of this idea is to create distributed measures of brain maturity based on a collection of measurements related to the extent of myelination, MRI measurements of fractional anisotropy which provides information on the development of neural fiber tracts, etc. Investigating brain maturation with MVA as outlined is particularly challenging as it is known that T1 & T2 imaging presentation inverts between the fetal/neonatal stage and the pediatric stage of development, largely due to myelination in the developing brain (Paus et al., 2001, Pujol et al., 2006, Deoni et al., 2012). Theoretically, MVA techniques should be able to overcome this challenge, however, configuring MVA to do so reliably is a major research challenge. Such a technology would have the potential to identify patients deviating from expected growth trajectories which may assist in the detection, characterization and differential diagnosis of a variety of neurodevelopmental disorders.
The scientific literature has seen enormous growth in developmental imaging of pre-adult populations making use of MVA technologies. However, the vast majority of this work has been focused in pediatric imaging with considerably less focus on neonatal and fetal imaging. Neonatal imaging is more challenging as brain size is considerably smaller than in pediatrics and it is more challenging for technicians to get a neonate to remain still during their imaging examination. Patient movement induces multiple types of imaging artifacts as discussed above, which make studies on neonate populations considerably more challenging. Fetal imaging is the greatest challenge of the three as the brain sizes are the smallest and movement remains a major issue. Furthermore, MRI technology is reliant on the spatial proximity of a radiofrequency (RF) coil (antenna) to the tissue/organ being imaged. Normal brain imaging benefits from a specialized head RF coil that is mounted immediately adjacent to the subject's cranium, however, in fetal imaging this is not possible and so coils located outside the mother's abdomen are necessary, inherently reducing image quality. Additional challenges exist in fetal imaging due to variations in tissue contrast observed in utero relative to later developmental ages. Regardless of the many challenges inherent in the use of MVA in the imaging of fetal, neonatal and pediatric populations, there is considerably large potential for ongoing research growth in this field.
5. Conclusions
Multivariate analysis (MVA) technologies can play a useful role in helping to answer questions about structural and functional organization and development in the brain. Furthermore, MVA techniques have the potential to better characterize a variety of medical conditions than a univariate technique can produce alone. MVA technologies have tremendous potential in the creation of the next generation of clinical diagnostic tests informed by the large amount of information acquired by magnetic resonance imaging (MRI). MVA technologies have exhibited enormous growth in developmental brain MRI in pre-adult populations with a strong emphasis on pediatric imaging. The technologies are very flexible and a wide range of potential applications have already been investigated, however, so many variations on MVA technologies are available in the scientific literature that ample research will need to be performed in order to thoroughly evaluate the tradeoffs imposed by the selection of a given MVA technique applied to a particular neurodevelopmental disorder. Future work will look at improving MVA techniques and adapting them to better characterize, diagnose and detect neurological developmental disorders in pre-adult populations as well as to assist in the identification of clinical variables associated with important aspects of patient outcome and disease etiology.
Acknowledgments
This article was supported financially by the National Institute of Health grants R01HD078561 and R03NS091587 to ET.
References
- Adluru N. Applications of epsilon radial networks in neuroimage analyses. Adv. Image and Video Technol. 2012;7087:236–247. doi: 10.1007/978-3-642-25367-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M.S. Anatomic brain magnetic resonance imaging of the basal ganglia in pediatric bipolar disorder. J. Affect. Disord. 2007;104(1–3):147–154. doi: 10.1016/j.jad.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Alayon S. A fuzzy system for helping medical diagnosis of malformations of cortical development. J. Biomed. Inform. 2007;40(3):221–235. doi: 10.1016/j.jbi.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Holou W.N. Prevalence and natural history of arachnoid cysts in children. J. Neurosurg. Pediatr. 2010;5(6):578–585. doi: 10.3171/2010.2.PEDS09464. [DOI] [PubMed] [Google Scholar]
- Altman N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am. Stat. 1992;46(3):175–185. [Google Scholar]
- Amarreh I. Individual classification of children with epilepsy using support vector machine with multiple indices of diffusion tensor imaging. NeuroImage: Clinical. 2014;4:757–764. doi: 10.1016/j.nicl.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . 5th ed. American Psychiatric Publishing; Arlington, VA: 2013. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- An M. Conference Record of the Forty Fourth Asilomar Conference on Signals, Systems and Computers. 2010. Multimodal MRI analysis of brain subnetworks in autism using multi-view EM; pp. 786–789. [Google Scholar]
- Anderson A. Non-negative matrix factorization of multimodal MRI, fMRI and phenotypic data reveals differential changes in default mode subnetworks in ADHD. NeuroImage. 2014;102(1):207–219. doi: 10.1016/j.neuroimage.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffero G.M. Prenatal ultrasound predictors of postnatal major cerebral abnormalities in fetuses with apparently isolated mild ventriculomegaly. Prenat. Diagn. 2015 doi: 10.1002/pd.4607. [DOI] [PubMed] [Google Scholar]
- Ball G.H., Hall D.J. A Novel Method of DATA Analysis and Classification. Stanford Research Inst; 1965. ISODATA. [Google Scholar]
- Batchelor P.G. Measures of folding applied to the development of the human fetal brain. IEEE Trans. Med. Imaging. 2002;21(8):953–965. doi: 10.1109/TMI.2002.803108. [DOI] [PubMed] [Google Scholar]
- Bray S. Applications of multivariate pattern classification analyses in developmental neuroimaging of healthy and clinical populations. Front. Hum. Neurosci. 2009;3:32. doi: 10.3389/neuro.09.032.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Bunge S.A. Immature frontal lobe contributions to cognitive control in children: evidence from fMRI. Neuron. 2002;33(2):301–311. doi: 10.1016/s0896-6273(01)00583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderoni S. Female children with autism spectrum disorder: an insight from mass-univariate and pattern classification analyses. NeuroImage. 2012;59(2):1013–1022. doi: 10.1016/j.neuroimage.2011.08.070. [DOI] [PubMed] [Google Scholar]
- Callen D.J.A. MRI in the diagnosis of pediatric multiple sclerosis. Neurology. 2009;72(11):961–967. doi: 10.1212/01.wnl.0000338629.01627.54. [DOI] [PubMed] [Google Scholar]
- Carbonell J.G. Machine Learning, an Artificial Intelligence Approach. Tioga Press; 1983. An overview of machine learning. [Google Scholar]
- Casey B.J. A developmental functional MRI study of prefrontal activation during performance of a Go-No-Go task. J. Cogn. Neurosci. 1997;9(6):835–847. doi: 10.1162/jocn.1997.9.6.835. [DOI] [PubMed] [Google Scholar]
- Cates J. Proceedings of Medical Image Computing and Computer Assisted Intervention. 11(Pt 1) 2008. Particle-based shape analysis of multi-object complexes; pp. 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-W. ADHD classification by a texture analysis of anatomical brain MRI data. Front. Syst. Neurosci. 2012;6:66. doi: 10.3389/fnsys.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. Neuropsychiatric Disorders and Pediatric Psychiatry. Vol. 69. 2011. Structural MRI in Autism Spectrum Disorder; pp. 63R–68R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.P. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage: Clinical. 2015;8:238–245. doi: 10.1016/j.nicl.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—a multi-center study. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2016;64:1–9. doi: 10.1016/j.pnpbp.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Cheng W. Individual classification of ADHD patients by integrating multiscale neuroimaging markers and advanced pattern recognition techniques. Front. Syst. Neurosci. 2012;6:58. doi: 10.3389/fnsys.2012.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Shin M.S. Neural network based automatic diagnosis of children with brain dysfunction. Int. J. Neural Syst. 2001;11(4):361–369. doi: 10.1142/S0129065701000825. [DOI] [PubMed] [Google Scholar]
- Colby J.B. Insights into multimodal imaging classification of ADHD. Front. Syst. Neurosci. 2012;6:59. doi: 10.3389/fnsys.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa A. Use of machine learning to identify children with autism and their motor abnormalitiesJ. Autism Dev. Disord. 2015;45(7):2146–2156. doi: 10.1007/s10803-015-2379-8. [DOI] [PubMed] [Google Scholar]
- Dai D. Classification of ADHD children through multimodal magnetic resonance imaging. Front. Syst. Neurosci. 2012;6:63. doi: 10.3389/fnsys.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley M. Hippocampal volume in childhood complex partial seizures. Epilepsy Res. 2006;72(1):57–66. doi: 10.1016/j.eplepsyres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- de Arruda G.F. Structure and dynamics of functional networks in child-onset schizophrenia. Clin. Neurophysiol. 2014;125(8):1589–1595. doi: 10.1016/j.clinph.2013.11.036. [DOI] [PubMed] [Google Scholar]
- Deoni S.C.L. Investigating white matter development in infancy and early childhood using myelin water faction and relaxation time mapping. NeuroImage. 2012;63(3):1038–1053. doi: 10.1016/j.neuroimage.2012.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G. Recursive cluster elimination based support vector machine for disease state prediction using resting state functional and effective brain connectivity. PLoS One. 2010;5(12):e14277. doi: 10.1371/journal.pone.0014277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G. Identification of neural connectivity signatures of autism using machine learning. Front. Hum. Neurosci. 2013;7:670. doi: 10.3389/fnhum.2013.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroye L., Gyorfi L., Lugosi G. Springer; 1996. A Probabilistic Theory of Pattern Recognition. (ISBN 0-3879-4618-7) [Google Scholar]
- Duarte J.V. Multivariate pattern analysis reveals subtle brain abnormalities relevant to the cognitive phenotype in neurofibromatosis type 1. Hum. Brain Mapp. 2014;35(1):89–106. doi: 10.1002/hbm.22161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunteman G.H. SAGE Publications Inc.; 1989. Principal Components Analysis. [Google Scholar]
- Ecker C. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. NeuroImage. 2010;49(1):44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Efron B., Tibshirani R.J. An Introduction to the Bootstrap. Chapman & Hall. 1994 [Google Scholar]
- Eloyan A. Automated diagnoses of attention deficit hyperactivity disorder using magnetic resonance imaging. Front. Syst. Neurosci. 2012;6:61. doi: 10.3389/fnsys.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D.A. Functional brain networks develop from a “local to distributed” organization. PLoS Comput. Biol. 2009;5(5):e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D.A. Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Front. Syst. Neurosci. 2012;6:80. doi: 10.3389/fnsys.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah R. Altered white matter microstructure underlies listening difficulties in children suspected of auditory processing disorders: a DTI study. Brain and Behav. 2014;4(4):531–543. doi: 10.1002/brb3.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.S. A practical definition of epilepsy. Epilepsia. 2014;55(4):475–482. doi: 10.1111/epi.12550. [DOI] [PubMed] [Google Scholar]
- Fox M.D. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier J.A. Structural brain magnetic resonance imaging of limbic and thalamic volumes in pediatric bipolar disorder. Am. J. Psychiatry. 2005;162(7):1256–1265. doi: 10.1176/appi.ajp.162.7.1256. [DOI] [PubMed] [Google Scholar]
- Froehlich C. Proceedings of the XXIV Brazilian Congress on, Biomedical Engineering. 2014. Classifying brain states for cognitive tasks: a functional MRI study in children with reading impairments; pp. 2476–2479. [Google Scholar]
- Fukuhara T., Luciano M.G. Clinical features of late-onset idiopathic aqueductal stenosis. Surg. Neurol. 2001;55(3):132–136. doi: 10.1016/s0090-3019(01)00359-7. [DOI] [PubMed] [Google Scholar]
- Galka A. Decomposition of neurological multivariate time series by state space modelling. Bull. Math. Biol. 2011;73(2):285–324. doi: 10.1007/s11538-010-9563-y. [DOI] [PubMed] [Google Scholar]
- Ghiassian S. Proceedings of 3rd NIPS Workshop on Machine Learning and Interpretation in NeuroImaging. 2013. Learning to classify psychiatric disorders based on fMR images: autism vs. healthy and ADHD vs. healthy. [Google Scholar]
- Gogtay N. Dynamic mapping of human cortical development during childhood through early adulthood. Proc. Natl. Acad. Sci. 2004;101(21):8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D. Using multivariate machine learning methods and structural MRI to classify childhood onset schizophrenia and healthy controls. Front. Psychiatry. 2012;3:53. doi: 10.3389/fpsyt.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths P.D. Anatomic localization of dyskinesia in children with “profound” perinatal hypoxic ischemic injury. Am. J. Neuroradiol. 2010;31:436–441. doi: 10.3174/ajnr.A1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J.S. Factor analysis of neuroanatomical and clinical characteristics of holoprosencephaly. Brain Dev. 2006;28(7):413–419. doi: 10.1016/j.braindev.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Harman H.H. University of Chicago Press; 1960. Modern Factor Analysis. [Google Scholar]
- Hart H. Pattern classification of response inhibition in ADHD: toward the development of neurobiological markers for ADHD. Hum. Brain Mapp. 2014;35(7):3083–3094. doi: 10.1002/hbm.22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart H. Predictive neurofunctional markers of attention-deficit/hyperactivity disorder based on pattern classification of temporal processing. J. Am. Acad. Child Adolesc. Psychiatry. 2014;53(5):569–578. doi: 10.1016/j.jaac.2013.12.024. (e1) [DOI] [PubMed] [Google Scholar]
- Hausler M. Inflammatory pseudotumors of the central nervous system: report of 3 cases and a literature review. Hum. Pathol. 2003;34(3):253–262. doi: 10.1053/hupa.2003.35. [DOI] [PubMed] [Google Scholar]
- Hoeft F. Morphometric spatial patterns differentiating boys with fragile X syndrome, typically developing boys, and developmentally delayed boys aged 1 to 3 years. J. Am. Med. Assoc. Psychiatry. 2008;65(9):1087–1097. doi: 10.1001/archpsyc.65.9.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F. Neural systems predicting long-term outcome in dyslexia. Proc. Natl. Acad. Sci. 2011;108(1):361–366. doi: 10.1073/pnas.1008950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekzema E. Regional volumes and spatial volumetric distribution of gray matter in the gender dysphoric brain. Psychoneuroendocrinology. 2015;55:59–71. doi: 10.1016/j.psyneuen.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Huang H.K. Image-matching as a medical diagnostic support tool (DST) for brain diseases in children. Comput. Med. Imaging Graph. 2005;29(2–3):195–202. doi: 10.1016/j.compmedimag.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Huang H. White and gray matter development in human fetal, newborn and pediatric brains. NeuroImage. 2006;33(1):27–38. doi: 10.1016/j.neuroimage.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A. John Wiley & Sons Inc.; 2004. Independent Component Analysis. [Google Scholar]
- Iannaccone R. European Child and Adolescent Psychiatry; 2015. Classifying adolescent attention-deficit/hyperactivity disorder (ADHD) based on functional and structural imaging. [DOI] [PubMed] [Google Scholar]
- Igual L. A fully-automatic caudate nucleus segmentation of brain MRI: application in volumetric analysis of pediatric attention-deficit/hyperactivity disorder. BioMedical Eng. OnLine. 2011;10:105. doi: 10.1186/1475-925X-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual L. Automatic brain caudate nuclei segmentation and classification in diagnostic of Attention-Deficit/Hyperactivity Disorder. Comput. Med. Imaging Graph. 2012;36(8):591–600. doi: 10.1016/j.compmedimag.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Igual L. Automatic internal segmentation of caudate nucleus for diagnosis of attention-deficit/hyperactivity disorder. Image Analysis and Recognition Lecture Notes on Computer Science. 2012;7325:222–229. [Google Scholar]
- Igual L. International Conference on High Performance Computing and, Simulation. 2012. Supervised brain segmentation and classification in diagnostic of Attention-Deficit/Hyperactivity Disorder; pp. 182–187. [Google Scholar]
- Ingalhalikar M. Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. NeuroImage. 2011;57(3):918–927. doi: 10.1016/j.neuroimage.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab A. Disrupted developmental organization of the structural connectome in fetuses with corpus callosum agenesis. NeuroImage. 2015;111(1):277–288. doi: 10.1016/j.neuroimage.2015.02.038. [DOI] [PubMed] [Google Scholar]
- Jiao Y. Predictive models of autism spectrum disorder based on brain regional cortical thickness. NeuroImage. 2010;50(2):589–599. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. Predictive models for subtypes of autism spectrum disorder based on single-nucleotide polymorphisms and magnetic resonance imaging. Adv. Med. Sci. 2011;56(2):334–342. doi: 10.2478/v10039-011-0042-y. [DOI] [PubMed] [Google Scholar]
- Juhasz C. White matter volume as a major predictor of cognitive function in Sturge–Weber syndrome. Arch. Neurol. 2007;64(8):1169–1174. doi: 10.1001/archneur.64.8.1169. [DOI] [PubMed] [Google Scholar]
- Juranek J. Association between amygdala volume and anxiety level: magnetic resonance imaging (MRI) study in autistic children. J. Child Neurol. 2006;21(12):1051–1058. doi: 10.1177/7010.2006.00237. [DOI] [PubMed] [Google Scholar]
- Kawakami N. World Automation Congress. 2010. A study on image features using intensity profile for cortical dysplasia degree estimation; pp. 1–6. [Google Scholar]
- Kearns M., Ron D. Algorithmic stability and sanity-check bounds for leave-one-out cross-validation. Neural Comput. 1999;11(6):1427–1453. doi: 10.1162/089976699300016304. [DOI] [PubMed] [Google Scholar]
- Kobashi S. Proceedings of the International Conference on Image Processing, Computer Vision, and Pattern Recognition. 2011. Fractal dimension based cortical dysplasia detection using MR images for children with epilepsy. [Google Scholar]
- Kohavi R. Proceedings International Joint Conference on Artificial Intelligence (IJCAI) 1995. A study of cross-validation and bootstrap for accuracy estimation and model selection. [Google Scholar]
- Krsek P. Predictors of seizure-free outcome after epilepsy surgery for pediatric tuberous sclerosis complex. Epilepsia. 2013;54(11):1913–1921. doi: 10.1111/epi.12371. [DOI] [PubMed] [Google Scholar]
- Lange H. Atypical diffusion tensor hemispheric asymmetry in autism. Autism Res. 2010;3(6):350–358. doi: 10.1002/aur.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levman J. Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. IEEE Trans. Med. Imaging. 2008;27(5):688–696. doi: 10.1109/TMI.2008.916959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Multivariate pattern analysis of DTI reveals differential white matter in individuals with obsessive–compulsive disorder. Hum. Brain Mapp. 2014;35(6):2643–2651. doi: 10.1002/hbm.22357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.-F. International Conference on Fuzzy Theory and it's Applications. 2012. Differentiation between resting-state fMRI data from ADHD and normal subjects: based on functional connectivity and machine learning; pp. 294–298. [Google Scholar]
- Lim L. Disorder-specific predictive classification of adolescents with attention deficit hyperactivity disorder (ADHD) relative to autism using structural magnetic resonance imaging. PLoS One. 2013;8(5):e63660. doi: 10.1371/journal.pone.0063660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M.V. Different functional neural substrates for good and poor language outcome in autism. Neuron. 2015;86(2):567–577. doi: 10.1016/j.neuron.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyek C. Bildverarbeitung für die Medizin, Informatik aktuell. Vol. 432. 2008. Detection of focal cortical dysplasia lesions in MRI using textural features. [Google Scholar]
- Mahanand B.S. AI 2013: Advances in Artificial Intelligence Lecture Notes in Computer Science. Vol. 8272. 2013. Computer aided diagnosis of ADHD using brain magnetic resonance images; pp. 386–395. [Google Scholar]
- Mandell J.G. Volumetric brain analysis in neurosurgery: part 2. Brain and CSF volumes discriminate neurocognitive outcomes in hydrocephalus. J. Neurosurg. Pediatr. 2015;15(2):125–132. doi: 10.3171/2014.9.PEDS12427. [DOI] [PubMed] [Google Scholar]
- Manton K.G. Wiley StatsRef: Statistics Reference Online; 2014. Cluster Analysis: Overview. [Google Scholar]
- Marzelli M.J. Neuroanatomical spatial patterns in Turner syndrome. NeuroImage. 2011;55(2):439–447. doi: 10.1016/j.neuroimage.2010.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh A.R. Spatial pattern analysis of functional brain images using partial least squares. NeuroImage. 1996;3:143–157. doi: 10.1006/nimg.1996.0016. [DOI] [PubMed] [Google Scholar]
- McLachlan G.J. Wiley Interscience; 2004. Discriminant Analysis and Statistical Pattern Recognition. [Google Scholar]
- Mesulam M.M. A cortical network for directed attention and unilateral neglect. Ann. Neurol. 1981;10:309–325. doi: 10.1002/ana.410100402. [DOI] [PubMed] [Google Scholar]
- Moosa A.N. Long-term functional outcomes and their predictors after hemispherectomy in 115 children. Epilepsia. 2013;54(10):1771–1779. doi: 10.1111/epi.12342. [DOI] [PubMed] [Google Scholar]
- Muzik O. Statistical parametric mapping: assessment of application in children. NeuroImage. 2000;12(5):538–549. doi: 10.1006/nimg.2000.0651. [DOI] [PubMed] [Google Scholar]
- Mwangi B. Prediction of pediatric bipolar disorder using neuroanatomical signatures of the amygdala. Bipolar Disord. 2014;16(7):713–721. doi: 10.1111/bdi.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelder J., Wedderburn R. Generalized linear models. J. R. Stat. Soc. Ser. A. 1972;135(3):370–384. [Google Scholar]
- Nielsen J.A. Multisite functional connectivity MRI classification of autism: ABIDE results. Front. Hum. Neurosci. 2013;7:599. doi: 10.3389/fnhum.2013.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paldino M.J. Independent contribution of individual white matter pathways to language function in pediatric epilepsy patients. NeuroImage: Clinical. 2014;6:327–332. doi: 10.1016/j.nicl.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh N.A. Perinatal factors and regional brain volume abnormalities at term in a cohort of extremely low birth weight infants. PLoS One. 2013;8(5):e62804. doi: 10.1371/journal.pone.0062804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T. Maturation of white matter in the human brain: a review of magnetic resonance studies. Brain Res. Bull. 2001;54(3):255–266. doi: 10.1016/s0361-9230(00)00434-2. [DOI] [PubMed] [Google Scholar]
- Peche S.S. A long-term follow-up study using IPMSSG criteria in children with CNS demyelination. Pediatr. Neurol. 2013;49(5):329–334. doi: 10.1016/j.pediatrneurol.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Peng X. Extreme learning machine-based classification of ADHD using brain structural MRI data. PLoS One. 2013;8(11):e79476. doi: 10.1371/journal.pone.0079476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzo D. Proceedings of Medical Image Computing and Computer Assisted Intervention. Vol. 8674. 2014. Detection of corpus callosum malformations in pediatric population using the discriminative direction in multiple kernel learning; pp. 300–307. [DOI] [PubMed] [Google Scholar]
- Pina-Camacho L. Predictors of schizophrenia spectrum disorders in early-onset first episodes of psychosis: a support vector machine model. Eur. Child Adolesc. Psychiatry. 2015;24(4):427–440. doi: 10.1007/s00787-014-0593-0. [DOI] [PubMed] [Google Scholar]
- Plitt M. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical. 2015;7:359–366. doi: 10.1016/j.nicl.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol J. Myelination of language-related areas in the developing brain. Neurology. 2006;66(3):339–343. doi: 10.1212/01.wnl.0000201049.66073.8d. [DOI] [PubMed] [Google Scholar]
- Rapoport J.L. Progressive cortical change during adolescence in childhood-onset schizophrenia. A longitudinal magnetic resonance imaging study. Arch. Gen. Psychiatry. 1999;56(7):649–654. doi: 10.1001/archpsyc.56.7.649. [DOI] [PubMed] [Google Scholar]
- Raudys S.J., Jain A.K. Small sample size effects in statistical pattern recognition: recommendations for practitioners. IEEE Trans. Pattern Anal. Mach. Intell. 1991;13(3):252–264. [Google Scholar]
- Reiss A.L. Brain development, gender and IQ in children. A volumetric imaging study. Brain. 1996;119(Pt. 5):1763–1774. doi: 10.1093/brain/119.5.1763. [DOI] [PubMed] [Google Scholar]
- Rencher A.C., Christensen W.F. John Wiley & Sons Inc.; 2012. Methods of Multivariate Analysis. [Google Scholar]
- Sanz-Cortes M. Automatic quantitative MRI texture analysis in small-for-gestational-age fetuses discriminates abnormal neonatal neurobehavior. PLoS One. 2013;8(7):e69595. doi: 10.1371/journal.pone.0069595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato J.R. Evaluation of pattern recognition and feature extraction methods in ADHD prediction. Front. Syst. Neurosci. 2012;6:68. doi: 10.3389/fnsys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmithorst V.J. Left ear advantage in speed-related dichotic listening is not specific to auditory processing disorder in children: a machine-learning fMRI and DTI study. NeuroImage: Clinical. 2013;3:8–17. doi: 10.1016/j.nicl.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z. A multivariate distance-based analytic framework for connectome-wide association studies. NeuroImage. 2014;93(1):74–94. doi: 10.1016/j.neuroimage.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. A multivariate surface-based analysis of the putamen in premature newborns: regional differences within the ventral striatum. PLoS One. 2013;8(7):e66736. doi: 10.1371/journal.pone.0066736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniatchkin M. Spatial filters and automated spike detection based on brain topographies improve sensitivity of EEG-fMRI studies in focal epilepsy. NeuroImage. 2007;37(3):834–843. doi: 10.1016/j.neuroimage.2007.05.049. [DOI] [PubMed] [Google Scholar]
- Smith H.F. A multivariate analysis of covariance. Biometrics. 1958;14:107–127. [Google Scholar]
- Subramanian K. A meta-cognitive interval type-2 fuzzy inference system and its projection based learning algorithm. Evol. Syst. 2014;5(4):219–230. [Google Scholar]
- Supekar K. Development of large-scale functional brain networks in children. PLoS Biol. 2009;7(7):e1000157. doi: 10.1371/journal.pbio.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeszko P.R. Brain structural abnormalities in psychotropic drug-naïve pediatric patients with obsessive–compulsive disorder. Am. J. Psychiatry. 2004;161(6):1049–1056. doi: 10.1176/appi.ajp.161.6.1049. [DOI] [PubMed] [Google Scholar]
- Talairach J., Tournoux P. Thieme Medical Publishers; New York: 1988. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System — An Approach to Cerebral Imaging. [Google Scholar]
- Tang J. Aberrant default mode functional connectivity in early onset schizophrenia. PLoS One. 2013;8(7):e71061. doi: 10.1371/journal.pone.0071061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K.M. Amygdala response to facial expressions in children and adults. Biol. Psychiatry. 2001;49(4):309–316. doi: 10.1016/s0006-3223(00)01066-0. [DOI] [PubMed] [Google Scholar]
- Uddin L.Q. Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biol. Psychiatry. 2011;70(9):833–841. doi: 10.1016/j.biopsych.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaadia E. Dynamics of neuronal interactions in monkey cortex in relation to behavioural events. Nature. 1995;373:515–518. doi: 10.1038/373515a0. [DOI] [PubMed] [Google Scholar]
- Vapnik V.N. Springer; 1995. The Nature of Statistical Learning Theory. [Google Scholar]
- Wang X. Eighth International Conference on Fuzzy Systems and Knowledge Discovery. 2011. Discriminative analysis of resting-state brain functional connectivity patterns of Attention-Deficit Hyperactivity Disorder using Kernel Principal Components Analysis; pp. 1938–1941. [Google Scholar]
- Wang J. Classification of fMRI patterns—a study of the language network segregation in pediatric localization related epilepsy. Hum. Brain Mapp. 2014;35(4):1446–1460. doi: 10.1002/hbm.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne R.T. A primer on multivariate analysis of variance (MANOVA) for behavioral scientists. Pract. Assessment, Res. Eval. 2014;19(17):1–10. [Google Scholar]
- Wee C.-Y. Diagnosis of autism spectrum disorders using regional and interregional morphological features. Hum. Brain Mapp. 2014;35(7):3414–3430. doi: 10.1002/hbm.22411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-J. Prediction of pediatric unipolar depression using multiple neuromorphometric measurements: a pattern classification approach. J. Psychiatr. Res. 2015;62:84–91. doi: 10.1016/j.jpsychires.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnanarayana B. PHI Learning Pvt. Ltd.; 2009. Artificial Neural Networks. [Google Scholar]
- You X. Proceedings of the International Conference on Image Processing, Computer Vision, & Pattern Recognition. 2009. Application of nonlinear classifiers with the principal component analysis in fMRI language activation pattern recognition in a multisite study for pediatric epilepsy. [Google Scholar]
- Youngstrom E.A. A primer on receiver operating characteristic curve analysis and diagnostic efficiency statistics for pediatric psychology: we are ready to ROC. J. Pediatr. Psychol. 2014;39(2):204–221. doi: 10.1093/jpepsy/jst062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. Multiparametric MRI characterization and prediction in autism spectrum disorder using graph theory and machine learning. PLoS One. 2014;9(6):e90405. doi: 10.1371/journal.pone.0090405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.Z. Medical Image Computing and Computer Assisted Intervention Lecture Notes in Computer Science. Vol. 3750. 2005. Discriminative analysis of brain function at resting-state for attention-deficit/hyperactivity disorder; pp. 468–475. [DOI] [PubMed] [Google Scholar]
- Zhu C.-Z. Fisher discriminative analysis of resting-state brain function for attention-deficit/hyperactivity disorder. NeuroImage. 2008;40(1):110–120. doi: 10.1016/j.neuroimage.2007.11.029. [DOI] [PubMed] [Google Scholar]