Abstract
Dead-end ultrafiltration (DEUF) has been reported to be a simple, field-deployable technique for recovering bacteria, viruses, and parasites from large-volume water samples for water quality testing and waterborne disease investigations. While DEUF has been reported for application to water samples having relatively low turbidity, little information is available regarding recovery efficiencies for this technique when applied to sampling turbid water samples such as those commonly found in lakes and rivers. This study evaluated the effectiveness of a DEUF technique for recoveringMS2 bacteriophage, enterococci, Escherichia coli, Clostridium perfringens, and Cryptosporidium parvum oocysts in surface water samples having elevated turbidity. Average recovery efficiencies for each study microbe across all turbidity ranges were: MS2 (66%), C. parvum (49%), enterococci (85%), E. coli (81%), and C. perfringens (63%). The recovery efficiencies for MS2 and C. perfringens exhibited an inversely proportional relationship with turbidity, however no significant differences in recovery were observed for C. parvum, enterococci, or E. coli. Although ultrafilter clogging was observed, the DEUF method was able to process 100-L surface water samples at each turbidity level within 60 min. This study supports the use of the DEUF method for recovering a wide array of microbes in large-volume surface water samples having medium to high turbidity.
Keywords: Dead-end ultrafiltration, Microbial recovery, Turbid water
1. Introduction
Hollow-fiber ultrafilters have become increasingly accepted as an efficient and cost-effective tool for recovering microbes and viruses from water (Knappett et al., 2011; Leskinen et al., 2010; Lindquist et al., 2007; Rhodes et al., 2011; Shapiro et al., 2010). Because of the small pore size of ultrafilter cartridges (~29–70 kDa), they can be used to recover a diverse array of microorganisms, including bacteria (Mull and Hill, 2009), viruses (Hill et al., 2007, 2010a, 2010b), and parasites (Hill et al., 2009). Commercially-available dialysis filters can currently be purchased for 16–30 USD per cartridge, making them a cost-effective alternative to other large-volume water sampling filters. The use of hollow-fiber ultrafilters is unique among water sampling techniques in that ultrafiltration (UF) enables simultaneous recovery of these diverse microbe types based on size exclusion rather than mechanisms such as adsorption–elution (Polaczyk et al., 2007). The first popularized UF methods were based on tangential-flow ultrafiltration (TFUF), in which a water sample in a container is pumped tangentially across ultrafilter membranes as it is recirculated through the ultrafilter cartridge under pressure (Oshima et al., 1995; Hill et al., 2005; Lindquist et al., 2007). The TFUF procedure is performed until the volume of the water sample in the container is reduced to a target volume; all water sample constituents larger than the pore-size of the ultrafilter are recovered in a “retentate” sample and the filters are often backflushed or eluted to recover additional microbes from the ultrafilter membranes.
Recently, dead-end ultrafiltration (DEUF) was reported as a simpler, field-deployable alternative to TFUF for recovering microbes from large-volume water samples when using hollow-fiber ultrafilters (Smith and Hill, 2009). The DEUF technique differs from TFUF in that one of the ultrafilter end ports is plugged so that the water sample cannot exit the ultrafilter through the end cap, but instead must pass perpendicularly through the hollow fiber membranes. Thus, particles and other water sample constituents larger than the pores in the ultrafilter membranes are trapped within the ultrafilter cartridge, as opposed to TFUF techniques which result in the production of a concentrated “retentate” sample (Hill et al., 2005). The DEUF method is simpler to perform than TFUF because the water sample passes through the ultrafilter cartridge only once, as occurs with other single-pass filtration methods such as US EPA Method 1623 filtration for parasite (oo) cysts using Envirochek® filters (US EPA, 2005c) or virus adsorption–elution filtration using NanoCeram® filters (Karim et al., 2009). When DEUF is completed, the ultrafilter cartridge can be capped and the ultrafilter shipped to a laboratory for recovery of microbes using either backflushing or elution. TFUF is less easily performed in the field, as it requires the water sample to be present in a container from which the sample is pumped for recirculation through the ultrafilter. For large-volume water samples (e.g., 100-L), multiple containers must be filled for processing using the TFUF method, whereas water sampled by the DEUF method can be pumped directly from the water source through the ultrafilter (or, if sampling a pressurized system, the system pressure can be used to force water through the ultrafilter without need for a pump).
While DEUF was reported by Smith and Hill (2009) to be effective for recovering diverse microbes in 100-L water samples having relatively low turbidity, few studies have investigated application of this technique to sampling turbid water samples (e.g., lake or river water). As the DEUF method employs a single-pass, perpendicular flow path through the ultrafilter, the method is subject to greater potential clogging than TFUF because during TFUF the horizontal flow of water over the membrane causes a scouring effect that helps to reduce the tendency for microbes and other particles to plug the ultrafilter membrane pores as water passes through the membranes (Kim and DiGiano, 2009). Thus, the main inherent issue associated with the use of ultrafilters is that non-target water constituents (e.g., soil particles, colloids, long-chain dissolved organics) are co-concentrated with microbes and viruses and may clog UF membranes at a faster rate than comparable filters with larger pore sizes (i.e., microfilters), and may also contain constituents that inhibit analytical procedures, especially molecular testing procedures such as PCR. The objective of this study was to expand upon the research reported by Smith and Hill (2009) by evaluating the DEUF method for recovering the same suite of microbes from surface water samples covering a range of turbidity values, and applying molecular assays to detect target analytes in DEUF concentrates. The parameters tested included the effect of turbidity on the filtrate rate, system pressure, and microbial recovery efficiencies. Water samples were assayed for a suite of five microbes (MS2 bacteriophage, enterococci, Escherichia coli, Clostridium perfringens, and Cryptosporidium parvum oocysts) to determine the recovery performance of the DEUF method.
2. Materials and methods
2.1. Water samples
Water samples were collected in Cubitainers® from Murphey Candler Park Lake in Atlanta, GA. To obtain water samples over a range of turbidity values [low (<20 NTU), middle (50 NTU), and high (100 NTU)], sampling days were chosen after rainfall events. All water samples were stored at 4 °C and used within two weeks of collection. The day before each experiment 40 L of water was removed from refrigeration and allowed to return to room temperature overnight. In order to evaluate potential water quality effects on the DEUF procedure, the physico-chemical quality of all water samples was characterized the day of the experiment using the following water quality parameters: temperature, pH, turbidity, specific conductance (SC), total hardness, total organic carbon (TOC), and total suspended solids (TSS). Temperature, pH, turbidity, SC, and TOC testing was performed as described previously (Hill et al., 2007). Total hardness was measured using the Hach total hardness test method 8213 and AL-DT digital titrator. TSS was determined using Standard Methods for the Examination of Water and Wastewater (Eaton et al., 2005).
2.2. Hydraulic performance of DEUF
Hydraulic characteristics of the DEUF method (filtrate rate and system pressure) were investigated to evaluate filter performance when processing water samples at three turbidity levels. Pressure changes were monitored with a 30 lb/in2 (206 kPa) oil-filled pressure gauge. Filtrate rates were measured manually with a graduated cylinder and clock. Water was pumped using a peristaltic pump at a nominal flow rate of 2900 mL/min; the corresponding filtrate rate and pressure were recorded every 5 min. Three replicate experiments were performed for the low turbidity range and four replicate for the two higher turbidity ranges.
2.3. Microbial recovery with DEUF
Recovery efficiencies for a suite of microbes were assessed using REXEED 25SX dialysis filters (Asahi Kasei, Japan) and low, middle, and high turbidity 100-L surface water samples. A 100-mL water quality sample and a 750-mL control sample were collected prior to filtration. The suite of analytes used for microbial recovery experiments consisted of MS2 bacteriophage (ATCC 15597-B1), C. parvum oocysts (Lot#15-07 Waterborne, Inc), C. perfringens, enterococci, and E. coli. There were sufficient concentrations of naturally occurring C. perfringens, enterococci, and E. coli in the surface water that additional seeding was not required. Background concentrations of MS2 and C. parvum were not detected.
Prior to each experiment, an MS2 bacteriophage culture stock containing 2.6 × 109 plaque forming units (PFU)/mL was diluted in amended PBS [0.01 M phosphate-buffered saline (Dulbecco'smodification; pH7.40) containing 0.01% (vol/vol) Tween 80 (Fisher), and 0.001% Y-30 antifoam emulsion (Sigma)], and then filtered using a 0.1-µmfilter to reduce the presence of microbe aggregates in the stock used to seed water samples. C. parvum oocysts at an initial concentration of 1 × 108 in 8 mL were diluted in deionized water and heat inactivated (30 min at 50 °C). The same quantity of microbes was seeded into both the 100-L input water sample and the 750-mL control sample. Seed levels were 1.2 × 105 to 4.7 × 105 PFU for MS2 and 4.1 × 105 to 8.8 × 105 C. parvum oocysts. Enterococci, C. perfringens, and E. coli were present at levels of 6.8 × 104 to 1.1 × 106 colony forming units (CFU), 7.6 × 104 to 1.3 × 106 CFU, and 1.8 × 104 to 1.4 × 106 CFU, respectively, in 100-L lake water samples used in this study.
The filtration unit setup is shown in Fig. 1 as previously described (Smith and Hill, 2009). Briefly, 100 L of water was pumped through the hollow-fiber ultrafilter and microbes captured within the ultrafilter cartridge. The filter was then backflushed with a 500-mL solution containing 0.5% Tween 80, 0.01% sodium polyphosphate (Sigma-Aldrich # 305553), and 0.001% Antifoam Y-30 Emulsion. The average final DEUF concentrate volumes were 532 ± 41 mL.
Fig. 1.
Schematic of dead-end ultrafiltration set-up.
Secondary processing of the DEUF concentrate and control samples was completed the same day as the DEUF procedure. The samples were assayed by immunofluorescence assay (FA) microscopy examination per EPA method 1623 for C. parvum (US EPA, 2005c) but the seed levels were high enough such that centrifugation and immunomagnetic separation (IMS) were not needed. MS2 was assayed by single-agar-layer plaque assay using EPA method 1602 with an E. coli F-amp host (ATCC700891) (US EPA, 2001). Enterococci were assayed by membrane filtration and mEI agar culture using EPA method 1600 (US EPA, 2005a). C. perfringens was assayed by membrane filtration and mCP agar culture (Bisson and Cabelli, 1979). E. coli was assayed by membrane filtration and modified mTEC agar culture using EPA method 1603 (US EPA, 2005b).
Three replicate experiments were performed using low range turbidity water, and four replicate experiments were performed for both the middle range and high range water samples. Percent recovery efficiencies were calculated by dividing the total number of each microbe measured in a DEUF concentrate sample by the total number of each microbe measured in the input sample for the experiment and multiplying the fraction by 100. One-way analysis of variance was used to test for significant differences between the water quality parameters and mean recovery efficiencies for each microbe at each turbidity range. Student's t-test was then used to identify significant differences in recovery efficiencies at different turbidity levels. The significant level α was set at 0.05 for all statistical tests.
2.4. Application of molecular testing to DEUF procedure
Three replicate experiments were performed to detect MS2 and C. parvum in low range turbidity water using real-time RT-PCR and PCR, respectively, following DEUF and secondary concentration procedures. MS2 and C. parvum oocysts were seeded into 100-L water samples for these experiments. Seed levels were 3.09 × 105 MS2 PFU and 1 × 103 C. parvum oocysts. Following DEUF, the concentrate sample was further concentrated by polyethylene glycol (PEG) precipitation (12% PEG 8000, 0.9 M NaCl, and 1% bovine serum albumin) for 2 h at 4 °C (Polaczyk et al., 2008). PEG-precipitated samples were centrifuged at 10,000 ×g for 30 min (4 °C) and the pelleted material was resuspended with amended PBS, resulting in average PEG concentrates of 10.3 ± 2.1 mL. Nucleic acid extraction was performed on 1.5 mL of the PEG concentrate by a previously reported procedure using a lysis buffer containing 4.5 M guanidinium isothiocyanate and bead beating using acid-washed ZrOx beads (Hill et al., 2007). The remaining PEG concentrate was processed to recover C. parvum oocysts by IMS. Following IMS, half of the sample was examined by FA microscopy per EPA method 1623 for C. parvum (US EPA, 2005c) and nucleic acid was extracted from the other half. Previously published real-time RT-PCR and PCR assays were used for molecular detection of MS2 and C. parvum, respectively (Hill et al., 2007). All samples were tested in duplicate on an iCycler iQ5 Real-time PCR Detection System (Bio-Rad, Hercules, CA). Reactions were carried out in a 50-µL final reaction mixture using TaqMan Environmental Master Mix 2.0 for PCR and TaqMan Fast Virus 1-step Master Mix for RT-PCR (Applied Biosystems, Foster City, CA), 250 nM of forward and reverse primers, 100 nM of FAM-labeled probes, 2.5 µL of a 20× mixture of non-acetylated BSA (Sigma-Aldrich, St. Louis, MO), and T4 gene 32 protein (New England Biolabs, Ipswich, MA). Samples were analyzed by PCR and RT-PCR using 10-µL and 2-µL DNA/RNA template volumes to account for potential assay inhibition. Real-time PCR cycling conditions were: denaturation at 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s, then annealing and fluorescence acquisition at 55 °C for 45 s. Real-time RT-PCR cycling conditions were: reverse transcriptase reaction at 50 °C for 5 min, denaturation at 95 °C for 20 s, followed by 45 cycles of denaturation at 95 °C for 3 s, annealing and fluorescence acquisition at 55 °C for 35 s.
3. Results
3.1. Water quality results
Using the nephelometric turbidity unit (NTU) data for the water samples, the samples were classified into three study groups based on relative differences in turbidity: low range turbidity (16 NTU), middle range turbidity (46 NTU) and high range turbidity (92 NTU) (Table 1). The turbidity data for the low, middle, and high range groupings were significantly different (<0.0001). There were no significant differences in lake temperature, pH, SC, and total hardness amongst the turbidity ranges. However, TSS concentrations (0.009, 0.036, 0.053 mg/L) and TOC concentrations (7.9, 9.9, 14.5 mg/L of C) correlated linearly with turbidity resulting in trend lines with R2 values of 0.938 and 0.989 respectively.
Table 1.
Water quality characteristics.
| Range | Field temp. (°C) | pH | Turbidity (NTU) | SC (µS/cm at 25 °C) | Total hardness (mg/L of CaCO3) | TSS (mg/L) | TOC (mg/L of C) |
|---|---|---|---|---|---|---|---|
| Low | 21 ± 3.0 | 7.3 ± 0.1 | 16 ± 2.4 | 87.3 ± 14.5 | 24 ± 2.4 | 0.009 ± 0.001 | 7.9 ± 1.0 |
| Middle | 21 ± 1.2 | 7.1 ± 0.1 | 46 ± 6.7 | 96.6 ± 16.9 | 24 ± 3.0 | 0.036 ± 0.012 | 9.9 ± 1.1 |
| High | 22 ± 1.5 | 7.1 ± 0.1 | 92 ± 7.9 | 85.5 ± 12.9 | 23 ± 4.9 | 0.053 ± 0.018 | 14 ± 2.3 |
3.2. Filtration hydraulic results
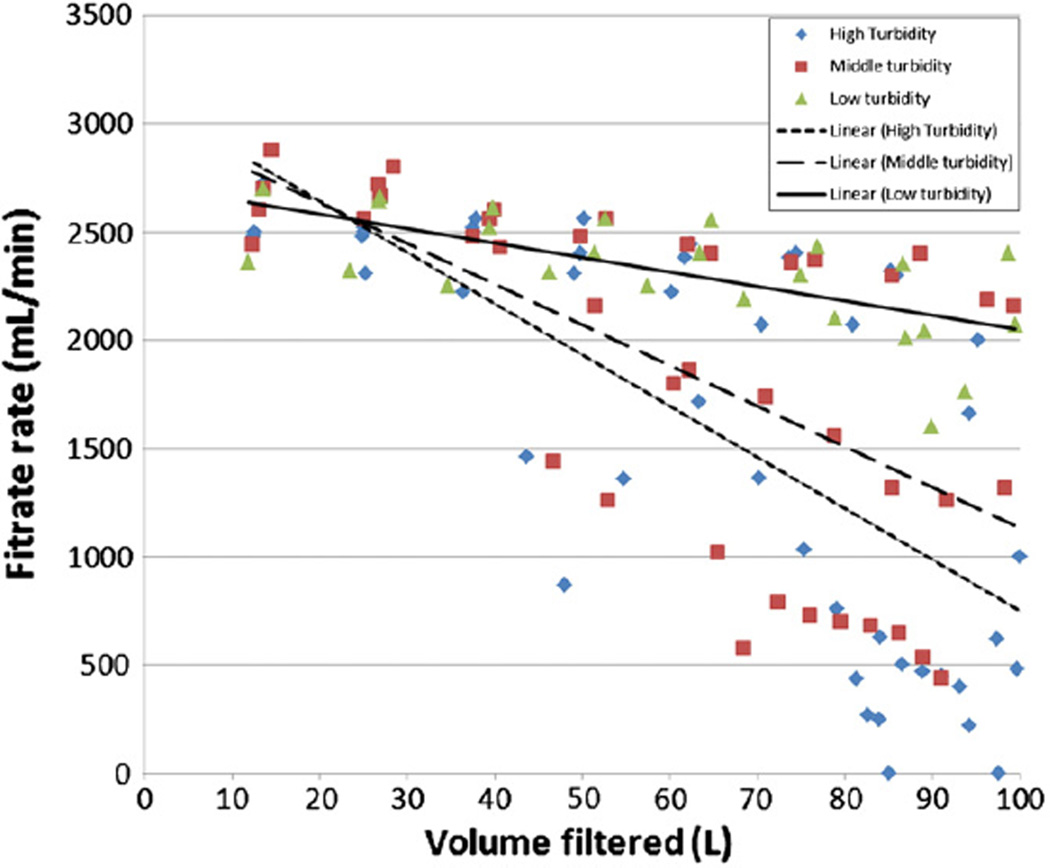
At a nominal input flow rate of 2900 mL/min, the average filtrate rates at the beginning of the low (16 NTU), middle (46 NTU), and high (92 NTU) range turbidity DEUF experiments (2590 mL/min, 2660 mL/min, 2550 mL/min, respectively) were not significantly different (p=0.67). After filtering 50 L of each water sample, filtrate rates were lower by an average of 6.1%, 18.3% and 13.3% for the low, middle and high turbidity ranges, respectively (Fig. 2). After filtering 80 L, average filtrate rates had decreased to 2180 mL/min, 1510 mL/min, and 1220 mL/min, for the low, middle, and high turbidity ranges, respectively. These filtrate rates for the low, middle, and high turbidity ranges were 16%, 43%, and 52% lower than at the beginning of the filtration experiments. At the end of the UF experiments, the last measured filtrate rates were an average of 2080 mL/min, 1530 mL/min, and 370 mL/min for the low, middle, and high turbidity levels, respectively. In two of the four high turbidity experiments, the filter clogged completely and the filtrate rate dropped to <1 mL/min after filtering 85 L and 97.5 L. For the DEUF method to filter 100 L of surface water it took on average 43 min for the low range, 50 min for the middle range, and 57 min for the high range turbidity experiments.
Fig. 2.
Association between water sample turbidity and UF filtrate rate.
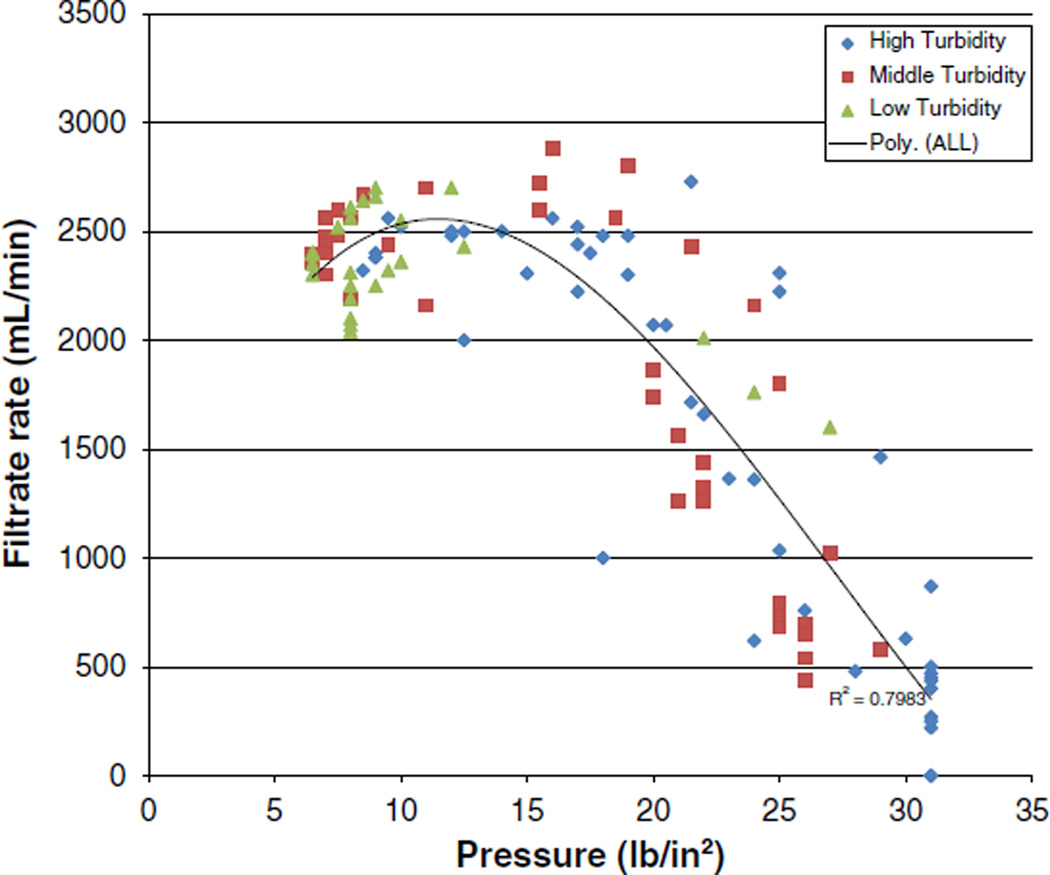
For all water turbidity levels combined, the correlation between pressure in the ultrafilters and filtrate rate exhibited a shoulder (no trend) effect between 2 L/min and 3 L/min filtrate rates followed by a linear, inversely proportional relationship (Fig. 3). The linear, inversely proportional effect is evidence of filter clogging when filtrate rates start to go below 2 L/min. Based on Fig. 3, clogging appeared to occur for all three of the turbidity levels when the filtrate rate decreased to less than 2 L/min and the pressure increased to 20 lb/in2 (138 kPa).
Fig. 3.
Correlation between filtrate rate and pressure in DEUF ultrafilter cartridges.
In order to more accurately estimate the volume at which clogging was observed at each turbidity level, we defined appreciable filter clogging as the point when the filtrate rate decreased by greater than 400 mL/min and pressure increased greater than 4 lb/in2 (27 kPa) in 5 min. Using this definition, appreciable filter clogging began when an average of 96 L, 75 L, and 72 L had been filtered for the low, middle, and high turbidity levels, respectively.
3.3. Microbial recovery
Average microbial recovery efficiencies for the DEUF method ranged from43% for C. parvum and C. perfringens in high turbidity water to 91% for C. perfringens in low turbidity water and 91% for enterococci in high-turbidity water (Table 2). Average recovery efficiencies for each study microbe across all turbidity ranges were: MS2 (66%), C. parvum (49%), enterococci (85%), E. coli (81%), and C. perfringens (63%). No significant differences in recovery efficiency were observed for E. coli, enterococci, or C. parvum at the turbidity levels studied. Average recovery efficiencies were significantly different for MS2 (p=0.008) and C. perfringens (p<0.0001) between turbidity levels. The MS2 recovery efficiencies decreased as the turbidity level increased, however the only significantly difference was determined between the low and high turbidity levels (p=0.003). The recovery efficiencies for C. perfringens were significantly different between all three turbidity levels: low and middle (p<0.0001), low and high (p<0.0001), and high and middle (p=0.02).
Table 2.
Recovery efficiency of DEUF for microbes seeded into 100 L of water at different turbidity levels.
| Turbidity (n) Range | Average recovery efficiency ± SD (%) | ||||
|---|---|---|---|---|---|
| MS2 | C. parvum | E. coli | C. perf. | Enterococci | |
| 5 NTU a | 73 ± 13 | 83 ± 21 | ND | 57 ± 21 | 78 ± 12 |
| 16 NTU (3) Low | 82 ± 9 | 49 ± 9 | 85 ± 7 | 91 ± 6 | 79 ± 4 |
| 46 NTU (4) Middle | 66 ± 9 | 56 ± 13 | 73 ± 11 | 54 ± 7 | 84 ± 11 |
| 92 NTU (4) High | 50 ± 11 | 43 ± 17 | 86 ± 7 | 43 ± 3 | 91 ± 14 |
Data from Smith and Hill using tap water amended with surface water (Smith and Hill, 2009).
3.4. Molecular detection of MS2 and C. parvum
Each of the three replicate experiments resulted in positive detections by real-time RT-PCR and PCR for MS2 and C. parvum, respectively (Table 3). Analysis of PEG-concentrated samples resulted in an average Ct value of 36.8 ± 0.7 for MS2 and 40.7 ± 1.4 for C. parvum. When IMS was performed to separate Cryptosporidium oocysts from potentially inhibitory constituents in the PEG concentrates, the resulting real-time PCR Ct values were substantially lower (34.2 ± 1.6). The average microbial recovery efficiencies associated with these DEUF experiments were 66% ± 14% for MS2 and 42% ± 3.7% for C. parvum.
Table 3.
Recovery and molecular detection of MS2 and C. parvum in 100-L surface water samples.
| Analyte | Recovery efficiencya (%) | PCR or RT-PCR analysis of PEG pellet (Ct value) |
|
|---|---|---|---|
| Direct PCR (no IMS) | With IMS processing | ||
| MS2 | 66 ± 14% | 36.8 ± 0.7 | NA |
| C. parvum | 42 ± 3.7% | 40.7 ± 1.4b | 34.2 ± 1.6b |
NA = not applicable.
For entire method, including DEUF and secondary processing procedures.
Assayed 2-µL DNA by direct PCR and 10-µL DNA from post-IMS sample without appreciable assay inhibition.
4. Discussion
DEUF has been shown in previous studies to effectively recover a diverse range of waterborne microbes from large-volume tap water and marine water samples having relatively low turbidity (Leskinen et al., 2009; Leskinen and Lim, 2008; Smith and Hill, 2009). The present study demonstrated that the DEUF method can also be an effective method for recovering diverse microbes and viruses from large volumes of high turbidity surface water and enabling sensitive detection using molecular testing. Data from this study indicate that water turbidity (and associated filter clogging) had a proportional impact on the recovery efficiencies for MS2 and C. perfringens, but was not observed for the other study analytes. The recovery efficiencies for C. parvum were similar for the three turbidity level categories used in this study, and were comparable to the C. parvum TFUF recoveries (42 ± 27%) that Simmons et al. reported for 10-L surface water samples having turbidities of 2.5 to 45 NTU (Simmons et al., 2001). However, the C. parvum recovery efficiencies reported in the present study were approximately 30% lower than previously reported in tap water using the DEUF method (Smith and Hill, 2009).
The recoveries of C. parvum oocysts in this study were also comparable to those reported for TFUF studies using 10 L of surface water. Reported recoveries of C. parvum oocysts in previous studies were 42% in 14.9 NTU, 45% in 56.2 NTU, and 27.5% in 159 NTU (Kuhn and Oshima, 2002; Morales-Morales et al., 2003; Simmons et al., 2001). Morales-Morales et al. also reported bacteria and bacteriophage recoveries (95% for E. coli, 73% T1 phage, and 62% PP7 phage) for TFUF processing of 10-L surface water samples (22.8 NTU) that are similar to the DEUF data reported in the present study (Morales-Morales et al., 2003). Even though TFUF likely maintains some of the microbes in suspension during filtration, as opposed to DEUF where all of the microbes are forced onto the filter fibers, the recovery efficiencies were not substantially different between the two methods in surface water or tap water.
One of the continuing uncertainties regarding application of UF for recovering microbes from environmental water samples has been the potential for method ineffectiveness due to filter clogging. This study demonstrated that the DEUF method was capable of filtering 96 L of 16 NTU water, 75 L of 46 NTU water and 72 L of 92 NTU water before appreciable filter clogging occurred. Although the nominal pore size of the REXEED 25SX ultrafilters used in this study was approximately 45 times smaller than traditional 0.45-µm bacteriological filters, suggesting a greater potential for clogging, these ultrafilters have a surface area of 2.5 m2. This large surface area (1440 times greater than a 47-mmdiameter filter) provides greater volumetric filtrate rates even when flux rates (the volume moving through a cross-sectional area per unit of time) are lower because of the small pore size of ultrafilters and potential clogging.
This study builds upon previous TFUF research that demonstrates that real-time PCR and RT-PCR can be effectively used to analyze UF concentrated water samples (Francy et al., 2009; Hill et al., 2010a, 2010b; Liu et al., 2012). In the present study, DEUF and secondary processing enabled consistent detection of MS2 and Cryptosporidium at concentrations of approximately 300 PFU/100 mL and 10 oocysts/L. The lower Ct values for C. parvum after IMS indicate that constituents in DEUF-concentrated water samples can inhibit PCR, but that purification procedures such as IMS can be effective for removing inhibitors to enable sensitive PCR detection.
The results from this study support and extend previous DEUF research findings, which indicate that this method can be effective for recovering and enabling sensitive detection of a wide array of microbes in diverse water types (Leskinen et al., 2010; Leskinen and Lim, 2008; Smith and Hill, 2009). The DEUF technique warrants further research with additional microbes and viruses (e.g., pathogenic viruses and bacteria) and water types (including treated wastewater). It represents a simple and cost-effective alternative for routine use in field studies for recovering pathogens and microbial indicators in environmental monitoring projects and waterborne outbreak investigations.
Acknowledgments
This work was supported by the CDC Office of Public Health Preparedness and Response. The findings and conclusions in this report are those of the authors and do not necessarily represent those of the CDC. Use of trade names and commercial sources is for identification only and does not imply endorsement by CDC or the US Department of Health and Human Services.
References
- Bisson JW, Cabelli VJ. Membrane filter enumeration method for Clostridium perfringens. Appl. Environ. Microbiol. 1979;37:55–66. doi: 10.1128/aem.37.1.55-66.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton AD, Rice EW, Greenberg AE. Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association; 2005. [Google Scholar]
- Francy DS, Bushon RN, Brady AMG, Bertke EE, Kephart CM, Likirdopulos CA, Mailot BE, Schaefer FW, III, Lindquist HDA. Comparison of traditional and molecular analytical methods for detecting biological agents in raw and drinking water following ultrafiltration. J. Appl. Microbiol. 2009;107:1479–1491. doi: 10.1111/j.1365-2672.2009.04329.x. [DOI] [PubMed] [Google Scholar]
- Hill VR, Polaczyk AL, Hahn D, Narayanan J, Cromeans TL, Roberts JM, Amburgey JE. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 2005;71:6878–6884. doi: 10.1128/AEM.71.11.6878-6884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill VR, Kahler AM, Jothikumar N, Johnson TB, Hahn D, Cromeans TL. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 2007a;73:4218–4225. doi: 10.1128/AEM.02713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill VR, Polaczyk AL, Kahler AM, Cromeans TL, Hahn D, Amburgey JE. Comparison of hollow-fiber ultrafiltration to the USEPA VIRADEL technique and USEPA method 1623. J. Environ. Qual. 2009;38:822–825. doi: 10.2134/jeq2008.0152. [DOI] [PubMed] [Google Scholar]
- Hill V, Jothikumar N, Vinjé J, Cromeans T. Project 3108. Water Research Foundation; 2010a. Sample Preparation Methods for Molecular Techniques for Drinking Water. [Google Scholar]
- Hill VR, Mull B, Jothikumar N, Ferdinand K, Vinje J. Detection of GI and GII noroviruses in ground water using ultrafiltration and TaqMan real-time RT-PCR. Food Environ. Virol. 2010b;2:218–224. [Google Scholar]
- Karim MR, Rhodes ER, Brinkman N, Wymer L, Fout GS. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 2009;75:2393–2399. doi: 10.1128/AEM.00922-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, DiGiano FA. Fouling models for low-pressure membrane systems. Sep. Purif. Technol. 2009;68:293–304. [Google Scholar]
- Knappett PSK, Layton A, McKay LD, Williams D, Mailloux BJ, Huq MR, Alam MJ, Ahmed KM, Akita Y, Serre ML, Sayler GS, van Geen A. Efficacy of hollow-fiber ultrafiltration for microbial sampling in groundwater. Ground Water. 2011;49:53–65. doi: 10.1111/j.1745-6584.2010.00712.x. [DOI] [PubMed] [Google Scholar]
- Kuhn RC, Oshima KH. Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. Can. J. Microbiol. 2002;48:542–549. doi: 10.1139/w02-049. [DOI] [PubMed] [Google Scholar]
- Leskinen SD, Lim DV. Rapid ultrafiltration concentration and biosensor detection of enterococci from large volumes of Florida recreational water. Appl. Environ. Microbiol. 2008;74:4792–4798. doi: 10.1128/AEM.00052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskinen SD, Harwood VJ, Lim DV. Rapid dead-end ultrafiltration concentration and biosensor detection of enterococci from beach waters of Southern California. J. Water Health. 2009;7:674–684. doi: 10.2166/wh.2009.086. [DOI] [PubMed] [Google Scholar]
- Leskinen SD, Brownell M, Lim DV, Harwood VJ. Hollow-fiber ultrafiltration and PCR detection of human-associated genetic markers from various types of surface water in Florida. Appl. Environ. Microbiol. 2010;76:4116–4117. doi: 10.1128/AEM.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist HDA, Harris S, Lucas S, Hartzel M, Riner D, Rochele P, DeLeon R. Using ultrafiltration to concentrate and detect Bacillus anthracis, Bacillus atrophaeus subspecies globigii, and Cryptosporidium parvum in 100-liter water samples. J. Microbiol. Methods. 2007;70:484–492. doi: 10.1016/j.mimet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Liu P, Hill VR, Hahn D, Johnson TB, Pan Y, Jothikumar N, Moe CL. Hollow-fiber ultrafiltration for simultaneous recovery of viruses, bacteria and parasites from reclaimed water. J. Microbiol. Methods. 2012;88:155–161. doi: 10.1016/j.mimet.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Morales-Morales HA, Vidal G, Olszewski J, Rock CM, Dasgupta D, Oshima KH, Smith GB. Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl. Environ. Microbiol. 2003;69:4098–4102. doi: 10.1128/AEM.69.7.4098-4102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mull B, Hill VR. Recovery and detection of Escherichia coli O157:H7 in surface water, using ultrafiltration and real-time PCR. Appl. Environ. Microbiol. 2009;75:3593–3597. doi: 10.1128/AEM.02750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima KH, Evansstrickfaden TT, Highsmith AK, Ades EW. The removal of Phages T1 and PP7, and Poliovirus from fiuids with hollow-fiber ultrafilters with molecular-weight cutoffs of 50000, 13000, and 6000. Can. J. Microbiol. 1995;41:316–322. doi: 10.1139/m95-044. [DOI] [PubMed] [Google Scholar]
- Polaczyk A, Roberts J, Hill V. Evaluation of 1MDS electropositive microfilters for simultaneous recovery of multiple microbe classes from tap water. J. Microbiol. Methods. 2007;68:260–266. doi: 10.1016/j.mimet.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Polaczyk AL, Narayanan J, Cromeans TL, Hahn D, Roberts JM, Amburgey JE, Hill VR. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods. 2008;73:92–99. doi: 10.1016/j.mimet.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Rhodes ER, Hamilton DW, See MJ, Wymer L. Evaluation of hollow-fiber ultrafiltration primary concentration of pathogens and secondary concentration of viruses from water. J. Virol. Methods. 2011;176:38–45. doi: 10.1016/j.jviromet.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Shapiro K, Mazet JAK, Schriewer A, Wuertz S, Fritz H, Miller WA, Largier J, Conrad PA. Detection of Toxoplasma gondii oocysts and surrogate microspheres in water using ultrafiltration and capsule filtration. Water Res. 2010;44:893–903. doi: 10.1016/j.watres.2009.09.061. [DOI] [PubMed] [Google Scholar]
- Simmons OD, Sobsey MD, Heaney CD, Schaefer FW, Francy DS. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 2001;67:1123–1127. doi: 10.1128/AEM.67.3.1123-1127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Hill VR. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl. Environ. Microbiol. 2009;75:5284–5289. doi: 10.1128/AEM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA. Method 1602: Male-specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL) Procedure. Washington, DC: Office of Water; 2001. [Google Scholar]
- US EPA. Method 1600: Enterococci in Water by Membrane Filtration Using membrane-Enterococcus Indoxyl-B-D-Glucoside Agar (mEI. Washington, DC: Office of Water; 2005a. [Google Scholar]
- US EPA. Method 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Eschericia coli Agar (Modified mTEC) Washington, DC: Office of Water; 2005b. [Google Scholar]
- US EPA. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. Washington, DC: Office of Water; 2005c. p. 68. [Google Scholar]