Abstract
Rheumatoid factors (RF) are autoantibodies that recognize epitopes in the Fc region of immunoglobulin (Ig) G and that correlate with the clinical severity of rheumatoid arthritis (RA). Here we report the X-ray crystal-lographic structure, at 3 Å resolution, of a complex between the Fc region of human IgG1 and the Fab fragment of a monoclonal IgM RF (RF61), derived from an RA patient and with a relatively high affinity for IgG Fc. In the complex, two Fab fragments bind to each Fc at epitopes close to the C terminus, and each epitope comprises residues from both Cγ3 domains. A central role in the unusually hydrophilic epitope is played by the side-chain of Arg355, accounting for the subclass specificity of RF61, which recognizes IgG1,−2, and −3 in preference to IgG4, in which the corresponding residue is Gln355. Compared with a previously determined complex of a lower affinity RF (RF-AN) bound to IgG4 Fc, in which only residues at the very edge of the antibody combining site were involved in binding, the epitope bound by RF61 is centered in classic fashion on the axis of the VH:VL β-barrel. The complementarity determining region-H3 loop plays a key role, forming a pocket in which Arg355 is bound by two salt-bridges. The antibody contacts also involve two somatically mutated VH residues, reinforcing the suggestion of a process of antigen-driven maturation and selection for IgG Fc during the generation of this RF autoantibody.
Keywords: autoantibody, autoantigen, rheumatoid factor, autoimmunity, X-ray crystallography
Introduction
Fc domains of immunoglobulin (Ig) G are not only the biological effector regions of the molecule, interacting with complement and Fc receptors, but also the targets of autoimmune recognition by rheumatoid factors (RF), autoantibodies associated particularly with rheumatoid arthritis (RA).1,2 Although not unique to that condition, high titers of RF in RA sera and synovia correlate with severe inflammatory joint damage, extra-articular pathology, and poor prognosis, indicative of a pathogenic role.3–7 Complex formation between RF and IgG, in the absence of any other antigen, may lead to activation of complement and other inflammatory mediators.8
The RF detected in RA patients can be of IgM, IgG, or IgA isotype. In individuals with no overt disease, the presence of RF is associated with antibody responses to bacteria and viruses in humans9 and experimental animals.10 These RF are mainly IgM and would facilitate the clearance of antigen by enhancing complement activation and phagocytosis. A distinction has therefore been made between “physiological” RF, which may serve a beneficial role in host defense, and “pathological” RF associated with RA and other systemic autoimmune diseases.11
RF affinity may well be important in a pathogenic role, because higher affinity (Kd ~10−7 to 10−8 M) IgM and IgG RF appear to correlate with disease.12,13 Another relevant consideration is the location of the epitope on Fc, because this will determine whether the complement or other receptor-binding sites on IgG is accessible in the RF/Fc complex. The locations of a number of distinct epitopes on IgG Fc have been deduced from the reactivity patterns of monoclonal RF from RA patients.14 They are most commonly located at the Cγ2–Cγ3 interface, but have also been found on the Cγ2 or Cγ3 domains alone or involving the carbohydrate on Cγ2.
The first direct visualization of RF/IgG binding was provided by the crystal structure of the complex between the Fab (antigen-binding fragment) of RFAN, a monoclonal IgM, and Fc of human IgG4.15,16 RF-AN has only a low affinity for IgG (Kd>10−5 M for Fab) and displays a common RF reactivity profile, binding only to IgG subclasses 1, 2, and 4; this is associated with a His-to-Arg substitution at residue 435 in the Cγ3 domain of IgG3 Fc. The crystal structure revealed two RF-AN Fabs binding to Fc, one to each heavy chain, at the predicted epitope spanning the junction of the Cγ2 and Cγ3 domains. Surprisingly, the antigen-binding site displayed a highly unconventional topology in which only residues along one edge of the potential combining site surface and directed away from the central axis of the VH/VL dimer were used, leaving much of the conventional site free. Although this suggested that RF-AN might have its primary specificity for another antigen, the involvement of somatically mutated residues in binding implied selection driven by Fc binding in its induction.
To test the generality of these observations, it is important to study RFs with higher affinity and different epitope specificity. To this end we investigated RF61, a monoreactive IgM RF secreted by an Epstein–Barr virus-transformed peripheral CD5+B cell from an RA patient.12 RF61 is of higher affinity (Kd~6×10−7 M) and has six somatically mutated amino acids in its complementarity determining regions (CDRs), indicative of antigen-driven selection and affinity maturation. Its specificity is for IgG1, −2, and −3, with ~10-fold lower affinity for IgG4, suggesting a different epitope recognition from that of RF-AN. Here we report the crystal structure of the RF61 Fab–IgG1 Fc complex. The two Fc epitopes are in an entirely novel location, namely the Cγ3–Cγ3 interface, including the C-terminal region of both heavy chains, and distant from the Cγ2–Cγ3 interface. Also in contrast with RF-AN, the interaction involves five of the six CDRs in the conventional manner of antigen–antibody recognition. The involvement of somatic mutations in the complex provides clear evidence for selection of RF through affinity maturation and selection by the autoantigen IgG Fc. This new structure provides further insights into the diversity and origin of RFs.
Results
Specificity of RF61
An ELISA was employed to measure the binding of RF61 IgM to human IgG subclasses. Titrations against IgG1, −2, and −3 were equivalent, whereas binding to IgG4 was about 10-fold weaker (Figure 1(a)). A competition ELISA to assess the inhibition of RF61 binding by Staphylococcus aureus protein A (SpA) showed no inhibition up to 10 μg/ml SpA, in contrast with RF-AN, which was >70% inhibitable at 10 ng/ml (Figure 1(b)).
Figure 1.
Binding specificity of RF61. (a) Binding of RF61 IgM to human IgG subclasses by ELISA demonstrates that the binding specificity is IgG1~IgG2~IgG3>IgG4. (b) Inhibition by Staphylococcus aureus protein A (SpA) of RF61 IgM and RF-AN IgM binding to human IgG by competition ELISA. Whereas the binding of RF-AN to IgG is inhibitable by SpA, that of RF61 is not.
Structure of the complex
The structure was solved and refined to the maximum resolution allowed by the crystals, namely 3 Å (see Table 1 for crystallographic data and refinement statistics) and contained a 2:1 complex with two IgM Fab fragments bound to the IgG Fc (Figure 2). The two Fc epitopes are located at the C-terminal end of the Cγ3 domains, distal to Cγ2 and close to the local 2-fold axis of the Cγ3 domain dimer. Their location is such that the VL domains of the two Fabs are almost brought into contact. Each epitope comprises regions of both heavy chains (Figure 3), including residues from loops AB, EF, and the C-terminal end of strand G of one Cγ3 domain, together with residues in strands A and G of the other (Figure 4). Thus, in the 2:1 complex each Cγ3 domain is contacted by both Fabs.
Table 1.
Data collection and refinement statistics for the RF61 Fab-IgG1 Fc complex
| Data collection ESRF, ID14-EH1 | |
| Space group | C2 |
| Unit cell (Å) | 242.0, 75.6, 102.4, β = 91.1° |
| Resolution (Å) | 25.0-3.0 (3.11-3.00) |
| Observations | 132,372 |
| Unique reflections | 36,545 |
| Completeness (%) | 97.9 (93.8) |
| I/σI | 14.1 (3.2) |
| Rmerge (%) | 8.1 (42.4) |
| Refinement statistics | |
| Work reflections/test reflections | 33,432/2563 (all with |F| >2 σ) |
| Rcryst. (%)/Rfree (%) | 22.4/28.8 |
| No. of protein atoms | 9819 |
| No. of carbohydrate atoms | 220 |
| No. of waters | 84 |
| Anisotropic B correction (Å2) | 37.8; –15.9; –21.9; 0.0; –6.6; 0.0 |
| R.m.s. deviation from ideal | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.4 |
| Dihedral angles (°) | 25.6 |
| Improper angles (°) | 0.95 |
| Fc-A CH2 | Fc-A CH3 | Fc-B CH2 | Fc-B CH3 | Fab-1 VHVL | Fab-1 CH1CL | Fab-2 VHVL | Fab-2 CH1CL | |
|---|---|---|---|---|---|---|---|---|
| Average B-factor (Å2) | 37.0 | 41.2 | 86.6 | 39.1 | 60.6 | 99.0 | 56.2 | 95.6 |
| Ramachandran quality: | ||||||||
| Most favored (%) | 88.8 | 84.8 | 70.7 | 87.8 | 77.9 | 57.0 | 80.5 | 56.7 |
| Additionally favored (%) | 11.2 | 15.2 | 28.3 | 12.2 | 18.4 | 33.3 | 16.8 | 32.2 |
| Generously allowed (%) | 0 | 0 | 1.1 | 0 | 2.1 | 6.7 | 1.6 | 8.8 |
| Disallowed (%) | 0 | 0 | 0 | 0 | 1.6 | 3.0 | 1.1 | 2.3 |
Figure 2.
Structure of the RF61 Fab–IgG1 Fc complex. The two chains of the Fc fragment, with the N-linked carbohydrate (ball-and-stick representation), are shown in dark and light-blue (chains A and B, respectively). For the two Fabs, bound symmetrically to the Cγ3 domains of the Fc, heavy chains are shown in red and green and light chains in orange and purple. Two cations and the cacodylate moiety are shown in yellow. (Figure produced by MolScript39).
Figure 3.
Location of the RF61 epitopes on Fc recognized by RF61 Fab. The Cγ3 domains of each of the two heavy chains (colored light and dark-blue) are viewed (a) in the same orientation as in Figure 2 and (b) at 90° to view (a), along the 2-fold axis looking onto the C-terminal end of the Fc. The Cγ3 residues involved in the interactions with each of the two Fabs are shown in yellow and purple; view (b) shows that each epitope spans the two Cγ3 domains. Residue Arg355 in each heavy chain is indicated.
Figure 4.
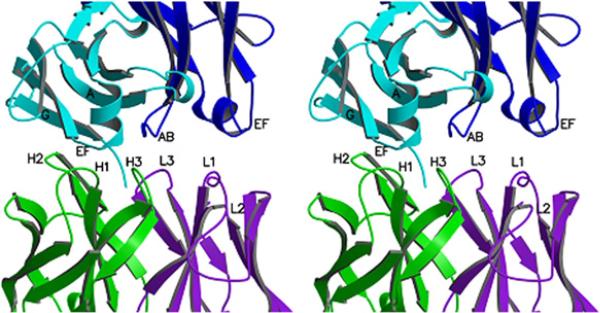
The RF61 Fab–IgG1 Fc contact region. The six CDRs of RF61, all of which make contact except L2, and the strands and loop regions that constitute the epitope on the Cγ3 domains of IgG Fc, are indicated in this stereo image. The VH and VL domains of RF61 are shown in green and purple, respectively, and the IgG Fc A and B chains are in light and dark-blue, respectively.
Figure 4 also shows the disposition of the CDRs in relation to the epitope. Five of the six CDRs contribute to the interaction, with no contacts from L2. The VH domain makes contacts with residues in both Cγ3 domains, with CDR H3 making cross-domain contacts, while VL contacts residues in one Cγ3 domain exclusively.
Nature of the RF/Fc interactions
Table 2 lists the 14 Fab and 13 Fc contact residues and the nature of their interactions, which include 13 H-bonds and two salt-bridges. Remarkably, all Fab contact residues involve hydrophilic groups, including the hydroxyls of four tyrosines, and most of the Fc contact residues, with the exception of Val422 and Leu441, are also hydrophilic. There are also four H-bonds mediated by ordered water molecules at the interface.
Table 2.
Contact residues in the RF61 Fab-IgG1 Fc interaction
| CDR residue (Kabat numbering) | Fc contact | Fc chain | Types of contact | |
|---|---|---|---|---|
| H1 | ||||
| Arg31 | Gly420 | A | HB | V1 |
| Val422 | A | V5 | ||
| Ser442 | A | HB | V2 | |
| Tyr35 | Asp356 | B | HB | V5 |
| H2 | ||||
| Tyr52 | Lys439 | A | V8 | |
| Ser440 | A | HB | V1 | |
| Tyr53 | Gln438 | A | HB | V6 |
| Ser440 | A | HB | V3 | |
| Ser54 | Gln438 | A | HB | V1 |
| H3 | ||||
| Asp98 | Arg355 | B | SB | V2 |
| Tyr100 | Thr350 | A | HB | V2 |
| Asp356 | B | V6 | ||
| Lys439 | A | V2 | ||
| Ser440 | A | HB | V3 | |
| Leu441 | A | V2 | ||
| Thr100a | Arg355 | B | HB | V2 |
| Asp100c | Arg355 | B | SB | V1 |
| L1 | ||||
| Ser30 | Gln418 | B | HB | V2 |
| Asn31 | Lys414 | B | HB | V3 |
| Tyr34 | Arg355 | B | HB | V3 |
| L2 | ||||
| None | ||||
| L3 | ||||
| Trp91 | Thr359 | B | V6 | |
| Asp93 | Lys414 | B | V3 | |
HB, hydrogen bond; SB, salt-bridge; Vn, number of van der Waals contacts.
The Fc binding surface contains four charged residues (Arg355, Asp356, Lys414, and Lys439), with four H-bonds and two salt-bridges made to these side-chains. A key role is played by Arg355, which is involved in salt-bridges to AspH98 and AspH100c on CDR H3 (Figure 5(a)). The major contribution of Arg355 to the interaction accounts well for the subclass specificity of RF61, because among the four human IgG isotypes, this residue is not conserved in IgG4, in which it is substituted by Gln.
Figure 5.
Details of the RF61 Fab/IgG1 Fc contact region. (a) Interactions of the Fc residue Arg355 with the RF61 combining site. The Fc polypeptide region is shown in cyan, and the heavy and light chains of the Fab are in yellow and orange, respectively (stereo image). Side chains of residues AspH99, TyrH100, and LeuH100b are omitted for clarity. (b) Interactions of RF61 somatic mutations ArgH31 and GlyH32 with Fc. ArgH31 and GlyH32 are somatic mutations in CDR-H1 of RF61. The side-chain of ArgH31 packs against ValH422 and H-bonds with Gly420 and Ser442 of the Fc. GlyH32 allows a local conformation of the main chain that would be disallowed for any other amino acid.
The majority of Fab contact residues (9/14) come from the VH domain and include four charged residues, ArgH31 (see Figure 5(b)), AspH98, AspH100c, and AspL93, between them accounting for two H-bonds and two salt-bridges. As observed in many other antibody–antigen complexes, CDR-H3 plays a major role in the interaction, contributing the largest number of contacts, as well as being topologically central and bridging both Cγ3 domains (Table 2). The CDR-H3 loop creates a pocket that accommodates the side-chain of Arg355, to which the salt-bridges noted above are made together with a H-bond to ThrH100a (Figure 5(a)). A central H3 residue TyrH100 interacts with both Cγ3 domains, contacting five Fc residues via two H-bonds and 15 van der Waals contacts.
Somatic mutation in contact residues
Comparison of the VH and VL sequences with their germline counterparts (Figure 6) identifies six probable somatic mutations, all in CDRs: Ser31Arg, Ser32Gly and Tyr34His (H1), Ser56Asn and Tyr59Phe (H2), and Ala90Thr (L3). To establish that they were correctly identfied as mutations, the corresponding closest VH germline segment, VH4.18, was earlier sequenced from the same patient. Of these substitutions, ArgH31 is an important contact residue that is closely packed against Val422 and H-bonds with Gly420 and Ser442 (Figure 5(b)), and GlyH32 permits a main-chain conformation (ϕ=90°, φ=5°) that would be disallowed for any other amino acid residue. The combined effect of these two mutations is potentially highly significant for the binding affinity. The other mutations are not Fc contact residues.
Figure 6.
Sequences of V regions of RF61 compared with germline genes. The corresponding reference germline gene segments are VH4.18 for VHIV and 1B9/F2 for Vλ1. Somatic mutations are identified in red, and residues making contact with the epitope are shown in boldface blue, with the exception of ArgH31 (shown in boldface red because it is a somatic mutation). Sequence data are from Ref. 12.
Discussion
Some RF antibodies are potentially important components in the pathogenesis of RA and occur at high levels in patient sera, whereas other physiological RFs arise as a normal consequence of antibody responses to exogenous antigens and may aid antigen clearance. We have undertaken structure determinations of two RFs derived from RA patients to define structural features that may account for the different roles these autoantibodies play. In this report we studied RF61, a human monoreactive IgM rheumatoid factor of relatively high affinity, secreted by an Epstein–Barr virus-transformed, CD5+B cell from an RA patient.12,13 The affinity of RF61 (Kd ~6×10−7 M) was indeed higher than that of other RFs isolated from the same patient, although it is lower than typical IgGs generated in an immune response. RFs such as this are characteristic of RA, compared with those in normal sera or paraproteinemias. The variable regions of RF61 comprise a VHIV gene in combination with a Vλ1 light chain. Sequencing has revealed the presence of six somatic point mutations located in the CDRs, five in VH and one in VL, indicative of affinity maturation (Figure 6). RF61 shows specificity for IgG1, −2, and −3, with weaker binding to IgG4. Here we describe the structure of the complex between the RF61 Fab fragment and Fc of IgG1 (the latter expressed as a fusion protein with part of the gD protein of Herpes simplex virus, which makes no contribution to the RF complex).
In previous work we determined the structure of RF-AN, another patient-derived RF, complexed with Fc of IgG4. The VH of RF-AN is VHIII-encoded with a VλIII-a light chain, and the affinity of the Fab fragment (K2d >10−5 M) is significantly lower than that of RF61. The subclass specificity of RF-AN also differs (IgG1, −2, −4 >> IgG3), but as with RF61, there are somatic mutations in CDRs of both VH (3 in H2) and VL (1 in L2). The structures of the RF-AN and RF61 complexes show a number of striking differences, which may relate to their affinities, origin, and functional role.
Epitope location
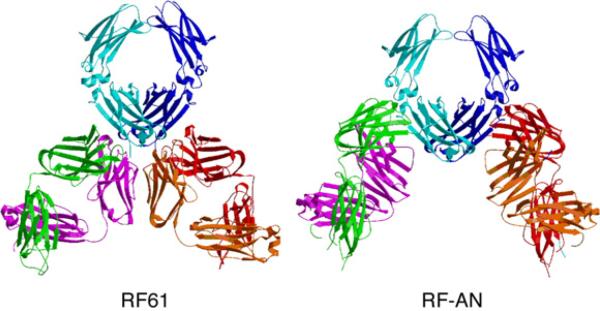
Whereas the RF-AN epitope is in the Cγ2–Cγ3 interface,15 that recognized by RF61 is located at the C-terminal (Cγ3) end of the Fc (Figure 7). Consistent with this, binding of RF-AN but not RF61 to IgG Fc is inhibitable by S. aureus protein A, which binds at the Cγ2–Cγ3 interface.17 Although in both complexes two Fabs are bound to one Fc, in the case of RF61 the Fabs are located as close to each other as possible without steric interference (Figure 7); an Fc epitope located any closer to the 2-fold axis of symmetry would allow only one Fab to bind at a time. The simultaneous binding of two Fabs may well be a necessary feature of RFs, enabling the generally low-affinity combining sites to form extended complexes. The RF61 epitope is almost as far as physically possible from the antigen-combining site of IgG and is also remote from the functional sites responsible for FcγR or C1q binding (in Cγ2) and from the N-linked carbohydrate, also in Cγ2 (Figure 2). The RF-AN epitope is less distal, but is also out of range of the FcR and C1q-binding sites and carbohydrate. Interestingly, both epitopes involve adjacent parts of two domains. Whereas RF-AN recognizes the structurally distinct Cγ2 and Cγ3 domains and binds to the cleft between them, each RF61 epitope comprises adjacent regions of the two identical Cγ3 domains (Figure 3).
Figure 7.
Comparison of RF61 and RF-AN complexes with human IgG Fc. The complexes of RF61 Fab and IgG1 Fc (this paper) and RF-AN Fab with IgG4 Fc15 are viewed with the Fc regions in a similar orientation (colored as in Figure 2). The very different locations of the epitopes and orientations of the Fabs are apparent. (Figure produced by MolScript39).
Interactions, subclass specificity, and affinity
The fact that a number of diverse ligands, including bacterial proteins A and G, FcαR, and FcRn, all interact with the Cγ2–Cγ3 cleft17–20 has been attributed to the hydrophobicity of this region.21 This is mirrored in the RF-AN complex, which involves a number of hydrophobic side-chains from both Fc epitope and Fab. In contrast, the character of the RF61 site is quite different: the epitope and binding interactions of RF61 are unexpectedly hydrophilic in nature, with a larger number of H-bonds as well as two salt-bridges. The latter involve two arginine residues, one on Cγ3 and the other on CDR-H1. Arg355, in Fc, is clearly a crucial RF61 residue, forming salt-bridges to two aspartic acids on CDR-H3, as well as two H-bonds to H3 and L1. The centrality of Arg355 can explain the subclass specificity of RF61, which binds approximately 10 times less avidly to IgG4 (in which Arg355 is replaced by Gln). This specificity pattern (IgG1, −2, −3>IgG4) is relatively unusual among RF, most of which bind with “Ga-related” specificity (IgG1, −2, −4 >> IgG3).2,14 In the case of RF-AN it was not possible to account clearly for subclass specificity from the X-ray structure; although a major role of His435 (Arg435 in IgG3) has been inferred from serology, this residue lay at the very edge of the interaction surface,15 and it was not clear how the substitution could affect RF-AN binding to IgG3. In contrast, for RF61 there is no doubt that subclass specificity is dependent on Arg355. Figure 1(a) also indicates a more subtle difference in binding affinity between IgG1 and IgG2/3. This may be explained by the involvement of Asp356 in hydrogen bonding to TyrH35 (Figure 5(a)), which would not be possible for the glutamic acid residue present in subclasses 2, 3, and 4. (All other Fc contact residues, with the exception of isoleucine for valine in IgG3 at position 422, are identical across the four subclasses).
Comparison of the two complexes reveals a number of features that are consistent with the greater binding affinity of RF61. The buried surface area of the complex is higher (~1600 Å2 cf 1280 Å2 for RF-AN) and more CDRs are engaged (5 cf 4), with more residues in contact with the epitope (14 cf 9), and a larger number of H-bonds (13 cf 3) and salt-bridges (2 cf 1).
Orientation of CDRs in the complex
The antigen-binding site of RF-AN displayed an unconventional orientation in relation to the epitope: rather than binding centrally onto the CDRs, only residues along one edge of the potential combining site surface and directed away from the central axis of the VH/VL dimer were used.15,16 Only 8 residues from four CDRs were engaged in the interaction, leaving much of the conventional site free and suggesting that RF-AN might display specificity for another antigen, perhaps even binding both simultaneously.15,16,22 Whereas all three VH CDRs contributed contacts, on VL only L2 was used. In contrast, the binding site of RF61 is engaged in a manner consistent with “classic” antigen–antibody complexes,23,24 utilizing five of six CDRs and 14 Fab contact residues, and with the autoantigen epitope occupying the center of the VH/VL β-barrel. In the light chain, RF61 utilizes L1 and L3, but not L2; this is again more typical of antigen–antibody complexes in which CDR-L2 is often the most distant from the bound epitope. Such a classic use of the binding site in RF61 would more readily allow the development of high-affinity antibodies.
Thus, the unusual topology of RF-AN remains unique to date and appears not to be a defining feature of RF autoantibody combining sites. Nevertheless, there are a number of points of similarity between the RF-AN and RF61 complexes. In both, VH plays a dominant role and the central location of the CDR-H3 loop in each complex is such that it interacts with both domains of the epitope (Cγ2 and Cγ3 for RF-AN, Cγ3 and Cγ3 for RF61) and provides more contacts than the other CDRs. Furthermore, in both complexes a tyrosine residue in CDR-H3 makes cross-domain interactions (TyrH98 in RF-AN, TyrH100 in RF61).
Role of somatic mutations
The RF-AN structure provided evidence for the involvement of a somatic mutation, and by implication an antigen-driven selection process, in the induction of RF, because an important contact residue in L2, ProL56, was a probable point mutation. The evidence is even more striking in RF61, where two of the six somatically generated residues make important contributions to the contact region. The substitution of Arg for germline-encoded SerH31 in CDR-H1 allows H-bond interactions with the main-chain carbonyl of Gly420 and the hydroxyl of Ser442, together with five van der Waals interactions to Val422, which would be impossible with the serine residue (Figure 5(b)). Moreover, mutation of the adjoining germline SerH32 to Gly is required for the correct main-chain conformation to allow those interactions to occur. Thus, even though both RFs are IgMs and may not have completed the highest degree of affinity maturation, somatic mutation is an important part of their autoreactivity.
In conclusion, our structural analyses of human RFs have revealed two types of binding interaction. RFs with a particularly low affinity, such as RF-AN, may use the binding site in an unconventional manner, whereas those of higher affinity are structurally classic. Previously we speculated that the low-affinity RFs arise as a form of cross-reaction with a foreign epitope, based on the topology of the RF-AN site, which left the central binding area accessible even when Fc was bound. In the case of RF61 there is no structural reason to consider the binding other than conventional and probably selected by Fc ab initio, suggesting that Fc is the true antigenic stimulus for pathogenic, somatically mutated IgM RFs generated in patients with RA. The classic use of the combining site as in RF61 would be better suited to development of higher-affinity antibodies, and by implication those involved in the disease process. Interaction with the Fc epitope is in both cases a combined result of germline residues, VDJ rearrangements, and somatic mutations in CDRs, rather than solely a property of unmutated germline genes.25,26
For both of these RFs, somatically mutated residues play an important role in the interaction, so that while they may have different origins, affinity maturation and selection by Fc appear to be a common feature.
Materials and Methods
RF61 IgM and Fab
The RF61 cell line was derived from CD5+ circulating cells from an RA patient and the secreted IgM was produced from monoclonal Epstein–Barr virus-transformed hybrid cells.12 RF61 cells were grown in roller culture in Dulbecco's modified Eagle's medium (Gibco) supplemented with 5% bovine fetal calf serum (FCS). Cell supernatants were dialyzed against 2 mM phosphate, pH 6, and the resulting euglobulin precipitate was dissolved in phosphate-buffered saline (PBS). The IgM was isolated by gel filtration through Biogel A-5M eluted with PBS. IgM in 0.1 M Tris–HCl, pH 8.3, was digested with trypsin (IgM:trypsin, 50:1) at 56 °C for 3 h to obtain the Fab fragment, as previously described.27
gD–Fc fusion protein
Coding sequences corresponding to the ectodomain of HSV-1 gD (strain Patton), residues 1–343, were fused with sequences corresponding to the constant region of human IgG1 (hinge, CH2–CH3) and the fusion protein expressed from the β-actin promoter in pHβAPr-1.28 A chinese hamster ovary cell line constitutively expressing the construct was established, producing approximately 1 μg/ml of secreted protein in tissue culture supernatant. Cells were grown in Iscove's medium supplemented with 5% FCS, which was passed through a protein A sepharose column prior to use to remove all protein A binding species. The gD–Fc fusion protein was then purified from tissue culture supernatant by adsorption to a protein A sepharose column and eluted with 0.1 M glycine buffer, pH 3, immediately adjusted to pH 7.5 with 1 M Tris–HCl. The protein was dialyzed against PBS and 10% glycerol and concentrated to 5 mg/ml.
ELISA
RF binding to human IgG subclasses
A 96-well ELISA plate was treated with 100 μl/well of human IgG1, IgG2, IgG3, or IgG4, all with κ light chains (The Binding Site Co.), at 5 μg/ml in PBS overnight at 4 °C. The plate was washed three times with PBS/Tween 20 (0.05%) and blocked with 200 μl 10% FCS for 2 h at 4 °C. Wells were rewashed and 100 μl RF61 IgM was added in doubling dilutions from 5 to 0.078 μg/ml in 10% FCS for 2 h at 4 °C. After being washed further, 100 μl anti-human μ-chain–horseradish peroxidase conjugate (Sigma), 1:10,000 dilution in 10% FCS, was added for 1 h at 4 °C. Wells were washed as above and tetramethylbenzidine substrate was added for 6 min before OD was determined at 450 nm.
Competition by protein A
A procedure similar to that described above was followed with wells coated with human IgG (5 μg/ml in PBS). A total of 100 μl protein A (Sigma) was added to wells in a 10-fold dilution series from 10 μg/ml to 1 pg/ml in 10% FCS and incubated for 3 h, followed by three washes, before the addition of RF at 2 μg/ml in 10% FCS for 1 h. Binding was developed with horseradish peroxidase anti-μ conjugate. The protein A-inhibitable RF-AN was used as positive control.
Preparation and crystallization of the complex
Crystallization of the complex has been described.29 Briefly, sitting drops were produced by mixing 1.5 μl of a reservoir solution consisting of 10% (wt/wt) monomethyl polyethylene glycol 5000, 3 mM zinc acetate, 3 mM cadmium chloride, 100 mM sodium cacodylate, pH 6.5, with 1.6 μl of RF61 Fab at 12 mg/ml and 0.8 μl gD–Fc at 10 mg/ml in 50 mM ammonium acetate, pH 6.0. Crystals for data collection were enlarged using streak seeding followed by macroseeding.30 Diffraction-quality crystals were obtained after a growth period of about 8 months.
Data collection
Data were recorded at ESRF beamline ID14-EH1 at cryogenic temperature (100 K) on a Quantum4 CCD detector and processed using the HKL package.31 Crystals were mounted in a nylon loop and plunged into liquid ethane for rapid cryocooling. The cryoprotectant solution used contained 14% (w/w) polyethylene glycol 5000, 27% ethylene glycol, 1.7 mM zinc acetate, 1.7 mM cadmium chloride, 55 mM sodium cacodylate, pH 6.5. The best crystals displayed measurable diffraction to 3 Å resolution and belong to space group C2 with unit cell dimensions a=242.0 Å, b=75.6 Å, c=102.4 Å, and β=91.1°.
Structure determination and refinement
The RF61 Fab/gD–Fc complex structure was determined by molecular replacement with AMoRe32 using atomic models from the Protein Data Bank. These structures displayed the highest sequence similarity to the crystallized molecules, as detected by a FASTA search within the Protein Data Bank. A clear solution was found only for the V domains of two Fabs and the Fc dimer.29 The asymmetric unit contained one Fc and the two Fabs. In the resulting model, the amino acids that differed were replaced with the correct sequence or substituted with alanine. The best initial electron density maps were obtained using the program Buster.35 The polypeptides (without carbohydrate) were used to generate initial phases and the calculated electron density maps clearly showed the two complex oligosaccharide chains of the Fc dimer attached to Asn297; nine sugar units, including galactose on the α(1–6) linked arm and a core fucose residue, were visible. Further refinement was carried out using either CNS36 or BUSTER/TNT35 using all data to 3 Å resolution with |F|>2σ (550 reflections were rejected from the unique set). Noncrystallographic symmetry constraints were imposed between the corresponding domains of Fc chains A and B (for domains Cγ2 and Cγ3) and between the corresponding variable and constant domains of the two Fabs. Model building and structure analysis were performed using the program O37 and the CCP4 suite of programs.38 After a few rounds of refinement the C domains of the two Fabs were successively added to the model. A final round of refinement without noncrystallographic symmetry restraints (using the conjugate gradient method in CNS) further reduced Rfree to 28.8%. Two cations, a Cd2+ and a Zn2+ ion, plus one cacodylate molecule that completes the coordination sphere of the Zn2+ ion, 84 water molecules, 4 acetate ions, and 4 methylpentanediol molecules were also added to the model. The final refinement statistics are shown in Table 1; the overall Rfree=28.8% and Rwork= 22.4% to 3 Å resolution.
The two interaction sites are virtually identical, but the “elbow” angles of the two Fabs are different, as are the orientations of the Cγ2 domains relative to Cγ3. The Fab C domains and two loops of one of the Cγ2 domains in Fc proximal to the IgG hinge (Figure 2, top), have higher overall B-factors. These regions (CH1, CL, and Cγ2 loops) lie distant from the interface region, which is well defined. The total buried surface areas for the two Fab interactions are 1536 and 1659 Å2, distributed approximately equally between Fab and Fc. No electron density was visible for the HSV-1 gD part of the fusion protein, which was linked to the N-terminal hinge region of the Fc, although it is possible that proteolytic cleavage had occurred.29
Protein Data Bank accession codes
The coordinates of the structure have been deposited in the Protein Data Bank with Accession Code 2j6e and the structure factors with Code r2j6esf. Other accession codes for the molecular replacement models used are as follows: 1DN221 for the Fc fragment, 1QLR33 for the Fab heavy chain, and 2FB434 for the λ light chain.
Acknowledgements
This work was supported by NIH Grants AR 40908, AI 45011, and AI 60573 to PC. BJS, MJT and SMF thank the Biotechnology and Biological Sciences Research Council (BBSRC, UK) and the Arthritis Research Campaign (UK) for support and Susanne Bell for expert technical assistance. We also thank Helena Browne (Department of Pathology, University of Cambridge) for preparation of the gD–Fc construct.
Abbreviations used
- CDR
complementarity determining region
- FCS
fetal calf serum
- Ig
immunoglobulin
- RA
rheumatoid arthritis
- RF
rheumatoid factors
- PBS
phosphate-bufferred saline
- SpA
Staphylococcus aureus protein A
References
- 1.Mannik M, Nardella FA, Sasso EH. Rheumatoid factors in immune complexes of patients with rheumatoid arthritis. Springer Semin. Immunopathol. 1988;10:2150–2230. doi: 10.1007/BF01857226. [DOI] [PubMed] [Google Scholar]
- 2.Jefferis R, Mageed RA. The specificity and reactivity of rheumatoid factors with human IgG. Monogr. Allergy. 1989;26:45–60. [PubMed] [Google Scholar]
- 3.Renaudineau Y, Jamin C, Saraux A, Youinou P. Rheumatoid factor on a daily basis. Auto-immunity. 2005;38:11–16. doi: 10.1080/08916930400022574. [DOI] [PubMed] [Google Scholar]
- 4.Rau R, Herborn G, Zueger S, Fenner H. The effect of HLA-DRB1 genes, rheumatoid factor and treatment of radiographic disease progression in rheumatoid arthritis over 6 years. J. Rheumatol. 2000;27:2566–2575. [PubMed] [Google Scholar]
- 5.Vaughan JH. Pathogenetic concepts and origins of rheumatoid factor in rheumatoid arthritis. Arthritis Rheum. 1993;36:1–6. doi: 10.1002/art.1780360102. [DOI] [PubMed] [Google Scholar]
- 6.Kalsi J, Isenberg D. Rheumatoid factor: primary or secondary event in the pathogenesis of RA? Int. Arch. Allergy Immunol. 1993;102:209–215. doi: 10.1159/000236528. [DOI] [PubMed] [Google Scholar]
- 7.Bridges SL. Update of autoantibodies in rheumatoid arthritis. Curr. Rheumatol. Rep. 2004;6:343–350. doi: 10.1007/s11926-004-0008-1. [DOI] [PubMed] [Google Scholar]
- 8.Male D, Roitt IM, Hay F. Analysis of immune complexes in synovial effusions of patients with rheumatoid arthritis. Clin. Exp. Immunol. 1980;39:297–306. [PMC free article] [PubMed] [Google Scholar]
- 9.Tarkowski A, Czerkinsky C, Nilsson LA. Simultaneous induction of rheumatoid factor and antigen-specific antibody-secreting cells during the secondary immune response in man. Clin. Exp. Immunol. 1985;61:379–387. [PMC free article] [PubMed] [Google Scholar]
- 10.Coulie PG, van Snick J. Rheumatoid factor (RF) production during anamnestic responses in the mouse. III. Activation of RF precursors is induced by their interaction with immune complexes and carrier-specific helper T cells. J. Exp. Med. 1985;161:88–97. doi: 10.1084/jem.161.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen PP, Carson DA. New insights on the physiological and pathological rheumatoid factors in humans. In: Coutinho A, Kazatchkine M, editors. Autoimmunity: Physiology and Disease. Wiley–Liss; New York: 1994. pp. 247–266. [Google Scholar]
- 12.Harindranath N, Goldfarb IS, Ikematsu H, Burastero SE, Wilder RL, Notkins AL, Casali P. Complete sequence of the genes encoding the VH and VL regions of low- and high-affinity monoclonal IgM and IgA1 rheumatoid factors produced by CD5+B cells from a rheumatoid arthritis patient. Int. Immunol. 1991;3:865–875. doi: 10.1093/intimm/3.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani L, Wilder RL, Casali P. Human rheumatoid B-1 (CD5+ B) cells make high affinity somatically hypermutated IgM rheumatoid factors. J. Immunol. 1993;151:473–488. [PMC free article] [PubMed] [Google Scholar]
- 14.Bonagura VR, Artandi SE, Davidson A, Randen I, Agostino N, Thompson K, et al. Mapping studies reveal unique epitopes on IgG recognised by rheumatoid arthritis-derived monoclonal rheumatoid factors. J. Immunol. 1993;151:3840–3852. [PubMed] [Google Scholar]
- 15.Corper AL, Sohi MK, Bonagura VR, Steinitz M, Jefferis R, Feinstein A, et al. Structure of a human IgM rheumatoid factor Fab in complex with its autoantigen IgG Fc. Nature Struct. Biol. 1997;4:374–381. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]
- 16.Sutton BJ, Corper AL, Bonagura VR, Taussig MJ. The structure and origin of rheumatoid factors. Immunol. Today. 2000;21:177–183. doi: 10.1016/s0167-5699(00)01589-9. [DOI] [PubMed] [Google Scholar]
- 17.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 18.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Crystal structure of the C2 fragments of streptococcal protein G in complex with the Fc domain of human IgG. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 19.Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 20.Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcαR1 and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- 21.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 22.Bouvet J-P, Dighiero G. Cross-reactivity and polyreactivity: the two sides of a coin. Immunol. Today. 2000;21:411–412. doi: 10.1016/s0167-5699(00)01679-0. [DOI] [PubMed] [Google Scholar]
- 23.Davies DR, Padlan EA, Sheriff S. Antibody–antigen complexes. Ann. Rev. Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 24.Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin. Struct. Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 25.Pascual V, Randen I, Thompson K, Sioud M, Forre O, Natvig J, Capra JD. The complete nucelotide sequences of the heavy chain variable regions of six monospecific rheumatoid facors derived from Epstein–Barr virus transformed B cells isolated from the synovial tissue of patients with rheumaotid arthritis. J. Clin. Invest. 1990;86:1320–1328. doi: 10.1172/JCI114841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Victor KD, Randen I, Thompson K, Sioud M, Forre O, Natvig J, Capra JD. Rheumatoid factors isolated from patients with autoimmune disorders are derived from germline genes distinct from those encoding the Wa, Po and Bla cross-reactive idiotypes. J. Clin. Invest. 1991;87:1603–1613. doi: 10.1172/JCI115174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohi MK, Corper AL, Wan T, Steinitz M, Jefferis R, Beale D, et al. Crystallization of a complex between the Fab fragment of a human immunoglobulin M (IgM) rheumatoid factor (RF-AN) and the Fc fragment of human IgG4. Immunology. 1996;88:636–641. doi: 10.1046/j.1365-2567.1996.d01-692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunning P, Leavitt J, Muscat G, Ng SY, Kedes L. A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc. Natl. Acad. Sci. USA. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stura EA, Taussig MJ, Sutton BJ, Duquerroy S, Bressanelli S, Minson AC, Rey FA. Scaffolds for protein crystallisation. Acta Crystallog. sect. D. 2002;58:1715–1721. doi: 10.1107/s0907444902012829. [DOI] [PubMed] [Google Scholar]
- 30.Stura EA. In: Seeding techniques. In Crystallization of Nucleic Acids and Proteins: A Practical Approach 2nd edit. Ducruix A, Giegé G, editors. Oxford University Press; Oxford, UK.: 1999. pp. 177–208. [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Sweet RM, editor. Macromolecular Crystallography Part A. Academic Press; London: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallog. sect. A. 1994;50:157–163. [Google Scholar]
- 33.Cauerhff A, Braden BC, Carvalho JG, Aparicio R, Polikarpov I, Leoni J, Goldbaum FA. Three-dimensional structure of the Fab fragment from a human IgM cold agglutinin. J. Immunol. 2000;165:6422–6428. doi: 10.4049/jimmunol.165.11.6422. [DOI] [PubMed] [Google Scholar]
- 34.Kratzin HD, Palm W, Stangel M, Schmidt WE, Friedrich J, Hilschmann N. The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I. Biol. Chem. Hoppe Seyler. 1989;370:263–272. [PubMed] [Google Scholar]
- 35.Roversi P, Blanc E, Vonrhein C, Evans G, Bricogne G. Modelling prior distributions of atoms for macromolecular refinement and completion. Acta Crystallog. sect. D. 2000;56:1313–1323. doi: 10.1107/s0907444900008490. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallog. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallog. sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallog. sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallog. 1991;24:946–950. [Google Scholar]