Abstract
Yellow fever virus (YFV) is endemic in tropical and sub-tropical regions of the world, with around 180,000 human infections a year occurring in Africa. Serologic testing is the chief laboratory diagnostic means of identifying an outbreak and to inform the decision to commence a vaccination campaign. The World Health Organization disseminates the reagents for YFV testing to African reference laboratories, and the US Centers for Disease Control and Prevention (CDC) is charged with producing and providing these reagents. The CDC M-antibody capture ELISA is a 2-day test, requiring titration of reagents when new lots are received, which leads to inconsistency in testing and wastage of material. Here we describe the development of a kit-based assay (YF MAC-HD) based upon the CDC method, that is completed in approximately 3.5 h, with equivocal samples being reflexed to an overnight protocol. The kit exhibits >90% accuracy when compared to the 2-day test. The kits were designed for use with a minimum of equipment and are stored at 4 °C, removing the need for freezing capacity. This kit is capable of tolerating temporary sub-optimal storage conditions which will ease shipping or power outage concerns, and a shelf life of >6 months was demonstrated with no deterioration in accuracy. All reagents necessary to run the YF MAC-HD are included in the kit and are single-use, with 8 or 24 sample options per kit. Field trials are envisioned for the near future, which will enable refinement of the method. The use of the YF MAC-HD is anticipated to reduce materials wastage, and improve the quality and consistency of YFV serologic testing in endemic areas.
Keywords: yellow fever virus, IgM, ELISA, kit, diagnostic
Introduction
Yellow fever virus (YFV) strain Asibi is the type species of the family Flaviviridae and was first isolated from a male patient in Ghana in 1927 (Stokes, 1928) . Chiefly transmitted by Aedes spp. Mosquitoes (Strode and Rockefeller Foundation. International Health Division., 1951), it is a viscerotropic virus, causing high fever, severe headaches, back and body aches, and nausea and vomiting, which typically present 3-6 days post-exposure. While the conditions of most patients improve after about 48 hours, around 15% will develop a more severe form of the disease that starts following a brief remission of symptoms. These individuals experience high fever, bleeding, jaundice, eventual shock and multiple organ failure (CDC, 2015). Yellow fever appeared in Barbados in 1647 as a result of the slave trade (McNeill, 2004) and the first outbreak in the US followed in 1668. Significant outbreaks occurred for more than 2 centuries in cities in the south and east of the country, with the last one recorded being in New Orleans in 1905 (Pierce, 2005). Once Aedes aegypti (formerly known as Stegomyia fasciatus) was identified as the chief vector of the disease (Reed and Carroll, 1901), mosquito control measures were adopted in developed countries that eliminated yellow fever as a public health concern. Today, yellow fever is most prevalent in tropical and subtropical Africa, but is also endemic in parts of S. America.
A widely-used vaccine is available to prevent yellow fever infections, which was derived from the Asibi strain (Theiler and Smith, 1937). The 17-D vaccine is generally considered safe; however, vaccine-related adverse events have been documented (McMahon et al., 2007). While vaccination campaigns are effective in limiting yellow fever outbreaks in Africa, the sylvatic cycle propagated by wild primates is not affected, allowing YFV to persist. Numbers are estimated to be around 180,000 infections per year in Africa, with more than 25,000 deaths (WHO, 2014). The efficacy of vaccination campaigns is dependent on a number of factors: the availability of vaccine; the percentage of the population that receives the vaccine; and the speed and accuracy with which the initial outbreak is identified. This last factor is critical because an outbreak can spread quite rapidly in the time between initial suspicion of yellow fever activity, and actual diagnosis. Molecular and serological tests form the basis of laboratory diagnosis, where serology is the most common and useful of the methods for most samples from a logistical standpoint. Diagnostic confirmation by serology comprises a screening IgM test (M-antibody capture ELISA, CDC MAC-ELISA) (Martin et al., 2000), followed by plaque-reduction neutralization test (PRNT)(Lindsey, Calisher, and Mathews, 1976). The PRNT requires specific cell culture facilities that are rarely available outside of reference laboratories, and the tests themselves usually take several days to a week to produce results. This limits the speed at which a diagnosis can be arrived at to inform the decision to vaccinate a population. Nevertheless, given the currently-accepted testing algorithm, a reliable and accurate screening method is important in the identification process to expedite the mobilization of vaccine stocks.
Many of the YF affected African countries have historically relied upon the Institut Pasteur de Dakar and the World Health Organization (WHO) to provide testing reagents for yellow fever virus, which are supplied to the latter by the US CDC. These reagents are intended for use with the CDC MAC-ELISA. However, the test takes over 2 days to perform, and because the reagents exhibit lot-to-lot variation, titration is required before use. Further, storage conditions can be suboptimal, and the quality of the results can be compromised. A robust test in kit format that is both rapid and accurate would therefore improve the quality of YF testing in affected countries. Desirable characteristics for a kit-format YF IgM assay are start-to-completion <1 working day; flexibility of storage conditions; all reagents supplied either in a ready-to-use format or dried and pre-measured, requiring only the addition of a specified volume of diluent included with the kit; stability of each component over a period of at least 6 months; sensitive and specific. This paper describes the development and validity testing of a kit designed to meet these requirements.
Materials and Methods
Serum specimens
Archived human serum specimens were obtained from the Arbovirus Diseases Branch Diagnostic and Reference Activity of the Division of Vector-Borne Diseases (DVBD), Centers for Disease Control and Prevention (CDC), and anonymized prior to use. The requirement for informed consent for use of the samples was exempted with protocol #6525.0 “Use of human specimens for laboratory research on arboviruses” by the CDC Human Research Protection Office. For development and validation of the test, serum samples totaled 243, comprising 139 negatives, 100 YF positives and 4 normal antigen (background) responders. Of the positives, 5 were from natural yellow fever infections (1 of Nigerian and 4 of Brazilian origin); 11 were from patients that experienced YF vaccine-related adverse events, and the remainder were from otherwise healthy YF vaccine recipients. The negatives were sera from patients suspected of having arboviral disease but which tested negative in the arboviral screening assays. Prior to use in the tests, samples were diluted 1:100 (based on initial evaluation in a <1 day protocol) in sample diluent (PBS/0.05% tween-20).
Determination of assay conditions
The CDC MAC-ELISA was used as the basis for kit development (Martin et al., 2000) and was considered the gold standard to which accuracy results were compared. The CDC MAC-ELISA consisted of the following: the inner 60 wells of microtiter plates were coated overnight at 4 °C with 75 μl/well goat anti-human IgM (Kirkegaard and Perry, Gaithersburg, MD) diluted 1:2000 in carbonate buffer pH 9.6, followed by a 30 minute blocking step using 200 μl/well 5% nonfat dried milk with 0.5% Tween-20. Plates were washed 5X, and serum samples diluted 1:400 in wash buffer (PBS/0.05% Tween-20) were added to 6 wells and incubated for 1 h at 37 °C. Plates were washed 5X and previously titrated viral and normal antigens were added in wash buffer to 3 wells each per sample, followed by an overnight incubation at 4 °C. After washing the plates 5X, a previously-titrated pan-flavivirus MAb 6B6C-1 HRP conjugate (Tsai et al., 1987) was added and incubated for 1 h at 37 °C. Plates were washed 10X and 75 μl/well Enhanced K-Blue TMB substrate (Neogen Corp,Lexington, KY) was added. Plates were incubated for 10 min at room temperature, after which the reaction was stopped using 50 μl/well 1N H2SO4. Reactions were read at 450 nm. A panel of eight serum samples consisting of 2 high, 1 medium, 1 low YF IgM-positive, 2 equivocals, 1 negative and 1 background responder (test panel) was used to monitor the development phase of the kit. The CDC MAC-ELISA was reformulated so that a <1 day test was feasible (the Comparative MAC-ELISA), and the concentrations of the assay components in this test provided a starting point for kit development. Each kit was intended to test 8 patient samples in triplicate, or 24 in singleplex. The Comparative MAC-ELISA used coating antibody as for the CDC MAC-ELISA and substituted 3% goat serum in PBS for the standard milk block. Control and test samples were diluted at 1:100 and incubated at 28 °C for 30 min, antigen was diluted at 1:50 and incubated at 4°C for 2 h, conjugate was diluted in block at 1:1500 and incubated for 30 min at 28 °C, substrate was added at 75 μl/well and KPL Stop Solution was added at 75 μl/well. Results were expressed as P/N values: the mean optical density (OD) of the sample reacted on viral antigen divided by the mean OD of the negative control reacted on the viral antigen. Tests were considered valid if the P/N of the plate positive control serum was ≥2.0, and the mean positive control OD reacted on viral antigen divided by the OD of the positive control reacted on the normal antigen was ≥2.0. For development of the yellow fever kit, each of the assay components were investigated individually and in combination with one another to determine the optimal conditions intended to meet the test requirements. The variables were: storage temperatures, component concentration, alternative buffers and stabilizing conditions, and brand of substrate. Not all variables were pertinent to all components, and specifics are listed below. Initially, each component was assayed at 0, 3, 6-8 and 12 months after preparation to measure stability under varying conditions. The test panel was used to investigate the viability of the potential kit components under these variable conditions by comparing results of the Comparative MAC-ELISA to those of tests where the component under investigation was substituted into the Comparative MAC-ELISA.
Individual components
Storage
All components were stored in the dark at either 4 °C or room temperature (RT) and were tested for activity at 0, 3, and 6 months after preparation; some were tested for up to a year. A subset of each component at each temperature was removed after 3 months and transferred to a 37 °C incubator for 2 weeks to simulate suboptimal shipping conditions.
Lyophilization of reagents
Lyophilization as noted below for specific reagents was performed using a Labconco Freeze Dry System (Labconco Corp., Kansas City, MO). Prepared reagents in stoppered borosilicate glass vials (Kimble Chase, Vineland, NJ) were frozen overnight at −20 °C. Vials were then placed in the freeze dry unit and frozen at −30 °C under vacuum for 18 hours, and then dried for 6 hours under vacuum at +30 °C.
Preparation of microtiter plates
Immulon 2 HB microtiter plates (Thermo Scientific, Rockford, IL) were coated using the CDC MAC-ELISA protocol. Plates were washed 5X using a plate washer, and commercial or laboratory-prepared stabilizing solutions were applied at volumes of 100 μl/well and incubated for 1 hour at room temperature (22.5 °C) as follows: Stabilguard (Surmodics, Inc., Eden Prarie, MN); Stabilcoat (Surmodics, Inc.); Immunoassay Stabilizer (Advanced Biotechnologies, Inc., Columbia, MD); and 6% polyvinyl alcohol (Sigma-Aldrich, St. Louis, MO) in H2O (Kolosova et al., 2006) after blocking for 45 min with 3% goat serum (Life Technologies, Grand Island, NY) in PBS. In addition, coated, blocked, unstabilized plates were also analyzed. Stabilizing solutions were removed using the aspirate function of the plate washer, plates were allowed to air dry at room temperature for 3 hours, and were stored in the dark in plastic bags containing a silica gel packet, and with air removed.
Positive and negative control sera
Yellow fever-positive control and negative control sera from the Diagnostic and Reference Activity, Arbovirus Diseases Branch, were prepared for performance analysis: 1:400 and 1:100 dilutions of positive control serum in Stabilzyme Select (Surmodics); 10% Stabilzyme Select in wash buffer; and a 1:100 dilution only in PBS/0.05% tween-20 (wash buffer). The negative control serum was prepared using the 3 diluents at 1:100 only. To increase the chances of being able to survive non-optimal storage and shipping temperatures, control sera were lyophilized into single-use aliquots. All aliquots were reconstituted in dH20 to the original volume of 400 μl.
Antigens
Two YF viral antigen lots (A and B) were used in these experiments. Lot A was made with wild-type (Asibi) virus and lot B was made with 17D vaccine strain. Both were propagated by infecting Vero cells where supernatants were harvested between days 4-6, adding media back to each flask after harvesting. The supernatants were clarified by centrifugation at 2400×g for 10 min at 4 °C, followed by the addition of 20% FBS (Atlas Biologicals, Fort Collins, CO). Lot A was inactivated with 5mRAD gamma irradiation, and then concentrated 20X using Centricon-70 filters (EMD Millipore, Darmstadt, Germany). Lot B was inactivated with 4.5 mRAD gamma irradiation followed by further inactivation with 0.025% β-propiolactone for 24h at 4 °C. Two forms of infectivity assays were performed in duplicate on the final products to confirm complete inactivation: plaque titration in Vero cells starting with undiluted material and inoculation into flasks of Vero cells passaged once a week for 3 weeks with checking for cytopathic effect (Goodman et al., 2014). Normal antigen was prepared in the same manner using uninfected Vero cells. The antigen stocks were frozen at −70 °C until use.
Lots A and B antigen stocks were diluted in 30% goat serum/10X wash buffer and lyophilized, so that the concentrations after reconstitution in dH20 were 1:50 and 1:35, respectively, in 3% goat serum/1X wash buffer. Additionally, lot B was diluted in 10X wash buffer and lyophilized so that the concentration after reconstitution in dH20 was 1:35 in 1X wash buffer. Lot B was also lyophilized in 2.5 % trehalose (Jain and Roy, 2010) in wash buffer. Normal antigens were treated identically to the viral antigens. The use of concentrated buffers was a practical step necessary to reduce the lyophilization volumes, which were sufficient for use with a single 96 well plate after reconstitution.
Conjugate
Flavivirus group-reactive monoclonal antibody 6B6C-1 conjugated to horseradish peroxidase (HRP) was used to detect the reactions. Lyophilized preparations were made in 30% goat serum/10X wash buffer so that the final conjugate dilution after reconstitution was 1:750 in 3% goat serum/1X PBS and Candor HRP Stabilizer (Boca Scientific, Boca Raton, FL). Liquid preparations were made at 1:1500 in Guardian Peroxidase Conjugate Stabilizer (Thermo Scientific, Rockford IL), Stabilzyme Select (Surmodics) and Stabilzyme HRP Stabilizer, and at a 1:23 dilution in Stabilzyme Select having a final dilution of 1:1500 in 3% goat serum/1X PBS, when the conjugate solution was added to reconstituted lyophilized 30% goat serum/10X PBS. All liquid preparations were stored in brown vials to protect the conjugate from light degradation.
Substrate
The following substrates were aliquotted into amber HDPE vials for use with single plates: Enhanced K-Blue TMB substrate (Neogen Corp), Surmodics BioFX TMB (Surmodics IVD, Eden Prairie, MN), Surmodics BioFX TMB Supersensitive (Surmodics IVD), and KPL Microwell Peroxidase 2 Component Substrate System (KPL, Gaithersburg, MD).
Stop solution
KPL Stop Solution was aliquotted into clear polypropylene vials for use with single plates.
Diluents
1X wash buffer (PBS/0.05% tween 20) was aliquotted into clear polypropylene vials to serve as a diluent for patient samples. Deionized water was aliquotted similarly and used to reconstitute lyophilized kit components.
Wash buffer
A 10X concentrate of wash buffer was sterile-filtered and placed in 2 sterile 50 ml centrifuge tubes.
Incubation conditions
To simulate laboratory conditions in tropical regions, tests were optimized with serum and conjugate incubations at 28 °C. Based upon previous experience that indicated the critical nature of the antigen incubation in the creation of a clean and sensitive signal, the antigen incubation step was investigated under temperature conditions of 4 °C, 28 °C and 37 °C for lengths of time varying from 30 min to overnight.
Stability studies
Each component stored at 4 °C and RT was substituted individually into the Comparative MAC-ELISA where all the other components were freshly-prepared. Results were compared to the Comparative MAC-ELISA. The “test panel” of sera was used to compare components that had been stored 0, 3, and 6 months or longer, and for 3 months followed by 2 weeks at 37 °C. Combinations of components that performed well in the individual stability studies were combined into “kits” and these were assayed using the “test panel” at 0 months after production. The best of these combinations was identified and used to determine the assay cutoff and accuracy of the kit. The kit was named “YF MAC-HD (yellow fever M-antibody capture-half day)”. The stabilities of the individual components chosen for use in the kit were also assayed at 12 months after production. The resulting kit was assayed for stability in the same manner as for the individual components using a test panel.
Determination of assay cutoff
Forty-eight anonymized serum samples encompassing 29 YF IgM-positives of a range of P/N’s from previous diagnostic IgM and PRNT results (including equivocals) and 19 negative samples were tested in the YF MAC-HD and in the YF MAC-HD with an overnight antigen incubation (YF MAC-ON) to determine the assay cutoff. All samples were retested in the CDC MAC-ELISA to confirm that the samples had not deteriorated during the time they were archived. Receiver operator characteristic (ROC) curves were computed for both protocols with the “ROCR” package in R, version 3.0.1 (RCoreTeam, 2014). Estimates of sensitivity and specificity computed using the ROC curve were used to guide the choice of cutoff, found as the value for which sensitivity was the closest to specificity. Estimates of apparent error rate (APER) were computed using leave-one-out cross validation of the data set. Ninety-five percent confidence intervals (CI) were calculated using Wilson’s score interval for proportions.
Accuracy and precision studies
For the accuracy and precision studies, the YF MAC-HD kits were used with 8 serum samples per plate, each sample tested in triplicate on both viral and normal antigens. Kits were prepared to test a panel of 196 previously-assayed serum samples at 0 months after production and at 6 months after storage at 4 °C. The panel did not include any of the samples used in the cut-off determination. The within-lab precision of the YF MAC-HD kits was determined using 6 anonymized YF IgM-positive and 2 negative serum samples. The positive samples encompassed high, medium and low P/N’s as determined by the CDC MAC-ELISA. These 8 samples were tested on 6 days spaced evenly over 1 month, and 2 tests per day were performed, spaced 2 hours apart.
Cross-reactivity with samples from other flaviviral infections and malaria
Freshly-prepared kits were assayed for cross-reactivity to IgM from dengue patients using 8 known high DEN IgM-positive sera from primary infections. In addition, 7 WNV IgM-positive, 8 Zika virus IgM-positive and 8 malaria-positive samples were assayed for cross-reactivity in the YF MAC-HD.
Results
Stability, choice of individual assay components, and incubation conditions
All assay components were used at the volumes specified in the CDC MAC-ELISA. Prior to choosing the individual components for kit compilation, each was tested for stability when stored at 4 °C and RT for at least 6 months and at 37 °C for 2 weeks after 3 months at either 4 °C or RT. The stability data for the individual components are presented in the tabs of the Supplemental Data file S1. The kit components were chosen based on stability, and compatibility with the other reagents. While many of the stabilizers used conferred good individual performance for the reagents when substituted into the Comparative ELISA, the cumulative effects of blockers and stabilizers in each of the components when assembled into a kit caused reduced signal. Hence, the number of components in which stabilizers were used was reduced to only those that were absolutely necessary to confer stability. The final kit components were as follows: Coated plates were stabilized with Stabilguard, dried, and stored in sealed bags with a silica gel packet; 1X wash buffer per the CDC MAC-ELISA protocol was used for all wash steps and as the sample diluent; sterile water for reconstitution of lyophilized components; positive and negative serum controls were lyophilized at 1:100 in 1X wash buffer; lot B viral and normal antigens were lyophilized in 10X wash buffer such that the reconstituted product was at a 1:35 dilution in 1X wash buffer; the conjugate comprised 2 components: 6B6C-1 HRP diluted 1:23 in Stabilzyme Select and stored in an amber vial, and lyophilized 30% goat serum/10X wash buffer for a final 1:1500 dilution of conjugate in 3% goat serum/1X wash buffer; the substrate was K-Blue TMB ELISA stored in an amber vial; KPL Stop Solution stored in a clear polypropylene vial. Overall there appeared to be slightly less variation in results when the reagents were stored at 4 °C as opposed to RT, although RT was still a feasible storage temperature (kit component by sample tab of Supplemental Data file S1). The antigen incubation times and temperatures showed that a 2 hour incubation at 4 °C was optimal. Signals diminished with shorter times and did not increase with longer times (data not shown). Alternative temperatures of 28 °C and 37 °C resulted in elevated normal antigen ODs and therefore were suboptimal. Limited data (not shown) suggested that serum and conjugate incubation steps could be successfully performed at temperatures that ranged between 21 °C and 37 °C, but that 4 °C was unsuitable, thus allowing for some flexibility in ambient temperature if an incubator was unavailable.
Stability of the YF MAC-HD kit
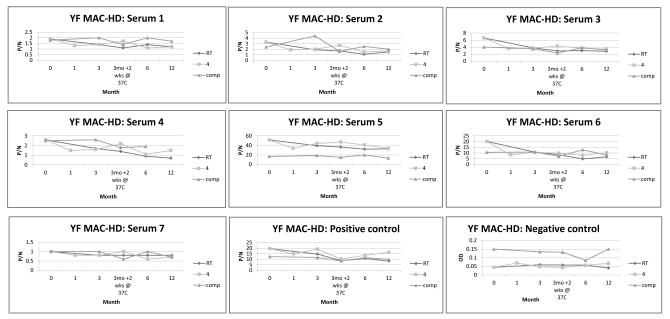
The assembled kit components were assayed over a period of 1 year after production to determine shelf life (Figure 1). The only major differences seen between the Comparative MAC-ELISA and the YF MAC-HD kit were the optical densities of the negative control across the time points, which maintained OD’s of close to 0.05 for the kit at both 4 °C and RT storage temperatures, whereas the Comparative MAC-ELISA OD’s varied between 0.085 and 0.15. The resulting P/N values for the test samples tested with the kit after storage at the 2 temperatures were similar to those yielded by the Comparative ELISA. A downward trend of the P/N’s was observed over time in which mean P/N reductions of 29% (RT) and 17% (4 °C) from month 0 to month 12 when the kit was compared to the Comparative MAC-ELISA. No obvious drop in the P/N’s was seen due to the 3 month storage at 4 °C or RT followed by 2 weeks at 37 °C (Figure 1) but significant reductions in OD were observed for serum 6 and the positive control under both conditions (Figure 2). When assayed with the other kit components, the positive control exhibited an OD of approximately 39% of the month 0 value when it was stored for 12 months at RT, whereas no deterioration was seen for the positive control when the kit was stored at 4 °C. The kit stored at 4 °C and tested at 12 months after preparation gave P/N’s and OD’s consistent with those measured at 6 months with the exception of sample 4 which showed a reduction in OD (Figure 2). By contrast, the kit that was stored at RT for 1 year showed gradual deterioration in overall OD’s across of the test serum panel (Figure 2), although the resulting P/N’s were not adversely affected.
Figure 1.
Stability of the YF MAC-HD kit over time.
The stability of the kit was measured at 0, 3, 6 and 12 months after production and storage at either RT or 4 °C using 8 serum samples, where sample 8 was a background responder (not plotted). A 2 week stress test at 37 °C was performed on kits after 3 months of storage. Test results were reported as P/N values and were plotted alongside those of the Comparative MAC-ELISA.
Figure 2.
Stability of the YF MAC-HD expressed as optical densities over time for kits stored at 4 °C and RT. Sample 8 was a background responder and was not plotted.
Determination of cut-off and testing algorithm
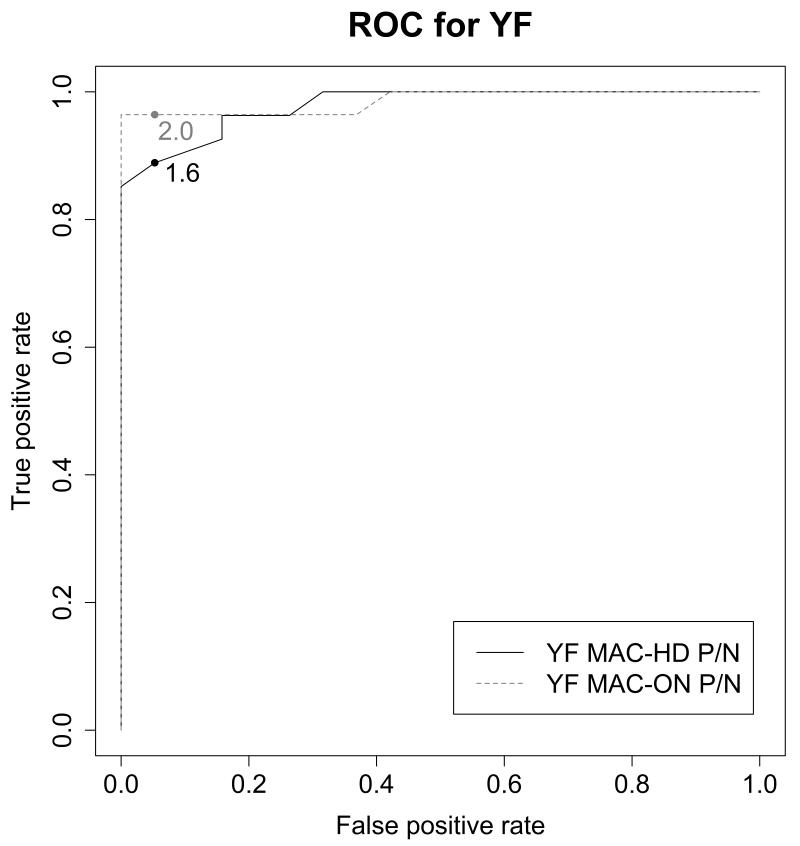
The cut-offs for the YF MAC protocols were informed by using ROC curve analysis, which considers the best combination of specificity and sensitivity by comparing the test results to those of a gold standard (in this case the CDC MAC-ELISA). Of the 48 samples, 2 were disqualified from the YF MAC-HD analysis and 1 for the YF MACON analysis due to uninterpretable results (see Supplemental data file 2). For the YF MAC-HD, a cut-off of P/N=1.6 was identified with a sensitivity of 88.9% and specificity of 94.7%. A cut-off of P/N=2.0 was determined for the overnight method, with a sensitivity of 96.4% and specificity of 94.7%. The ROC curves and cut-off points are shown in Figure 3. Based upon these data, an equivocal zone was established for each test: greater than or equal to 1.2, but less than 2.0 for the 2 h test; greater than or equal to 2.0, but less than 3.0 for the YF MAC-ON. When only the YF MAC-HD results that fell outside of the equivocal zone were considered, the proportion of correctly classified positive samples was 100% and the proportion of correctly classified negative samples was 100%. For the YF MAC-ON results that fell outside of the equivocal zone, the proportion of correctly classified positive samples was 95.5% and was 100% for negative samples. A statistical summary is shown in Table 1. Of the remaining 9 samples that landed within the equivocal zone for YF MAC-HD (and therefore were not classified as correct or incorrect), 6 gave correct results when tested in the YF MAC-OD (not shown). The testing algorithm that suggested the best combination of efficient use of time and most accurate results, was to use the YF MAC-HD as the initial screen, and to reflex any equivocal or uninterpretable results to the YF MAC-ON.
Figure 3.
Receiver operator characteristic curves to determine P/N cut-off values in the YF MAC-HD and the YF MAC-ON. The optimal balance between sensitivity and specificity based upon the results with true positives and negatives was P/N of 1.6 for YF MAC-HD and 2.0 for YF MAC-ON.
Table 1.
Statistical summary of YF MAC-HD and YF MAC-ON methods using cut-offs derived by ROC analysis.
| Method | N | Sensitivity (95% CI)* | Specificity (95% CI) | APER (95% CI) |
|---|---|---|---|---|
| YF MAC-HD | 46 | 88.9 (68.9, 96.7) | 94.7 (71.5, 99.2) | 13.0 (6.1, 25.7 |
| YF MAC-HD excluding equivocals | 37 | 100 (81.1, 100) | 100 (68.7, 100) | 0.0 (0.0, 10.4) |
| YF MAC-ON | 47 | 96.4 (79.3, 99.5) | 94.7 (71.5, 99.2) | 4.3 (1.1, 14.3) |
| YF MAC-ON excluding equivocals | 38 | 95.5 (74.7, 99.3) | 100 (73.6, 100) | 2.8 (0.5, 14.2) |
Confidence intervals derived using leave-one-out cross-validation analysis
Accuracy and Precision
The sample set used to test the YF MAC-HD for accuracy was intentionally compiled so that it included some “true” equivocals and background responders, based upon results of the CDC MAC-ELISA, to make the set as realistically challenging as possible. The set consisted of 4 background responders, 10 equivocals, 65 positives and 117 negatives. Freshly-prepared YF MAC-HD kits were used to test the 196 samples (month 0). The same was done using kits that were stored for 6 months at both 4 °C and RT prior to use. Results were analyzed using the cut-off criteria of P/N < 1.2 = NEG, 1.2 ≤ P/N < 2.0 = EQUIV, and P/N ≥ 2.0 = POS. Any POS or EQUIV samples for which the viral antigen OD was less than 2X of the normal antigen was classed as a background responder (BKGD). The YF MAC-ON protocol was used to resolve any equivocal or background results. The sensitivity and specificity of the YF MAC-HD were calculated, where “true” equivocals and background responders were not included (because they were neither true positives nor true negatives), and where equivocal and background results for true positives or true negatives in the YF MAC-HD were counted as incorrect. Sensitivity of the YF MAC-HD was 84.6% (95% CI 72.2, 92.1) and specificity was 86.3% (95% CI 77.7, 92.0). Accuracy (where “true” EQUIV and BKGD samples were included) was 82.1% (95% CI 76.2, 86.9). After retesting any samples that resulted as EQUIV or BKGD by using the YF MAC-ON with cut-off criteria of P/N < 2 =NEG; 2.0 ≤ P/N < 3.0 = EQUIV and P/N ≥ 3.0 = POS, resulted in an increase in the overall accuracy (88.3%, 95% CI 83.0, 92.1) of the test. At 6 months after kit preparation and storage at 4 °C, the sensitivity was 89.7% (95% CI 78.6, 95.4), specificity was 95.7% (95% CI 89.2, 98.3), and overall accuracy was 89.8% (95% CI 84.8, 93.3). After reflexing EQUIV and BKGD results to the YF MAC-ON, overall accuracy improved to 90.8% (95% CI 86.0, 94.1).
The between test precision of the YF MAC-HD was determined using 8 samples that varied between high positive to low negative that were tested twice per day, 2 h apart, on each of 6 days spaced evenly over 1 month. Precision expressed as %CV was a mean of 11.1% across all samples, with a range of 7-14.5%.
Cross-reactivity
Of the 8 sera with high P/N values from primary dengue infections, 7 gave positive YF IgM results in both the YF MAC-HD and the YF MAC-ON. However, the P/Ns were an average of 9-fold higher for the YF MAC-HD and 7 – fold higher for the YF MAC-ON (Table 2) compared to the P/Ns for dengue IgM. Two each of the 7 WNV and 8 Zika virus infections showed cross-reactivity in the YF MAC-HD. One malaria-positive sample gave a positive result in the YF MAC-HD. This sample was tested further using CDC MAC-ELISA for DENV and WNV (data not shown) gave negative results for those viruses. A yellow fever virus PRNT indicated this was a false-positive result; however, positive and equivocal results of YF IgM immunofluorescence assay and YF CDC MAC-ELISA, respectively, suggested the presence of low amounts YF IgM.
Table 2.
YF MAC-HD results of serum samples from confirmed flavivirus and Malaria infections.
| Sample | Confirmed Dx | Dx result* | YF MAC-HD P/N |
|---|---|---|---|
| 1 | DEN | 62.1 | 3.7 |
| 2 | DEN | 58.9 | 1.1 |
| 3 | DEN | 11.6 | 2.2 |
| **4 | DEN | 15.1 | 14.1 |
| 5 | DEN | 19.3 | 4.1 |
| 6 | DEN | 19.4 | 9 |
| 7 | DEN | 16.7 | 2.9 |
| 8 | DEN | 11.5 | 3.2 |
| 9 | WN | 9 | 5.7 |
| 10 | WN | 4.7 | 1 |
| 11 | WN | 6.4 | 0.9 |
| 12 | WN | 21.2 | 12.6 |
| 13 | WN | 22.4 | 0.5 |
| 14 | WN | 3 | 1 |
| 15 | WN | 7.4 | 0.9 |
| 16 | ZIKA | 12.1 | 0.7 |
| 17 | ZIKA | 11.1 | ***1.2 |
| 18 | ZIKA | 6.6 | 0.6 |
| 19 | ZIKA | 8.1 | 3.6 |
| 20 | ZIKA | 36.1 | 0.6 |
| 21 | ZIKA | 4.4 | 0.7 |
| 22 | ZIKA | 15.6 | 0.6 |
| 23 | ZIKA | 13.3 | 7.2 |
| 24 | Malaria | POS | 0.8 |
| 25 | Malaria | POS | ***bkgd |
| 26 | Malaria | POS | 1 |
| 27 | Malaria | POS | 0.6 |
| 28 | Malaria | POS | 0.6 |
| 29 | Malaria | POS | 0.8 |
| 30 | Malaria | POS | ***bkgd |
| 31 | Malaria | POS | ****4.2 |
Diagnostic results presented are IgM ELISA P/N for DEN, WN and ZIKA where P/N 2.0>=3.0 is equivocal and P/N>=3.0 is positive; CareStart rapid antigen test was used for malaria
Sample 4 is a secondary DEN infection as determined by PRNT where there is a <4-fold difference in titer between results for each of the 4 serotypes
Samples 17, 25 and 30 were confirmed as negative by YF MAC-ON
Sample 31 was negative by YF and DEN1/2 PRNT; positive by YF IgM IFA, positive YF MAC-ON, equivocal by YF CDC MAC-ELISA and negative by WN and DEN CDC MAC-ELISA
Discussion
The YF MAC-HD was designed as a single-use test for up to 8 samples tested in triplicate, or up to 24 samples tested singly. The YF MAC-HD proved to be sensitive, specific, and precise. When equivocal or background responders were identified by the YF MAC-HD they were reflexed to the YF MAC-ON, which resulted in an overall accuracy of >90% even after the kits had been stored for 6 months at 4 °C and where the serum set of 196 included purposefully challenging borderline samples. The initial ROC data indicated that the YF MAC-ON was capable of greater sensitivity and specificity than the YF MAC-HD. In the interests of efficient turnaround time for the majority of samples, an equivocal zone was established where samples giving equivocal (or background) results in the YF MAC-HD were reflexed to the YF MAC-ON which improved accuracy for these borderline samples.
The kit that was stored at RT for 1 year showed gradual deterioration in overall OD’s across the samples, but interestingly, the OD’s and P/N’s of the individual components stored at RT for 12 months did not reveal the source of this deterioration, which points to a combinatorial effect of 2 or more of the components. While accuracy studies were not performed on kits used 12 months after production, the results for the single kit stored at 4 °C provided an indication that a 12 month shelf life was achieved. The results also indicated that transient non-optimal storage conditions, such as a 37 °C for 2 weeks as might be experienced during shipment, would not adversely affect the P/N results of the test samples. The positive control only would likely show a drop in OD if the kits were otherwise stored correctly at 4 °C, but not so much as to render the tests invalid. Thus, the kits would still be of use after being subjected to non-optimal shipping conditions or storage at RT for a limited time, and the use of cold packs during shipment would mitigate against potential deterioration.
The YF MAC-HD is intended for use in a laboratory setting that has standard equipment such as a refrigerator, calibrated pipettors and a plate reader. The use of a plate washer is preferable, but the use of a squirt bottle to fill the wells and removal of buffer using a pipettor is an acceptable alternative. The test was purposefully optimized with incubation temperatures of 28 °C to simulate the ambient temperatures of laboratories at tropical latitudes. If a 28 °C incubator is available this would afford the greatest consistency of results, but incubation on a benchtop would be an acceptable alternative. Because YFV is a BSL-3 agent, safe handling procedures such as use of personal protective equipment are needed regardless of the laboratory conditions. If a safety cabinet is unavailable for use with this test, samples can be heat inactivated at 56 °C for 30 min (Robinson et al., 2010), under which conditions arboviruses of all species are inactivated.
A limitation to this assay is that because of the cross-reactive nature of the whole viral YF antigen with flaviviral antibodies, serum samples that produce positive results should at least be checked using a DENV IgM ELISA, for which kits are commercially available (Hunsperger et al., 2014), and ideally with tests for other endemic flaviviruses, such as WNV IgM and Zika viruses. The cross-reactivity observed with the YF MAC-HD kit represented a worst-case scenario in which the DEN-positive samples were known to have high P/N ratios to DENV. Cross-reactivity was observed to a lesser extent with WN and Zika viruses, but this serves to illustrate the care with which serodiagnosis must be made. Cross-reactivity among flaviviruses is well-recognized and the only feasible way to avoid such cross-reactivity in ELISA would be to employ a recombinant antigen in which any cross-reactive epitopes were engineered to be reduced or absent (Chiou et al., 2008). Such an assay is urgently-needed; however, the kit developed here was not intended or anticipated to be an improvement over the existing CDC MAC-ELISA in terms of specificity; rather, the intent was to improve the usability of the existing test. The performance of true confirmatory testing by PRNT is only possible in reference laboratories equipped with cell culture facilities, and in areas such as Africa, access to these is both limited and often time-prohibitive when an outbreak is at hand. In addition, even confirmatory PRNTs are unlikely to yield definitive results where a secondary flaviviral infection is present, a very acute infection is present, or recent vaccination has occurred. Ideally, acute samples should be tested by RT-PCR methodology, where positive results are definitive, especially when a yellow fever outbreak is suspected. The lack of availability of such tests in African regional laboratories however, makes the practical application of molecular diagnosis difficult.
Another limitation is that the kit is designed for the analysis of only 8 samples, each tested in triplicate on the viral and normal antigens. This allows for a greater level of accuracy in results, and the avoidance of false-positives caused by occasional discrepant wells. While the recommended protocol for this kit is for 8 samples, a 24-sample option where only the controls are run in triplicate will make this more cost-efficient if necessary. The importance of testing the sample on the normal antigen to identify false-positives due to direct nonspecific interaction between IgM and MAbs has been documented (Prince and Yeh, 2013). A point-of-care test such as a dipstick assay would be desirable as an indicator of yellow fever, but to date is not available. A test capable of detecting viral non-structural protein-1 in acute sera might prove a useful adjunct to the YF-MAC-HD in a similar capacity to those designed for dengue virus (Bessoff et al., 2010).
Recently-vaccinated patients will produce positive reactions in the YF MAC-HD; indeed, most of the samples used to validate the method were from vaccinees. Therefore, it is imperative to gather the vaccination history of the patient to aid in interpretation of results. Interpretation must also incorporate information regarding date of onset of symptoms and date of sample acquisition. In our experience, samples taken less than 9 days post-onset of symptoms can sometimes produce false-negative IgM results; therefore the acquisition of a follow-up sample would be required for a definitive interpretation. This was illustrated by a malaria sample that appeared to be potentially cross-reactive in the YF MAC-HD, which was difficult to interpret with certainty without knowing the vaccination history of the patient. The lack of serum from natural infections limited statements regarding the usefulness of the YF MAC-HD with these infections; however, one sample from Nigeria and 4 from Brazil indicate that the YF MAC-HD is capable of detecting antibodies directed to African and South American viruses. The YF MAC-HD was validated using serum samples only and the utility of the test is unknown for CSF or alternate sources of antibodies.
The development, stability and validation of the kit has been performed here under a controlled laboratory environment using samples of known pedigree. Further evaluation of the assay under the more challenging circumstances of laboratories in YF-endemic areas will be undertaken in future field trials. In this way, further experience can be gained and the kit will be refined as necessary before it is considered for use in the capacity intended. If the field trials are successful, we plan to have kits manufactured in larger numbers. Currently the only commercial source of YF ELISA kits is in Peru (Tariki Fiebre Amarilla, Instituto Nacional de Salud).
In conclusion, the YF MAC-HD shows great promise as a means to improve the ease of use and quality of serological testing for YFV infections in tropical environments, and should mitigate the need for on-site standardization of reagents and remove the need for freezing of test components. The details of the kit development contained in this manuscript will hopefully be of help to others in the field, as this type of information is generally lacking in the literature due to the commercial nature of most kits.
Supplementary Material
Acknowledgements
The authors wish to thank Dr. Ana Maria Bispo (Istituto Oswaldo Cruz, Fiocruz, Brazil) for providing serum samples from naturally-acquired infections; and Kennedy Odhiambo Ojwang, Plague Lab, Uganda Virus Research Institute, Arua Uganda, for the malaria samples.
References
- Bessoff K, Phoutrides E, Delorey M, Acosta L, Hunsperger E. Utility of a commercial nonstructural protein 1 antigen capture kit as a dengue virus diagnostic tool. Clinical and vaccine immunology. 2010;17:949–53. doi: 10.1128/CVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC 2015 http://www.cdc.gov/yellowfever/symptoms/index.html.
- Chiou S-S, Crill W, Chen L-K, Chang G-J. Enzyme-linked immunosorbent assays using novel Japanese encephalitis virus antigen improve the accuracy of clinical diagnosis of flavivirus infections. Clinical and vaccine immunology. 2008;15:825–35. doi: 10.1128/CVI.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CH, Russell BJ, Velez JO, Laven JJ, Nicholson WL, Bagarozzi DA, Moon JL, Bedi K, Johnson BW. Development of an algorithm for production of inactivated arbovirus antigens in cell culture. Journal of virological methods. 2014;208:66–78. doi: 10.1016/j.jviromet.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsperger E, Yoksan S, Buchy P, Nguyen V, Sekaran S, Enria D, Vazquez S, Cartozian E, Pelegrino J, Artsob H, Guzman M, Olliaro P, Zwang J, Guillerm M, Kliks S, Halstead S, Peeling R, Margolis H. Evaluation of commercially available diagnostic tests for the detection of dengue virus NS1 antigen and anti-dengue virus IgM antibody. PLoS neglected tropical diseases. 2014;8:e3171. doi: 10.1371/journal.pntd.0003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain NK, Roy I. Coligan John E., editor. Trehalose and protein stability. Current protocols in protein science. 2010 doi: 10.1002/0471140864.ps0409s59. editorial board. [et al.] Chapter 4, Unit 4 9. [DOI] [PubMed] [Google Scholar]
- Kolosova A, Shim W-B, Yang Z-Y, Eremin S, Chung D-H. Direct competitive ELISA based on a monoclonal antibody for detection of aflatoxin B1. Stabilization of ELISA kit components and application to grain samples. Analytical and bioanalytical chemistry. 2006;384:286–94. doi: 10.1007/s00216-005-0103-9. [DOI] [PubMed] [Google Scholar]
- Lindsey HS, Calisher CH, Mathews JH. Serum dilution neutralization test for California group virus identification and serology. Journal of clinical microbiology. 1976;4:503–10. doi: 10.1128/jcm.4.6.503-510.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. Journal of clinical microbiology. 2000;38:1823–6. doi: 10.1128/jcm.38.5.1823-1826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AW, Eidex RB, Marfin AA, Russell M, Sejvar JJ, Markoff L, Hayes EB, Chen RT, Ball R, Braun MM, Cetron M, Yellow Fever Working G. Neurologic disease associated with 17D-204 yellow fever vaccination: a report of 15 cases. Vaccine. 2007;25:1727–34. doi: 10.1016/j.vaccine.2006.11.027. [DOI] [PubMed] [Google Scholar]
- McNeill JR. “Yellow Jack” and Geopilitics: Environment, Epidemics, and the struggles for Empire in the American Tropics, 1650-1825. OAH Magazine of History. 2004;18:9–13. [PubMed] [Google Scholar]
- Pierce J, Writer J. Yellow Jack: How yellow fever ravaged America and Walter Reed discovered its deadly secrets. John Wiley & Sons; Hoboken, NJ: 2005. p. p3. [Google Scholar]
- Prince H, Yeh C. Reactivity of human IgM binding murine monoclonal 6B6C1 (IgG2a) with other murine monoclonal IgG antibodies. Journal of clinical laboratory analysis. 2013;27:27–30. doi: 10.1002/jcla.21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCoreTeam . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. http://www.R-project.org/ [Google Scholar]
- Reed W, Carroll J. The Prevention of Yellow Fever. Public health papers and reports. 1901;27:113–29. [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Featherstone D, Vasanthapuram R, Biggerstaff BJ, Desai A, Ramamurty N, Chowdhury AH, Sandhu HS, Cavallaro KF, Johnson BW. Evaluation of three commercially available Japanese encephalitis virus IgM enzyme-linked immunosorbent assays. The American journal of tropical medicine and hygiene. 2010;83:1146–55. doi: 10.4269/ajtmh.2010.10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes AB, Johannes H, Hudson P. Experimental transmission of yellow fever into laboratory animals. Am. J. Trop. Med. 1928;8:103–164. [Google Scholar]
- Strode GK, Rockefeller Foundation. International Health Division . Yellow fever. 1st ed McGraw-Hill; New York: 1951. [Google Scholar]
- Theiler M, Smith HH. The Effect of Prolonged Cultivation in Vitro Upon the Pathogenicity of Yellow Fever Virus. The Journal of experimental medicine. 1937;65:767–86. doi: 10.1084/jem.65.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TF, Bolin RA, Montoya M, Bailey RE, Francy DB, Jozan M, Roehrig JT. Detection of St. Louis encephalitis virus antigen in mosquitoes by capture enzyme immunoassay. Journal of clinical microbiology. 1987;25:370–6. doi: 10.1128/jcm.25.2.370-376.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Yellow fever. 2014. Fact Sheet No. 100.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.