Abstract
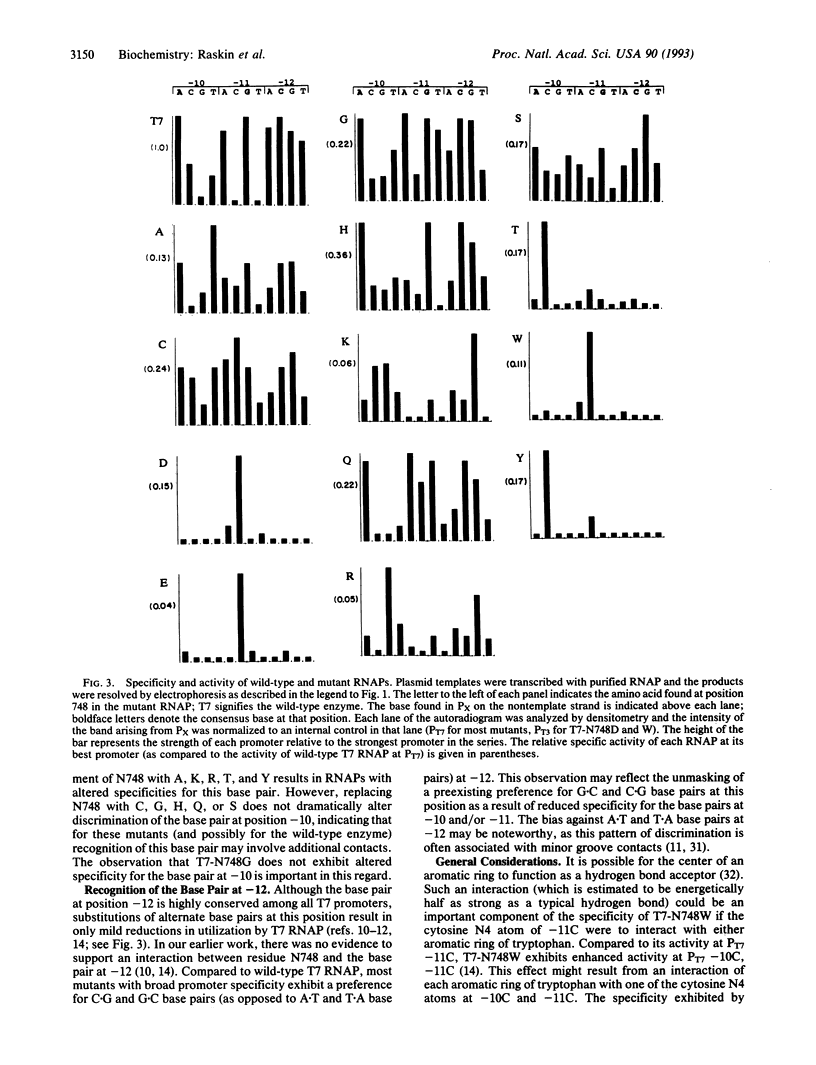
The amino acid at position 748 in T7 RNA polymerase (RNAP) functions to discriminate base pairs at positions -10 and -11 in the promoter. We have constructed a series of T7 RNAP mutants having all possible amino acid substitutions at this position. Surprisingly, most (13/19) substitutions result in active RNAPs, and many of these exhibit altered promoter specificities. Identification of mutant RNAPs with altered specificities expands the repertoire of highly specific phage RNAPs that are available for use in phage RNAP-based transcription systems and highlights the complexity of sequence-specific DNA recognition.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck P. J., Gonzalez S., Ward C. L., Molineux I. J. Sequence of bacteriophage T3 DNA from gene 2.5 through gene 9. J Mol Biol. 1989 Dec 20;210(4):687–701. doi: 10.1016/0022-2836(89)90102-2. [DOI] [PubMed] [Google Scholar]
- Brown J. E., Klement J. F., McAllister W. T. Sequences of three promoters for the bacteriophage SP6 RNA polymerase. Nucleic Acids Res. 1986 Apr 25;14(8):3521–3526. doi: 10.1093/nar/14.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985 Jul 5;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Dalma-Weiszhausz D. D., Gartenberg M. R., Crothers D. M. Sequence-dependent contribution of distal binding domains to CAP protein-DNA binding affinity. Nucleic Acids Res. 1991 Feb 11;19(3):611–616. doi: 10.1093/nar/19.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz A., Weisser H. J., Kössel H., Hausmann R. The gene for Klebsiella bacteriophage K11 RNA polymerase: sequence and comparison with the homologous genes of phages T7, T3, and SP6. Mol Gen Genet. 1990 Apr;221(2):283–286. doi: 10.1007/BF00261733. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Krippl B., Bernstein K. E., Westphal H., Studier F. W. Targeting bacteriophage T7 RNA polymerase to the mammalian cell nucleus. Gene. 1988 Sep 7;68(2):259–266. doi: 10.1016/0378-1119(88)90028-5. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Ebright R. H. Identification of amino acid-base pair contacts by genetic methods. Methods Enzymol. 1991;208:620–640. doi: 10.1016/0076-6879(91)08032-d. [DOI] [PubMed] [Google Scholar]
- Ebright R. H. Use of "loss-of-contact" substitutions to identify residues involved in an amino acid-base pair contact: effect of substitution of Gln18 of lac repressor by Gly, Ser, and Leu. J Biomol Struct Dyn. 1985 Oct;3(2):281–297. doi: 10.1080/07391102.1985.10508417. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross L., Chen W. J., McAllister W. T. Characterization of bacteriophage T7 RNA polymerase by linker insertion mutagenesis. J Mol Biol. 1992 Nov 20;228(2):488–505. doi: 10.1016/0022-2836(92)90837-a. [DOI] [PubMed] [Google Scholar]
- Gunasekera A., Ebright Y. W., Ebright R. H. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem. 1992 Jul 25;267(21):14713–14720. [PubMed] [Google Scholar]
- Joho K. E., Gross L. B., McGraw N. J., Raskin C., McAllister W. T. Identification of a region of the bacteriophage T3 and T7 RNA polymerases that determines promoter specificity. J Mol Biol. 1990 Sep 5;215(1):31–39. doi: 10.1016/S0022-2836(05)80092-0. [DOI] [PubMed] [Google Scholar]
- Klement J. F., Moorefield M. B., Jorgensen E., Brown J. E., Risman S., McAllister W. T. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depends primarily upon a three base-pair region located 10 to 12 base-pairs upstream from the start site. J Mol Biol. 1990 Sep 5;215(1):21–29. doi: 10.1016/s0022-2836(05)80091-9. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. Tinkering with enzymes: what are we learning? Science. 1987 Jun 5;236(4806):1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- Kotani H., Ishizaki Y., Hiraoka N., Obayashi A. Nucleotide sequence and expression of the cloned gene of bacteriophage SP6 RNA polymerase. Nucleic Acids Res. 1987 Mar 25;15(6):2653–2664. doi: 10.1093/nar/15.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B. Bending of synthetic bacteriophage 434 operators by bacteriophage 434 proteins. Nucleic Acids Res. 1991 Aug 11;19(15):4115–4119. doi: 10.1093/nar/19.15.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Carlson P. DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature. 1992 Jan 2;355(6355):89–91. doi: 10.1038/355089a0. [DOI] [PubMed] [Google Scholar]
- Koudelka G. B., Harbury P., Harrison S. C., Ptashne M. DNA twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 repressor. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4633–4637. doi: 10.1073/pnas.85.13.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Harrison S. C., Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. 1987 Apr 30-May 6Nature. 326(6116):886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease--DNA interaction. Science. 1990 Nov 9;250(4982):776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- Levitt M., Perutz M. F. Aromatic rings act as hydrogen bond acceptors. J Mol Biol. 1988 Jun 20;201(4):751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., Carter A. D. Regulation of promoter selection by the bacteriophage T7 RNA polymerase in vitro. Nucleic Acids Res. 1980 Oct 24;8(20):4821–4837. doi: 10.1093/nar/8.20.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw N. J., Bailey J. N., Cleaves G. R., Dembinski D. R., Gocke C. R., Joliffe L. K., MacWright R. S., McAllister W. T. Sequence and analysis of the gene for bacteriophage T3 RNA polymerase. Nucleic Acids Res. 1985 Sep 25;13(18):6753–6766. doi: 10.1093/nar/13.18.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt B. A., Dunn J. J., Studier F. W. Nucleotide sequence of the gene for bacteriophage T7 RNA polymerase. J Mol Biol. 1984 Feb 25;173(2):265–269. doi: 10.1016/0022-2836(84)90194-3. [DOI] [PubMed] [Google Scholar]
- Morris C. E., Klement J. F., McAllister W. T. Cloning and expression of the bacteriophage T3 RNA polymerase gene. Gene. 1986;41(2-3):193–200. doi: 10.1016/0378-1119(86)90098-3. [DOI] [PubMed] [Google Scholar]
- Muller D. K., Martin C. T., Coleman J. E. Processivity of proteolytically modified forms of T7 RNA polymerase. Biochemistry. 1988 Jul 26;27(15):5763–5771. doi: 10.1021/bi00415a055. [DOI] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Raskin C. A., Diaz G., Joho K., McAllister W. T. Substitution of a single bacteriophage T3 residue in bacteriophage T7 RNA polymerase at position 748 results in a switch in promoter specificity. J Mol Biol. 1992 Nov 20;228(2):506–515. doi: 10.1016/0022-2836(92)90838-b. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. DNA in profile. Trends Biochem Sci. 1991 Dec;16(12):467–470. doi: 10.1016/0968-0004(91)90181-t. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Walter J., Huber R., Sjölin L. Structure of bovine pancreatic trypsin inhibitor. Results of joint neutron and X-ray refinement of crystal form II. J Mol Biol. 1984 Dec 5;180(2):301–329. doi: 10.1016/s0022-2836(84)80006-6. [DOI] [PubMed] [Google Scholar]