Abstract
The interplay of active and repressive histone modifications is assumed to play a key role in the regulation of gene expression. In contrast to this generally accepted view, we show that transcription of genes temporally regulated during fly and worm development occurs in the absence of canonically active histone modifications. Conversely, strong chromatin marking is related to transcriptional and post-transcriptional stability, an association that we also observe in mammals. Our results support a model in which chromatin marking is associated to stable production of RNA, while unmarked chromatin would permit rapid gene activation and de-activation during development. In this case, regulation by transcription factors would play a comparatively more important regulatory role.
Post-translational modifications of histones define an evolutionarily conserved “code” that governs differential gene expression1. Trimethylation of histone H3 at lysine 4 (H3K4me3) and at lysine 36 (H3K36me3), for instance, correlate with active transcription, whereas H3K9me3 and H3K27me3 are usually linked to transcriptional repression2, 3. The combinatorial behavior of histone modifications along regulatory regions—reflecting and/or influencing the specific arrangement of transcription factors—modulates the expression levels of genes, conferring them with a unique temporal and spatial transcriptional program. Computational models have been developed that can predict gene expression from histone modifications with great accuracy4, 5.
A number of recent reports, however, indicate that expression of certain genes may occur in absence of histone modifications canonically associated to active genes. The modENCODE project reported that some expressed genes lacked H3K4me36. Hödl and Basler found that cells that lack H3K4 methylation, respond to developmental signaling pathways by activating target gene expression in Drosophila wing imaginal discs7. Chen et al. observed that pre-midblastula transition (pre-MBT) genes have particularly low levels of H3K4me38. More recently, Zhang et al. reported that genes within yeast heterochromatic regions can be transcribed in absence of active histone marks9. Here, we show that active transcription in the absence of chromatin marking is actually a general feature of genes that are strongly regulated during development. We analyzed data produced by modENCODE in whole animals and tissues in fly and worm, characterized the fly transcriptome by RNASeq and the epigenome by ChIPSeq in two spatially well-defined and relatively homogeneous developmental fly tissues, and carried out targeted experimental validations in isolated cells. All these analyses strongly suggest that expression of genes regulated during fly development can occur in the absence of marks typically associated with active genes, and, indeed, this expression does not seem to be affected by perturbations of the histone methyltransferase system. Conversely, we found that chromatin marking is associated not only to transcriptional levels, but also to transcriptional and post-transcriptional stability—an association that appears to be conserved through metazoan evolution.
Results
Expression without histone modifications during development
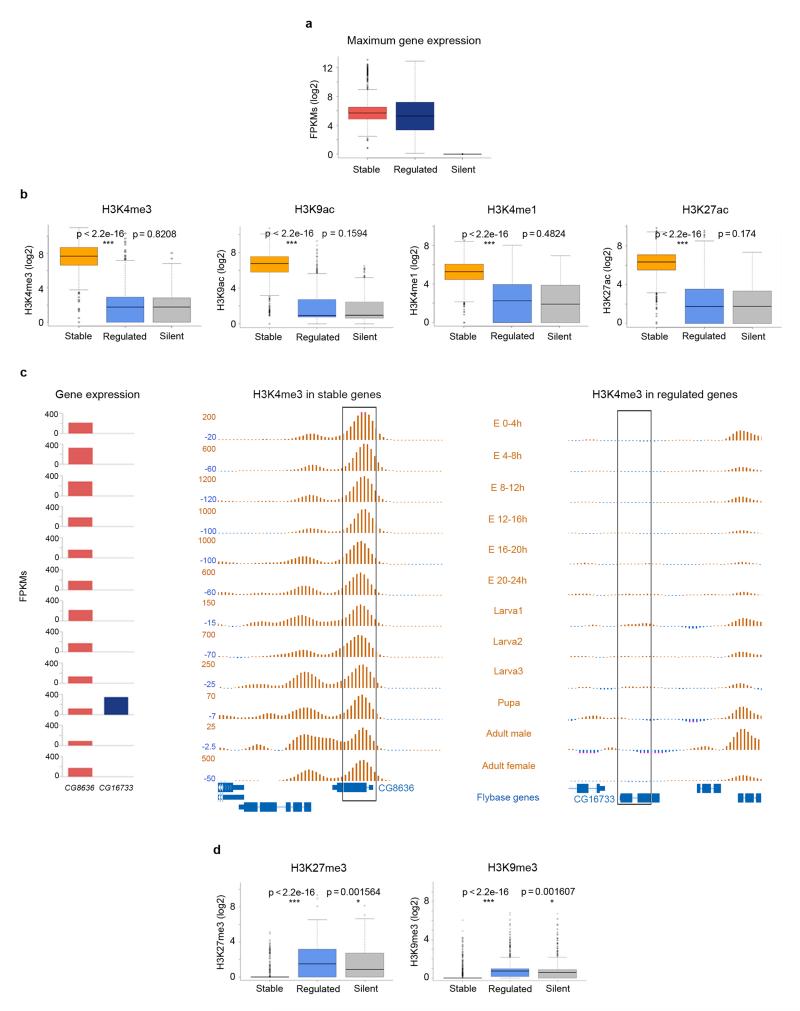
To investigate the dynamics of chromatin marking in genes regulated during development, we analyzed data produced within the Drosophila melanogaster modENCODE project6, 10. We specifically analyzed RNASeq and ChIPSeq data for H3K4me3, H3K9ac, H3K4me1, H3K27ac, H3K27me3 and H3K9me3 on whole animals (Supplementary Fig. 1a). To measure transcriptional stability, we computed the coefficient of variation of gene expression over 12 developmental time points (Methods and Supplementary Fig. 1b)—lower values corresponding to higher transcriptional stability. The distribution of the coefficient of variation uncovers a large class of genes that show constant expression during development, and two other minor classes containing genes whose expression is highly variable—often restricted to a limited number of stages (Supplementary Fig. 1c, d). We arbitrarily selected the 1,000 genes with the highest coefficient of variation, and defined them as developmentally regulated, because of their variable pattern of expression along time. Conversely, we selected the 1,000 genes with the lowest coefficient of variation, and defined them as developmentally stable. For each gene, we determined the time point at which its expression is the highest. At this time point, we did not observe strong differences between the expression of stable and regulated genes (Fig. 1a). At the same time point we measured the levels of histone modifications for each gene (Methods). We found that at the point of highest expression, stable genes are strongly marked by histone modifications typically associated to active transcription, H3K4me3 and H3K9ac, and also to enhancers: H3K4me1 and H3K27ac. Unexpectedly, however, regulated genes show very low levels of these modifications, comparable to those of silent genes (Fig. 1b, Supplementary Fig. 2). In Figure 1c we compare the pattern of H3K4me3 along fly development in CG8636, a gene stably expressed during development, and in CG16733, a gene specifically expressed in pupa. CG8636 shows a strong H3K4me3 peak downstream from the transcription start site whereas CG16733 lacks any marking, even at the pupa stage, where it is expressed at higher levels than CG8636. (See also Supplementary Fig 3.) This contrasting pattern of histone marking is not only apparent when comparing genes with extreme behavior, but it is a distinct feature of the partition of the entire set of fly genes in two major classes according to transcriptional stability (Supplementary Fig. 4). For the histone modifications typically associated to inactive genes, H3K27me3 and H3K9me3, we observed that regulated genes showed levels higher than those of stable ones, and similar to those of silent genes (Methods, Fig. 1d). The levels of these marks, however, are generally low compared to the levels of active marks, even for genes silent during development—a large proportion of which lack any evidence of them (Supplementary Fig. 5a, b), as it has already been previously reported11. We found only a weak relationship between the level of repressive marks and gene expression (Supplementary Fig. 5c, d).
Figure 1. Distribution of histone modification levels in stable, regulated and silent genes during fly development.
a, Expression of stable, regulated, and silent genes during fly development at the time point of maximum expression for each gene. Gene expression was computed as FPKMs by the modENCODE consortium. The bottom and top of the boxes are the first and third quartiles, and the line within, the median. The whiskers denote the interval within 1.5 times the Inter Quartile Range (IQR) from the median. Outliers are plotted as dots. b, Normalized levels of H3K4me3, H3K9ac, H3K4me1 and H3K27ac at the time point of maximum expression during D. melanogaster development. These values represent the maximum height of the ChIPSeq peak within the gene body. P-values were computed using the Wilcoxon text (two-sided). c, Profiles of H3K4me3 during the 12 fly developmental time points in CG8636, a gene stably expressed during fly development, and CG16733, a pupa-specific gene. The expression (measured as FPKMs) along these points for the two genes is given on the left. d, Levels of H3K27me3 and H3K9me3 at the time point of maximum expression, computed as the average height of the ChIPSeq signal within the gene body, in stable, regulated and silent genes.
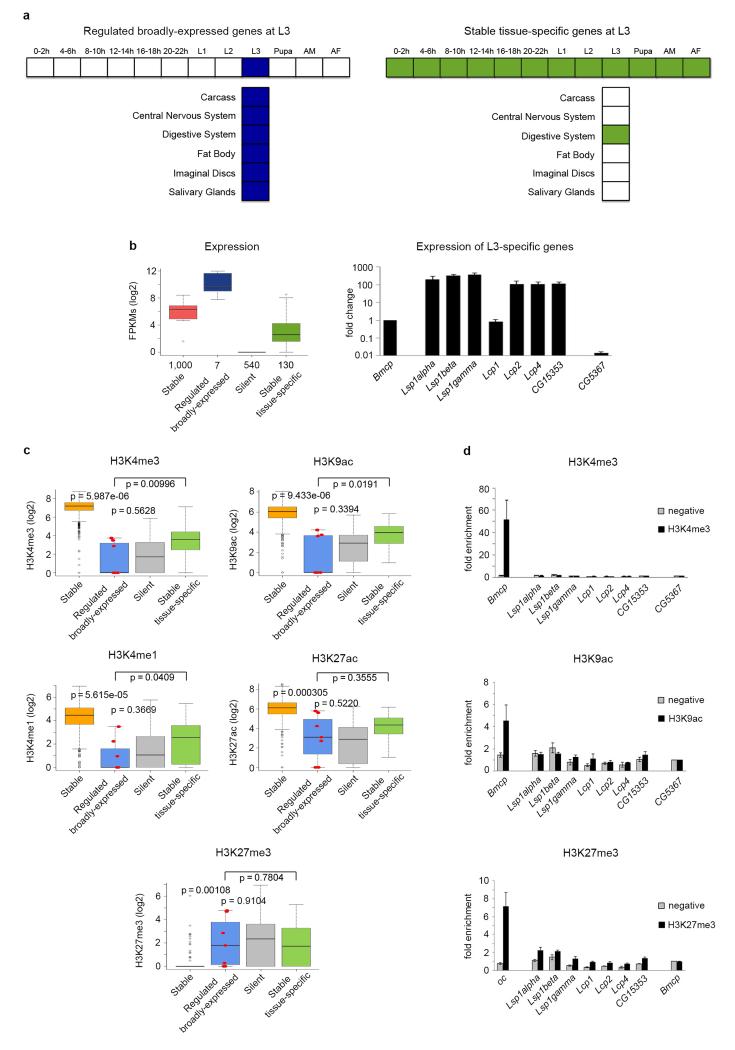
Given that developmental chromatin maps produced in the modENCODE project are on whole organisms, it could be argued that apparent lack of chromatin marking is the consequence of the expression of regulated genes being spatially confined to specific organs, tissues or subtissular domains. While, indeed, regulated genes show in general a spatially restricted pattern of expression, chromatin marking can actually be detected in stable genes that exhibit also a restricted expression pattern comparable to that in regulated genes (Supplementary Fig. 3). To further investigate the potential effect of restricted expression in the ability to detect chromatin marking, we used tissue-specific RNASeq data from modENCODE12. Third instar larva (L3) is the time point with the largest number of tissues available: carcass, central nervous system, digestive system, fat body, imaginal discs and salivary glands. Using L3 tissue-specific RNASeq data, we identified seven regulated genes expressed in all six available tissues at L3 (“Regulated broadly-expressed” Fig. 2a, left panel). Conversely, we identified 130 stable genes specifically expressed in only one of the aforementioned tissues in L3 (“Stable tissue-specific”, Fig. 2a, right panel). Regulated broadly-expressed genes have much higher expression levels than stable tissue-specific genes when measured in the whole body (almost four-fold, Fig. 2b), as well as, in general, when measured on individual tissues (Supplementary Fig. 6). They have also higher expression levels than stable genes overall. However, the levels of H3K4me3, H3K9ac, H3K4me1, and H3K27ac in regulated broadly-expressed genes are significantly lower than in stable genes, even than in stable tissue-specific genes, and comparable to those in silent genes (Fig. 2c). We confirmed both gene expression and levels of H3K4me3 and H3K9ac by qPCR (Fig. 2b) and ChIP-qPCR (Fig. 2d), respectively.
Figure 2. Gene expression and histone modifications in regulated broadly-expressed and stable tissue-specific genes at third instar larvae.
a, Diagrams of developmentally regulated genes broadly-expressed across multiple tissues at third instar-larvae L3 (left panel), and stable genes expressed in only one tissue at L3 (right panel). b, Gene expression levels at L3 measured by whole organism RNASeq (left panel). The number of genes in each category is given under the boxplots. The bottom and top of the boxes are the first and third quartiles, and the line within, the median. The whiskers denote the interval within 1.5 times the IQR from the median. Outliers are plotted as dots. Validation by qPCR of the expression at L3 of regulated broadly-expressed genes compared to a stable gene (Bmcp) and a silent gene (CG5367) (right panel). Error bars represent the Standard Error of the Mean (SEM) from three independent replicates. c, Levels of H3K4me3, H3K9ac, H3K4me1 and H3K27ac on whole L3 individuals. The seven regulated genes broadly-expressed at L3 are depicted as red dots within the boxplots. P-values were computed using the Wilcoxon test (two-sided). d, Validation by individual ChIPs and qPCR of H3K4me3 and H3K9ac in regulated genes broadly-expressed at L3. H3K4me3 and H3K9ac ChIPs are represented as enrichment of the marks over the silent gene (CG5367). Error bars represent the SEM from three independent replicates.
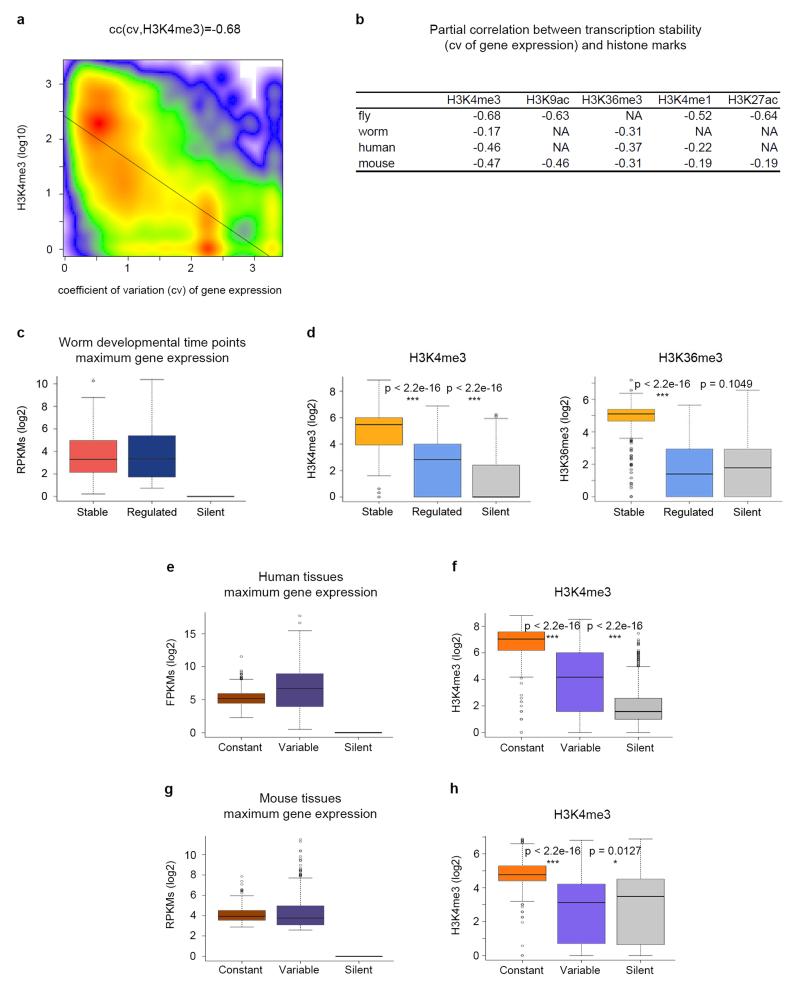
All these results strongly suggest that activation of genes regulated during development occurs mostly in the absence of histone modifications canonically linked to active genes. Our results also point to strong chromatin marking association not only with transcriptional levels, but also with transcriptional stability. We calculated the coefficient of correlation (cc) across all genes between the coefficient of variation of gene expression across developmental time points as computed above, and the level of histone modifications at the developmental time point of highest expression. We used partial correlations to control for a potential confounding effect of gene expression levels (see Methods). For all active histone modifications, the partial correlations are negative and significant (as low as cc = −0.68 for H3K4me3, Fig. 3a, b, Supplementary Fig. 7), strongly supporting association between transcriptional stability and active chromatin marking.
Figure 3. Association between histone modifications and transcription stability in metazoans.
a, Scatterplot of H3K4me3 levels at the time point of highest expression during fly development and transcriptional stability measured as the coefficient of variation of gene expression across time points. The correlation is computed as the partial correlation given gene expression. b, Partial correlations between active marks and transcription stability (the coefficient of variation). Correlations are computed controlling for gene expression. All correlations are statistically significant (p-value < 2.2e–16). P-values were computed using Student’s t-test (two-sided). c, Expression of stable, regulated and silent genes during worm development at the time point of maximum expression. The bottom and top of the boxes are the first and third quartiles, and the line within, the median. The whiskers denote the interval within 1.5 times the IQR from the median. Outliers are plotted as dots. d, Levels of H3K4me3 and H3K36me3 at the time point of maximum expression during worm development. e, Expression of genes with constant and variable expression at the tissue/cell line of highest expression across multiple samples from the Roadmap Epigenomics Mapping Consortium. f, Levels of H3K4me3 at the tissue of maximum expression. g, Expression of genes with constant and variable expression at the tissue of highest expression across ten mouse tissues from the mouse ENCODE project. h, Levels of H3K4me3 at the tissue of maximum expression. These levels correspond to the maximum height of the ChIPSeq peak within the gene body. P-values were computed using Wilcoxon text (two-sided).
To investigate whether lack of chromatin marking in regulated genes and the association between chromatin marking and transcriptional stability are conserved in other metazoans, we first analyzed RNASeq-based gene expression on seven time points through C. elegans development13 and ChIP-chip data on two histone modifications available for these time points in modENCODE: H3K4me3 and H3K36me3. While both, the resolution and the reliability of the chromatin data obtained through ChIP-chip are lower in worm than in the fly ChIPSeq, we observed the same trend: the expression level at the time point of maximum expression is very similar in regulated and stable genes (Fig. 3c), while regulated genes show lower levels of H3K4me3 and H3K36me3, more similar to those of silent genes (Fig. 3d). As in flies, there is a significant association between transcriptional stability and active histone marking (Fig. 3b).
Unfortunately, genome-wide transcriptomic and epigenetic developmental maps of the resolution of those from modENCODE are not yet available for mammalian (or vertebrate) systems. Nevertheless, using transcriptomic and epigenomic data across multiple tissues and cell lines in human and mouse, we did find that active chromatin marking is associated to transcription stability also in mammalian systems. We used RNASeq and ChIPSeq data for H3K4me3, H3K36me3 and H3K4me1 for 56 human adult and fetal tissues, primary cells and cultured cell lines from the Roadmap Epigenomics Mapping Consortium14. We found strong negative correlation between the coefficient of variation of gene expression across these samples, and histone levels (Fig. 3b). The gene set with highest variation of expression across human tissues is likely to show some enrichment in regulated genes. Thus, we selected the 1,000 genes with the highest coefficient of variation as variably expressed genes, and the 1,000 genes with the lowest as constantly expressed. In the cell type in which the expression of each gene is highest, variable genes show higher expression than constant genes (Figure 3e). Yet, the levels of active histone modifications in these cell types are much lower in variable than in constant genes (Fig. 3f, Supplementary Fig. 8a). Very similar results are obtained in mouse when using ENCODE data15 (Fig. 3b, g, h and Supplementary Fig. 8b).
Expression without histone modifications in imaginal discs
Data generated by the modENCODE projects monitor complex systems encapsulating great cellular heterogeneity. To investigate the dynamics of chromatin marking during development in a more homogeneous cellular environment, we characterized the transcriptome by RNASeq (Supplementary Fig. 9a, b and Supplementary Table 1) and the epigenome by ChIPSeq in two D. melanogaster third instar larval tissues: Wing and Eye-antenna imaginal discs (WID and EID, respectively). We specifically monitored H3 and the active marks H3K4me3, H3K9ac, H3K4me1, and H3K27ac, plus the transcription elongation mark H3K36me3 (Supplementary Fig. 9c). Both, WID and EID, are epithelial tissues in early differentiation stages, and differentially expressed genes are likely to be under temporal developmental control. While WID and EID epigenomes and transcriptomes are very similar (Supplementary Fig. 9d-e), differentially expressed genes do exhibit functions strongly consistent with the known biology of these tissues (Supplementary Tables 2, 3 and Supplementary Fig. 9f).
We then investigated the marking of regulated and stable genes in WID and EID. To focus on genes under stronger regulation, we identified 55 developmentally regulated genes expressed in EID, but not in WID, and 10 regulated genes expressed in WID, but not in EID. We also identified a set of 284 stable genes highly expressed both in EID and WID, as well as a set of 30 genes silent in both (Supplementary Tables 4-7 and Methods).
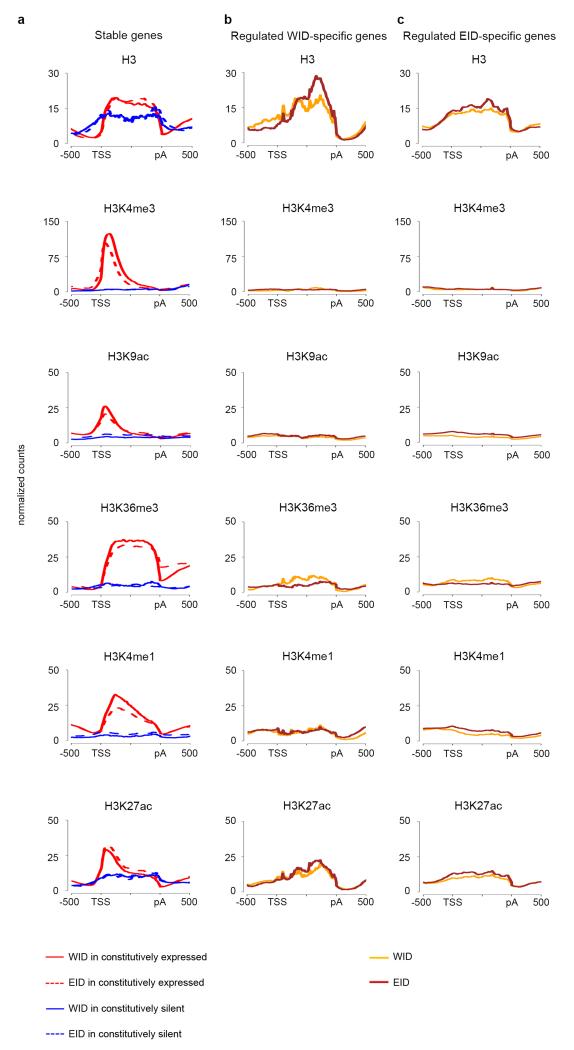
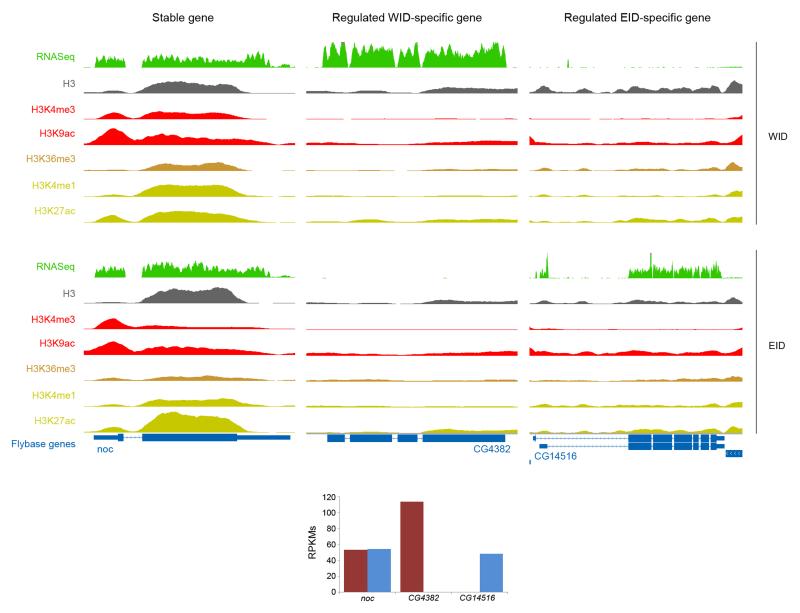
We next compared marking of stable, silent, and regulated WID- and EID-specific genes (from now on simply, WID- and EID-specific). Consistent with previous observations16, 17, the WID- and EID-profiles of stable genes are very similar, as are those of silent genes (Fig. 4a). Stable and silent genes are both characterized by higher stable nucleosome occupancy than nearby intergenic regions, but the genic nucleosome (H3) enrichment is larger for stably expressed than for silent genes. Stable genes are also strongly marked by H3K4me3, H3K9ac, H3K36me3, and also, as observed in modENCODE, by H3K4me1 and H3K27ac. Silent genes mostly lack these histone modifications. Regulated tissue-specific genes exhibit, however, a contrasting behavior. As expected, WID-specific genes lack active modifications in EID (Fig. 4b), and, conversely, EID-specific genes are not marked in WID (Fig. 4c). Unexpectedly, but consistently with the behavior that we observed in modENCODE data, WID-specific genes are not marked in WID either, nor EID-specific genes in EID. Absence of active histone marking cannot be attributed to the lack of nucleosomes because H3 is observed in these genes (Fig. 4b, c). It is unlikely that it originates either from higher nucleosome turnover in regulated genes since, at least in Drosophila S2 cells18, nuclear turnover is similar for stable and regulated genes (Supplementary Fig. 10). Lack of histone marking is not due, either, to the relative low expression level of WID- or EID-specific genes, since even when these genes have high levels of expression, comparable to those of constitutively expressed genes, there is no marking by active modifications. This is illustrated in Figure 5 (see Supplementary Fig. 11 for more examples). The WID-specific gene CG4382 and the EID-specific gene CG14516 have similar levels of expression than the stable gene noc. This gene, however, is strongly marked by histone modifications in both WID and EID, while CG4382 and CG14516 are marked in neither. Lack of chromatin marking cannot be attributed to the restricted expression of tissue-specific genes, since the expression of noc is also restricted to specific regions both in WID and EID19, 20. H3 levels of tissue-specific and stable genes are comparable and only depend weakly on the expression status of genes (Fig. 5).
Figure 4. Profiles of H3 and histone modifications in Wing (WID) and Eye-antenna (EID) imaginal discs.
a, Profiles on stable and silent genes in WID and in EID. b, Profiles on regulated WID-specific genes in WID and EID. c, Profiles on regulated EID-specific genes in WID and EID.
Figure 5. Profiles of RNA expression, H3 and histone modifications in Wing (WID) and Eye-antenna (EID) imaginal discs.
Noc is a gene stably expressed in WID and EID; CG4382 a WID-specific and CG14516, an EID-specific gene. Levels of gene expression (as RPKMs) are depicted at the bottom of the panels. Screenshots have been obtained through the UCSC Genome Browser56.
Active transcription without histone modifications
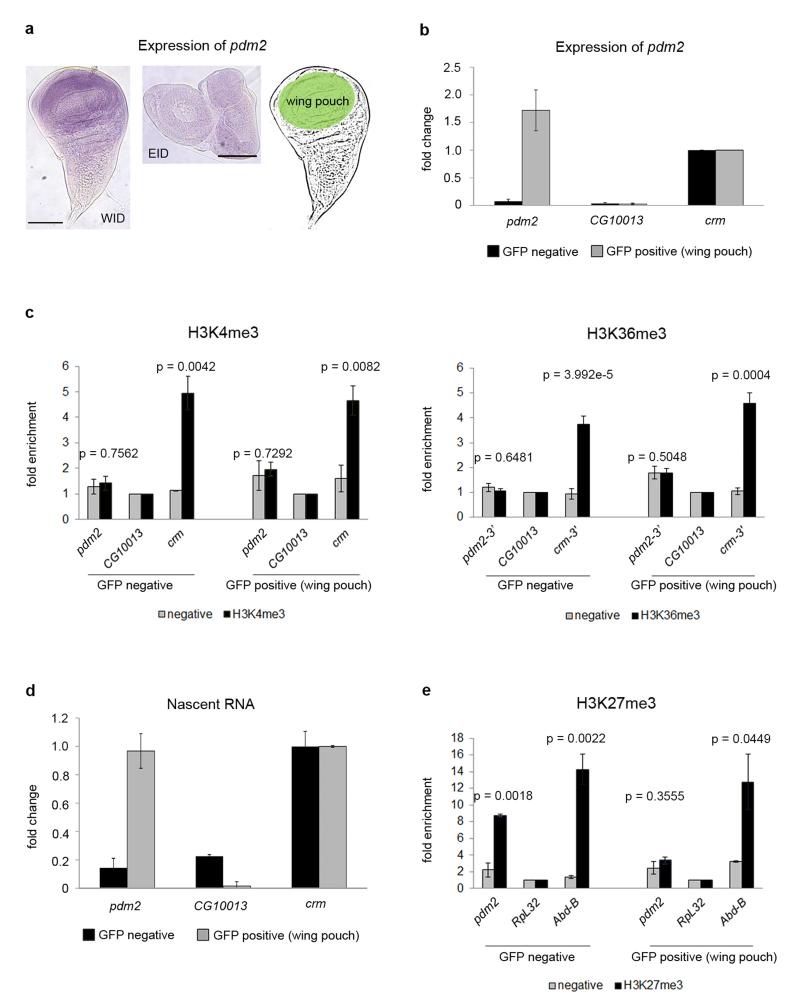
While WID and EID are relatively homogeneous tissues, they already show cellular sub-specialization at third instar larvae. For instance, the WID-specific gene POU domain protein 2 (pdm2), like nubbin (nub)21, with strong temporal and spatial regulation during development, is only expressed in the wing primordium (wing pouch) at third instar larva (Fig. 6a). To unequivocally demonstrate lack of chromatin marking in developmentally regulated genes, we took advantage of the nub-GAL4 construct to drive expression of GFP only in the wing pouch, where pdm2 is expressed. Thus, we collected all cells expressing pdm2 and investigated chromatin marking for this gene only in the cells in which it is expressed. More specifically, dissection and dissociation of wing discs followed by cell-sorting analyses allowed the isolation of two populations of cells: the wing pouch (nub domain, GFP positive) and the rest of the wing (GFP negative) (Fig. 6a and Methods). By using qPCR we found that the expression of pdm2, restricted to sorted GFP positive cells, is even higher than the expression of crm, a gene expressed at the same level throughout the WID (Fig. 6b). ChIP assays followed by qPCR on sorted cells showed that the levels of H3K4me3, and H3K36me3 in pdm2 are significantly lower than in crm, and comparable to those in CG10013, a gene silent in the whole WID (Fig. 6c). High RNA levels of pdm2 in the wing pouch (Fig. 6b) do not necessarily demonstrate active transcription, since transcription could have occurred at an earlier time point. To assess active gene expression we directly measured newly transcribed RNA (nascent RNA) in sorted cells. As shown in Figure 6d, pdm2 active transcription in GFP positive cells is as high as transcription of the control gene crm.
Figure 6. Active transcription of pdm2 without chromatin modifications.
a, Expression of pdm2 in WID (left panel) and EID (middle panel) labeled with a pdm2-specific probe. The gene is only expressed in the wing pouch of the WID, highlighted in green. The scale bars represent 100 μm. b, Expression of pdm2 in sorted cells analyzed by qPCR. Gene expression is normalized by the control gene crm. Error bars represent the SEM from three biological replicates. c, ChIP analysis of H3K4me3, H3K36me3 and of negative controls without antibody on sorted cells. ChIPs are represented as enrichment of the marks over a silent gene non-marked with H3K4me3 and H3K36me3 (CG10013). Crm is used as positive control for these modifications. Error bars represent the SEM from at least three biological replicates. P-values were computed using the Student’s t-test (two-sided). d, Newly transcribed RNA of GFP-sorted cells. Nascent RNA is normalized by the control gene crm. Error bars represent the SEM of four biological replicates. e, ChIP analysis of H3K27me3 and of negative controls without antibody on sorted cells. H3K27me3 ChIPs are represented as enrichment of the mark over a constitutively expressed gene non-marked with H3K27me3 (RpL32). Abd-B is used as positive control for this modification. Error bars represent the SEM from at least three biological replicates.
To investigate marking by repressive histone modifications in expressed genes (Fig 1d), we monitored the levels of H3K27me3 in pdm2, a gene exhibiting this modification at L3 when measured in the whole organism. We performed individual ChIP-qPCR in sorted cells and found that pdm2 is indeed marked by H3K27me3 in WID, but only outside the wing pouch. No marking was observed in the wing pouch, where pdm2 is expressed (Fig. 6e). This suggests that the repressive modifications detected in whole organisms in regulated genes (Fig. 1d) could originate from organs or tissues in which these genes are not expressed.
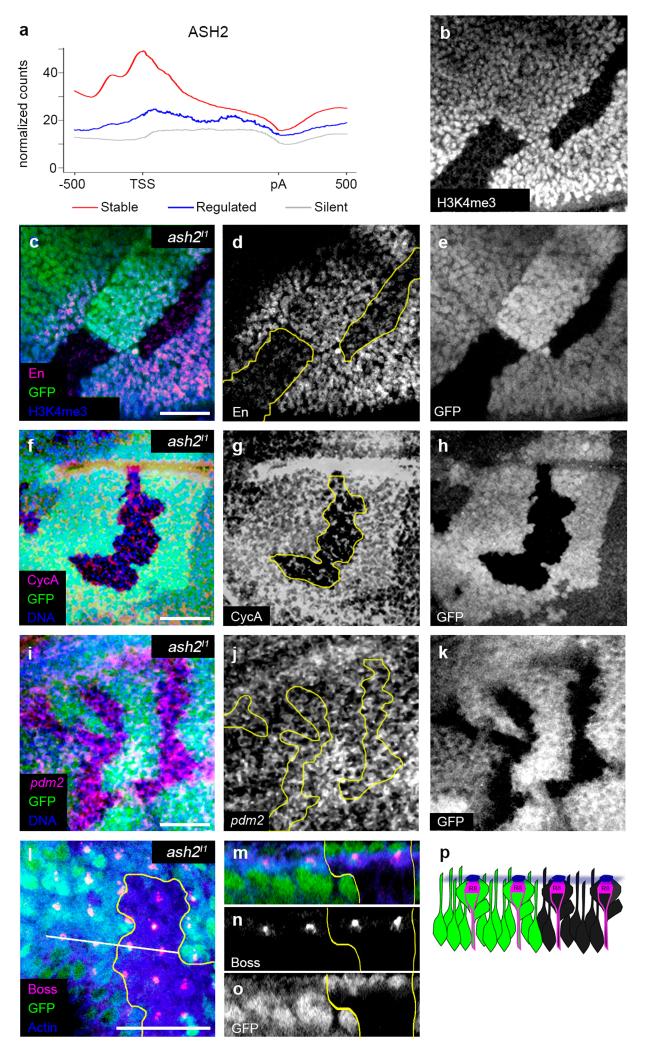
Lack of active marking suggests that genes regulated throughout development may not respond to histone modification systems. Therefore, we specifically investigated the response of regulated genes to the lack of ASH2 (Absent, small or homeotic disc 2), a key co-factor for H3K4 methylation22. First we characterized ASH2 occupancy along fly genes using ChIPSeq data obtained in WID17 and found a very strong depletion of ASH2 binding to the promoters of regulated genes compared to those of stable genes (Fig. 7a). Second, we used the ash2I1 mutant allele to interfere with H3K4me3. Since this allele is lethal in late third-instar larvae/early pupae23, we performed clonal analyses in WID and EID. We specifically analyzed two stable genes: engrailed (en), expressed in the posterior compartment of the WID, and Cyclin A (CycA), ubiquitously expressed in the WID, as well as two regulated genes: pdm2, expressed in the wing pouch, and bride of sevenless (boss), expressed in the differentiated photoreceptor R8 cell of the EID. We confirmed lack of H3K4me3 in ash2I1mutant clones (Fig. 7b), and observed a clear reduction in the levels of En and CycA, while the expression of Boss and pdm2 was not affected (Fig. 7c-p).
Figure 7. Reduction of H3K4me3 does not affect expression of regulated genes.
a, Distribution of ASH2 binding in stable (red), regulated (blue) and silent genes (grey). b, H3K4me3 is strongly decreased in ash2I1 mutant clones in WID. c, En immunostaining in WID (merged). The scale bar represents 20 μm. d, The levels of the stable gene En are reduced in mutant clones. e, GFP negative cells indicate ash2I1 mutant cells in c and d. f, CycA immunostaining in WID (merged). The scale bar represents 20 μm. g, CycA is decreased in ash2I1 mutant clones. h, GFP negative cells indicate ash2I1 mutant cells in f and g. i, pdm2 fluorescence in situ hybridization in ash2I1 mutant clones in WID (merged). The scale bar represents 20 μm. j, No changes in pdm2 expression are observed in ash2I1 mutant clones. k, GFP negative cells indicate ash2I1 mutant cells in i and j. l, Boss immunostaining in EID. The scale bar represents 20 μm. m, Optical cross-section (white line in l) showing Boss in all R8 photoreceptor cells (merged). n, No changes in Boss expression are observed in ash2I1 mutant clones. o, GFP negative cells indicate ash2I1 mutant cells in m and n. p, Diagram summarizing the result in m–o. Green cells express the wild-type ash2 allele and black cells correspond to homozygous ash2I1 mutant cells. Boss (magenta cap) localizes in the apical side of R8.
Genome organization of regulated genes
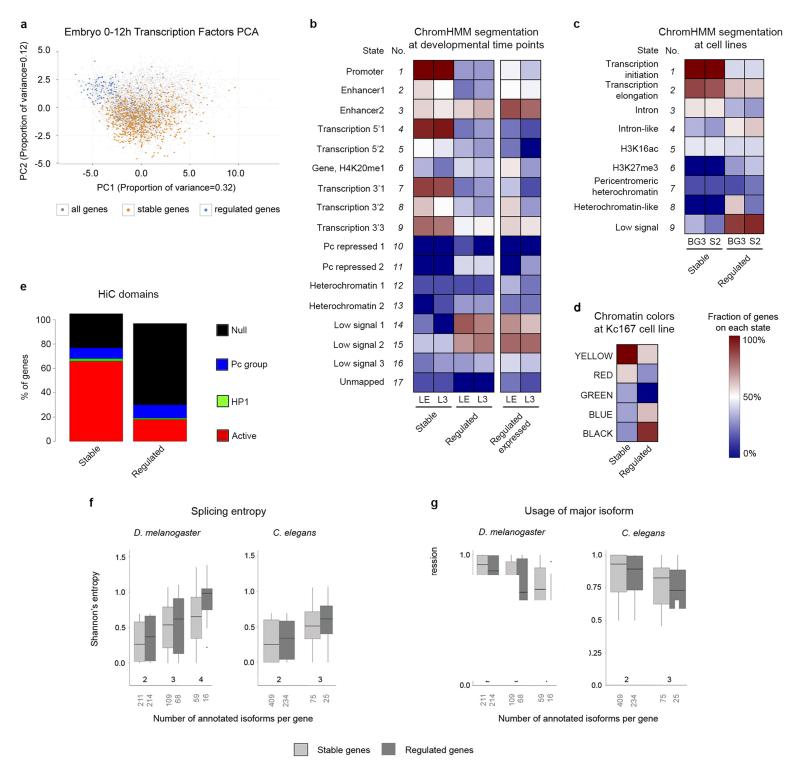
It has been suggested that developmental control genes are under a characteristic regulatory program24. They tend to harbor increased number of transcription factor binding sites25 and are characterized by “peaked” (or narrow) promoters, compared to housekeeping genes which are associated to more “dispersed” (or broad) promoters26-29. Using the promoter classification of Ni et al.30, we found that stable genes are strongly enriched in broad (and weak) promoters compared to regulated genes (444 vs. 12). In contrast, the proportion of peaked promoters is similar in stable and regulated genes (42 vs. 38). Overall, however, our set of regulated genes exhibits most of the characteristics that have been reported for developmental and/or peaked promoter genes in Drosophila and other species (see Lenhard et al.31 for a review). Thus, promoters of regulated genes show stronger conservation29-31, particularly in predicted transcription factor binding motifs (Supplementary Fig. 12). They are depleted in DNA Replication related Element (DRE) sequences, which are associated to disperse initiation of transcription8 (15% of regulated compared to 39% of stable genes), and enriched in TATA Binding Protein (TBP) boxes, characteristic of tighter gene regulation32, 33 (49% vs 15%). In contrast, promoters of stable genes overlap modENCODE High Occupancy Target (HOT) regions, associated to open chromatin and ubiquitous expression34, 35, more often than promoters of regulated genes (67% vs. 8%). We also found that the overall pattern of transcription factor binding clearly separates regulated from stable genes, as revealed by Principal Component Analysis (PCA) based on ChIP-chip data for 20 transcription factors in fly embryos (Fig. 8a). Finally, analyses of published data36-40 of knockdowns or overexpression of several transcription factors have frequently larger impact on the expression of regulated than of stable genes (Supplementary Table 8).
Figure 8. Promoter architecture and genome organization in stable and developmentally regulated genes.
a, Principal Component Analysis (PCA) of genes expressed in the Drosophila embryo between 0 and 12h, based on ChIP-chip binding profiles of twenty transcription factors. b, Fraction of stable and regulated genes in different states from chromatin segmentations in Late Embryo (LE) and L343. Right, proportion of regulated genes when considering only genes expressed in LE or L3. c, The same as in b, for segmentations in BG3 and S2 cell lines42. d, The same as in b, for the segmentation in Kc16741. BLACK chromatin corresponds to repressive chromatin. YELLOW and RED chromatin is typical of transcriptionally active regions. GREEN and BLUE chromatin correspond to repressive chromatin. e, Proportion of stable and regulated genes mapping to spatial chromatin domains, considering the 1,169 domains inferred by HiC in fly embryos44. f, Distribution of Shannon’s entropy of splicing in stable and regulated genes. Shannon’s entropy is computed at the developmental time point in which gene expression is the maximum. The number of genes of each category appears below the X-axis. The bottom and top of the boxes are the first and third quartiles, and the line within, the median. The whiskers denote the interval within 1.5 times the IQR from the median. Outliers are plotted as dots. g, Distribution of the relative usage of the major isoform. The Y-axis is the fraction of the total transcriptional output of the gene that is captured by the most abundant isoform.
Regulated genes also exhibit a characteristic genome organization. We mapped our sets of stable and regulated genes to a number of genome segmentations, representing epigenomic domains, recently obtained in Drosophila cell lines (Kc16741, BG3 and S242) and developmental time points (late embryo, LE, and L343). We systematically found that regulated genes tend to occur in chromatin states that are depleted in histone modifications (Figure 8b-d), even when considering only regulated genes expressed in the developmental time point at which the segmentation has been obtained (Figure 8b). Epigenomic domains in turn, spatially organize into well-defined physical domains within the nucleus44. Silent chromatin regions, in particular, fold into modular chromosomal entities, which we found enriched in regulated genes (Fig. 8e). The nuclear lamina plays a key role in this physical organization, through the interaction with large continuous chromosomal domains. These Lamina Associated Domains (LADs) are generally depleted of chromatin marks45, and we consistently found that regulated genes are strongly enriched in LADs (52% compared to 5% of stable genes in 412 LADs from Kc167 cells46).
Histone modifications and alternative splicing
Beyond its role in primary RNA production, chromatin structure has also been implicated in subsequent steps of RNA processing. In particular, a number of studies have uncovered a relationship between nucleosome occupancy and exon-intron structure47, 48 and between specific histone modifications and alternative splicing49-51. We found in fly WID and EID that highly included exons are characterized by higher H3 occupancy when compared to lowly included ones, as previously reported in mammals48 (Methods, Supplementary Tables 9, 10 and Supplementary Fig. 13a, b), and that the correlation between H3 occupancy and exon inclusion peaks very close to the acceptor site (Supplementary Fig. 13c, d).
We speculated, thus, that strong chromatin marking might not be only associated to more stable RNA production, but also to a tighter regulation of alternative splicing. To measure alternative splicing complexity, we computed the Shannon’s entropy on the relative abundance of a gene’s alternative splicing isoforms (Methods). The splicing entropy grows with the number of isoforms and with the evenness of their relative abundances. Higher entropic values can be interpreted as tight regulation of alternative splicing, while lower values would correspond to more stochastic production of alternative isoforms. As hypothesized, splicing entropy, measured at the time point of maximum gene expression, is lower for strongly marked stable genes than for unmarked developmentally regulated genes (Fig. 8f). Further supporting tighter regulation of splicing, we also found that the major isoform captures a larger fraction of the total transcriptional output in stable than in regulated genes (Fig. 8g).
Discussion
Cell type specific transcriptional regulation is crucial to maintain cell identity throughout the lifetime of organisms, yet it must be flexible enough to allow for responses to endogenous and exogenous stimuli. This regulation is mediated by specific molecular factors (e.g. cell type specific transcription factors, and chromatin modifications), as well as by the topological organization of the genome. In particular, modifications occurring on DNA and on histones regulate gene expression by establishing and maintaining specific chromatin states52, 53. The association of certain modifications with transcriptional activation or repression has become widely accepted. Nevertheless, expression of genes in the absence of chromatin marks has also been reported6-9. Here we found that transcription in the absence of most canonically active chromatin marks is actually a characteristic feature of genes that are regulated during fly and worm development. These are not necessarily equivalent to developmental control genes, many of which are known to be marked11, 52.
Analyses of tissue-specific gene expression data, as well as our targeted validation experiments, support that our observations do not arise from the expression of developmentally regulated genes being low or confined to small cell populations, from limited detection sensitivity, and/or from persistence in the cell of RNA molecules transcribed at some earlier standpoint. Thus, while factors not accounted for cannot be completely ruled out, our observations appear to reflect a true biological property of genes regulated throughout development—maybe a consequence of these genes being partially unresponsive to histone modifications systems.
We also found that strongly marked chromatin state is associated to more tightly controlled transcriptional and post-transcriptional regulation, in particular to splicing. This is consistent with earlier observations54 of simultaneous enrichment in the expression of chromatin modifying enzymes and splicing factors in cell-enriched testis, and with the higher levels of H3K36me3 found by de Almeida et al.49 in mammalian constitutive exons compared to alternative exons.
Overall, our results lead us to hypothesize that the relative contribution of transcription factors and histone modifications to regulation of gene expression differentiates the transcriptional programs of stable and regulated genes. In stable genes that are constitutively expressed, strong chromatin marking leads to transcriptional stability and tightly controlled RNA production. In these genes, regulation by transcription factors would play a comparatively smaller role. In contrast, genes regulated during development that need to be rapidly activated and de-activated are characterized by an unmarked chromatin state. In these other genes, transcription factors binding to chromatin would play the predominant regulatory role. These distinct regulatory programs would be reflected in the topological organization of the chromatin fiber within the nucleus, with regulated genes located in silent chromosomal modular domains that physically interact with the nuclear lamina55.
While we found evidence for this model of transcriptional regulation specifically in the fly, preliminary results suggest that it may be generalizable to other metazoans. Although detailed transcriptional, epigenetic, and topological maps of genomes are being produced in an increasing number of cell lines and tissues, developmental maps are still sparse in mammalian species. Exhaustive monitoring through a much larger variety of conditions, differentiation states and developmental stages is required to fully understand the layer of epigenetic regulation that mediates between genome sequence and RNA production.
Online Methods
Drosophila strains
The strains used were: Canton S as a wild type and nub-GAL4/+; UAS-GFP/+. Flies were kept on standard media at 25°C.
Tissue disaggregation and cell sorting
Wing imaginal discs (WID) from nub-GAL4/+; UAS-GFP/+ flies were dissected in PBS and incubated for 1h in a 10x trypsin solution (Sigma T4174) at room temperature in a rotating wheel. Cells were vigorously pipetted and kept on ice in Schneider’s insect medium. To discard dead cells, DAPI was added to the sample at 1 μg/mL final concentration. Cells were sorted in a FACSAria (BD) with the 85 μm nozzle. We were able to recover around 2.5·106 GFP negative and 2·106 GFP positive cells from 400 WIDs. An independent sorting experiment was done per each replicate, both for ChIPs and gene expression analyses.
RNA extraction, retrotranscription and Real-Time PCR
As starting material, 120 WID and 250 eye-antenna imaginal discs (EID) were used for RNASeq. For pdm2 gene expression analysis, WIDs from 400 nub-GAL4/+; UAS-GFP/+ flies were disaggregated. RNA from sorted cells was extracted with ZR-RNA MicroPrep Kit from Zymo Research. For L3-specific genes expression, 5 third instar larvae were frozen and RNA was extracted with Quick-RNA MiniPrep Kit, from Zymo Research. Retrotranscriptions and qPCRs were performed as described previously17. For quantification of RNA amounts, standard curves of each pair of primers were performed and the efficiency of amplification was calculated. The Cts obtained from the qPCR were corrected according to the amplification efficiency of the primers. Primers used for Real-Time PCR are listed in Supplementary Table 11.
Genetic mosaics
Clones mutant for ash2I1 were obtained by mitotic recombination using the FLP/FRT technique57. yw;FRT82Bash2I1/TM6C flies were crossed with ywhsflp;FRT82BGFP/TM6B and wing and eye imaginal discs from third instar Tubby+ larvae were dissected. Heat shock was carried out for 45 minutes at 37°C [52 ± 4 hours after egg laying (AEL)] to induce clone generation.
In situ hybridizations and immunohistochemistry
In situ hybridizations and immunostaining were carried out according to standard protocols. The cDNA for pdm2 was PCR amplified using primers listed below and cloned into a pBSK+/− vector at EcoRI restriction site. Riboprobe was synthesized using T7 polymerase and digoxigenin labeled ribonucleotides (Roche). Alkaline phosphatase conjugated with anti-digoxigenin (Roche) and NBT and BCIP (Roche) were used to develop in situ hybridization. Peroxidase conjugated anti-digoxigenin and Tyramide signal amplification (TSA, Life Technologies) was used for fluorescent in situ hybridization (FISH). WIDs and EIDs were analyzed with a DMLB microscope and SPE confocal microscope (Leica). Primary antibodies used were: rabbit anti-H3K4me3 (1:1,000, Abcam/ab8580), mouse anti-En (1:25, DSHB/4D4) and mouse anti-CycA (1:100, DSHB/A12), mouse anti-BOSS (1:1,000)58 and rabbit anti-GFP (1:1,000, Santa Cruz Biotechnology/sc-8334). Fluorescently labeled secondary antibodies were from Life Technologies and Jackson Immunochemicals. Discs were mounted in SlowFade (Life Technologies) supplemented with 1 μM TO-PRO-3 (Life Technologies) to label nuclei. For all in situs and immunostainings around 10 imaginal discs were analyzed. All experiments were performed twice.
Chromatin immunoprecipitation
Third instar larva WID or EID isolated from Canton S flies were fixed, pooled in 700 μL and processed as described17. Around 300 imaginal discs were used in these experiments. Trypsin treated cells from GFP transgenic flies were fixed after sorting for 10 minutes at room temperature and sonicated in a Diagenode Bioruptor for 15 minutes at high power in lysis buffer (1% SDS, 10 mM Tris HCl ph 8.0 and 2mM EDTA). Immunoprecipitations were performed in RIPA buffer. For L3 ChIPs and Imaginal Discs ChIPSeq experiments we used 1 μg of the corresponding antibody. For ChIPs in sorted cells we used 0.45 μg of anti-H3K4me3, 0.3 μg of anti-H3K36me3, 0.33 μg of anti-H3K27ac and 1 μg of anti-H3K27me3. For L3 time-specific ChIPs, 5 Canton S wall-wandering third instar larvae were disrupted, fixed and sonicated as indicated above. . Immunocomplexes were recovered with Invitrogen ProteinA magnetic beads for 2h. The beads were washed three times in RIPA or IP buffer, once in LiCl buffer and twice in TE17. Primers used for Real-Time PCR are listed in Supplementary Table 11. The antibodies used for ChIP were: H3 (Abcam/ab1791); H3K4me3 (Abcam/ab8580) (Millipore-Upstate/07-473), H3K9ac (Abcam/ab4441), H3K36me3 (Abcam/ab9050), H3K4me1 (Diagenode/CS-037-100), H3K27ac (Abcam/ab4729) and H3K27me3 (Upstate-Millipore/07-449).
Nascent RNA
For Nascent RNA assays, 400 WIDs nub-GAL4/+; UAS-GFP/+ were dissected and disaggregated as described above. Click-IT® Nascent RNA Capture Kit from Molecular Probes (C10635) was used according to the manufacturer’s instructions. Briefly, disaggregated cells were incubated with 0.5 mM 5-ethynil uridine (EU) in Schneider’s Insect Medium for 1 h at room temperature. Total RNA was extracted and biotinylated with 0.25 mM biotin-azide for 30 minutes at room temperature. Biotinylated RNA was precipitated overnight at −80°C and purified with Streptavidin conjugated beads for 30 minutes at room temperature. Nascent RNA was eluted in 0.1 % SDS 5 minutes at 99°C and retrotranscription was carried out as described above. Four biological replicates were performed. Primers used for Real-Time PCR are listed in Supplementary Table 11.
Solexa/Illumina sequencing
Solexa/Illumina sequencing was carried out at the Ultrasequencing Unit of the Centre for Genomic Regulation (CRG, Barcelona, Spain). All protocols for Solexa/Illumina ChIPSeq and for RNASeq analysis were carried out following the manufacturer’s protocol. For ChIPSeq, 10 ng of each sample were used and fragments between 300 and 350 bp were size selected before sequencing. For RNASeq, 5 μg of total RNA were used to sequence.
Drosophila melanogaster genome and annotation
We used the FlyBase12 annotation release 5.12 for the genome version dm3.
RNASeq and ChIPSeq read mapping
Reads of 36 and 40 bp obtained from single-end RNASeq and ChIPSeq sequencing from WID and EID-cells were aligned using GEM59 allowing up to two mismatches to the D. melanogaster genome (version dm3) and, for RNA, to all possible junctions of 5′-3′-ordered exon pairs occurring within the same annotated gene. ChIPSeq and RNASeq raw data and profiles of read counts were deposited in the NCBI-GEO repository under the accession number GSE56551.
Gene and transcript expression analysis
Reads mapping uniquely to the genome were used to quantify genes and transcripts separately in each tissue using the FluxCapacitor60. Expression levels are given in Reads Per Kilobase per Million mapped reads (RPKM). Linear regression analysis between log transformed WID and EID RPKMs gave a highly significant slope and intercept. Thus, we identified 628 genes at least one unit above the linear regression line (differentially expressed genes in EID) and 184 genes at least one unit below (differentially expressed genes in WID). To build our collection of regulated tissue-specific genes from each differentially expressed gene set, we required coefficient of variation >= 1.2 and at least 1.5 RPKMs in one tissue and less than 0.1 RPKM in the other one (55 EID-specific genes and 10 WID-specific genes, respectively, resulted from this criterion). Finally, those genes with coefficient of variation < 1.2 that are expressed in both tissues (> 2.3 RPKMs) with a difference in expression of less than 20% were selected as stable expressed in the two tissues (284 genes) and the genes whose expression in both tissues is 0 RPKMs were considered to be silent (30 genes).
ChIPSeq analyses
ChIPSeq reads for H3, H3K4me3, H3K9ac, H3K36me3, H3K4me1 and H3K27ac were extended to the full average fragment length in the corresponding experiment. For each position in the genome the number of extended ChIPSeq reads overlapping this position was recorded. Each sample was normalized by the total number of sequenced reads and the average fragment length. The genome-wide correlation between WID and EID samples was computed using the UCSC Table browser on windows of 1,000 nucleotides56. To compute the correlation between ChIPSeq samples and RNASeq expression data, we assigned to each gene the highest peak of the corresponding ChIP signal within the gene body and correlated this value to the expression of the gene. To produce the graphical distribution of reads for each sample around a particular site (Transcription Start Sites, TSS, polyAdenylation Sites, pAS and splice Acceptor Sites, AS), we calculated the weighted number of reads on each position from −500 bp to +500bp of each TSS, pA and AS, according to FlyBase. To graphically represent an idealized gene, we normalized the location of the reads within the gene using a window of 100 units, and calculated the mean at each point. We extended this representation 500 bps upstream and downstream of the gene. To compare WID and EID samples, we calculated the weighted number of reads on each position in the normalized ChIPSeq profiles.
ENCODE and Roadmap Epigenomic analyses
Stable and developmentally regulated genes in D. melanogaster
To define the transcriptional stability of genes, we calculated the coefficient of variation of gene expression, as reported by the modENCODE consortium10, for each protein-coding gene that has detectable expression in 12 selected developmental time points (Supplementary Fig. 1a). From the full ranking of 13,635 genes, we defined the bottom 1,000 genes with lowest variation of expression during development as stable, and the top 1,000 genes with highest variation as developmentally regulated genes. In addition, at each time point we selected the same number of silent genes than regulated genes expressed at that time point, for a total of 1,000 silent genes. For these genes, we measured the strength of the highest peak (measured as the log of the number of reads reported by modENCODE) within the gene body at the time point in which its expression is maximum for H3K4me3, H3K9ac, H3K4me1, H3K27ac, and the average signal within the gene body for H3K27me3 and H3K9me3 modENCODE ChIPSeq profiles (NCBI GEO accession: GSE16013). Due to data issues with ChIPSeq for three samples: H3K9ac (Adult male) and H3K9me3 (L3 and Adult male), we used ChIP-chip data in these cases instead. The Wilcoxon test (two-sided) was used to evaluate the statistical significance of the difference between ChIP values for stable, regulated and silent genes on each sample. To build the subsets of low, medium and high regulated genes, we ranked the top 1,000 regulated genes by their expression (in the time point of maximum expression) and we classified them into three groups of the same number of genes. Partial correlations between the coefficient of variation and the histone marking of genes, with the effect of the expression of such genes removed, were calculated with the ggm R package.
L3-specific genes analysis
To compare the expression and histone modification marking levels in regulated broadly expressed and stable tissue-specific genes we used anatomy RNASeq data from modENCODE consortium available in Flybase12. We used the gene sets previously defined for modENCODE analysis to create new subgroups of genes:
Stable: the 1,000 genes with the lowest coefficient of variation of gene expression across modENCODE time points
Silent: genes identified as silent in L3 stage (RPKM = 0)
Regulated broadly-expressed at L3: developmentally regulated genes that are detected in L3 whole body data, and that are furthermore expressed with at least 1 RPKM in each of the 6 tissues with L3 tissue-RNASeq available
Stable tissue-specific at L3: from the set of extended stably expressed genes (P1 in Supplementary Fig 4) we selected the genes that, using L3 tissue-RNASeq, are detected as expressed with at least 10 RPKM in 1 of the tissues and not higher than 1 in all the other remaining tissues. We identified 26 carcass-specific genes, 8 central nervous system-specific genes, 36 digestive-specific genes, 21 fat body-specific genes, 36 imaginal disc-specific genes and 4 salivary glands-specific genes.
The expression and histone modification levels were calculated using L3 data from modENCODE following the methodology of the previous analysis.
Stable and developmentally regulated genes in C. elegans
We estimated H3K4me3 and H3K36me3 levels in 7 developmental stages (Early Embryo, Late Embryo, Larvae L1, L2, L3, L4 and Young Adult) from array signal files in Gerstein et al35. To define developmentally stable and regulated genes, we also used the same procedure as in fly. To obtain gene and transcript quantifications, we mapped the RNASeq reads from modENCODE C. elegans35 to the Wbcel215.68 version of the genome using GEM59, and used the FluxCapacitor60 to produce the quantifications. Partial correlations between the coefficient of variation and the histone marking of genes, with the effect of the expression of such genes removed, were calculated with the ggm R package.
Human and mouse analyses
To define the transcriptional stability of human genes, we calculated the coefficient of variation of gene expression, as reported by the Roadmap Epigenomics Consortium, for each protein-coding gene that has detectable expression in the set of 56 consolidated epigenomes14. From the full ranking of 18,064 genes, we defined as constant genes the bottom 1,000 genes with lowest variation of expression across the 56 tissues and cell lines, and the top 1,000 genes with highest variation as variable genes. In addition, at each epigenome we selected the same number of silent genes than variable genes expressed at that tissue, for a total of 1,000 silent genes. For these genes, we measured the strength of the highest peak (measured as the log of the number of reads reported by the Roadmap Epigenomics consortium) within the gene body at the tissue in which its expression is maximum for H3K4me3, H3K36me3 and H3K4me1. The Wilcoxon test (two-sided) was used to evaluate the statistical significance of the difference between ChIP values for constant, variable and silent genes on each sample. Partial correlations between the coefficient of variation and the histone marking of genes, with the effect of the expression of such genes removed, were calculated with the ggm R package.
The same protocol was applied in the analysis of the mouse ENCODE15 RNASeq and ChIPSeq (H3K4me3, H3K9ac, H3K36me3, H3K4me1 and H3K27ac) data in ten adult tissues for which RNASeq data and ChIPSeq data all modifications are available: Cerebellum, Cortex, Heart, Kidney, Liver, Placenta, Small intestine, Spleen, Testis and Thymus.
Nucleosome turnover
Using the provided Nascent RNA signal tracks in S2 cells18 with no treatment we calculated the average signal in the gene body of the previously defined stable, regulated and silent gene sets. Stable and regulated genes with signal over 1 were kept (986 stable and 56 regulated) and silent genes with signal equal 0 were also kept (258 genes). In these remaining genes we calculated the nucleosome turnover rate as the average CATCH-IT signal, within the gene body, in S2 cells with no treatment18. The Wilcoxon test (two-sided) was used to evaluate the statistical significance of the signal among the gene sets.
Promoter analyses
To measure the conservation of the promoters of regulated and stable genes across 12 Drosophilids, we computed the average of the UCSC PhastCons multiz15way track56 along the promoter sequences of each gene set (promoter length: 200 bp). To characterize the promoters of regulated and stable genes, we used the MatScan program61 with the full collection of 827 predictive matrices available in Jaspar and Transfac62, 63. From each initial pool of predictions, we removed those binding sites within genome regions in the UCSC genome browser that presented on average a probability lower than 0.95 to be conserved across the 12 flies PhastCons multiz15way alignments64. The Wilcoxon test (one-sided) was used to evaluate the statistical significance of the difference for stable and developmentally regulated gene sets on each comparison (PhastCons scores and number of conserved sites). For the identification of focused/dispersed initiation sites8, 33, we searched for putative binding sites of TBP and DRE in the promoter sequence of the top 1,000 stable and the top 1,000 regulated genes (promoter length: 100 bp). We selected TBP as a marker of focused initiation and DRE as a representative of dispersed initiation. The weight matrix for TBP is from Jaspar62 and for DRE is from Fly Factor Survey65.
Principal Components Analysis (PCA) was performed based on the ChIPSeq levels of 20 Transcription Factors in Embryos at 0–12h in the promoter regions of genes with expression above 10 as measured by tilling arrays at this time point6.
Genome segmentations
To match the states of a particular map of genome segmentation and our sets of stable and regulated genes, we counted how many genes of these two groups overlap with the segments of each state. To annotate our collection of genes, we used the modENCODE ChromHMM66 maps of BG3 and S2 cell lines42, the hiHMM maps of Late Embryo and L343 and the chromatin types identified by Filion and colleagues41. To annotate the topological information of stable and regulated genes, we conducted a 4similar analysis on the HiC genome domains previously identified on Late Embryo44 and the Lamina Associated Domains reported in Kc cells46.
Trancription factor perturbation analysis
To study the effects of transcription factors in stable and regulated genes we analyzed publicly available data on knock-down or overexpression of various Drosophila transcription factors36-40. First we checked how many stable and regulated genes were expressed in the tissue/cell type used in each study before the perturbation of the transcription factor, using published expression data on brain L336, Kc cells67, S2 cells67 and our L3 eye imaginal disc. Genes with RPKM > 1 were considered expressed. Then, we intersected the stable and regulated expressed genes with the genes identified as differentially expressed in each study.
Splicing entropy
For each gene, we computed the Shannon’s entropy (or diversity index) based on the relative frequencies of the gene’s annotated isoforms in a given cell line. Let g be a gene with n annotated isoforms with relative frequencies p1, …,pn, in a given condition, the entropy of g, H(g), is computed as
H(g) growths with the number of annotated isoforms and with the evenness of their frequencies. H(g) is zero when there is only one expressed isoform (which would correspond to tight regulation of isoform expression), and it is maximum when all isoforms are equally expressed (which would correspond to lack of splicing regulation and stochastic production of alternative splicing isoforms). Based on transcript quantifications produced by the modENCODE project for the fly, and computed by us for the worm (see Methods), we calculated the splicing entropy of each gene at the developmental time point in which its expression is at its maximum. The boxplots in Figure 8f display the distribution of H(g), separately for genes with different number of isoforms.
Supplementary Material
Acknowledgements
We thank D. Gonzalez-Knowles, A. Breschi and M. Melé, for help with data analysis, F. Serras, M. Morey and A. Kornblihtt for insightful suggestions and G. Cavalli for discussing data before publication, as well as the anonymous reviewers for their critical input. We thank the modENCODE, ENCODE (human and mouse) and the Roadmap Epigenomics Mapping consortiums for granting open access of these resources to the scientific community. We also thank the Ultrasequencing Unit of the CRG (Barcelona, Spain), for sample processing and the Confocal Unit of the CCiTUB (Universitat de Barcelona, Spain). This work was performed under the financial support of the Spanish Ministry of Economy and Competitiveness with grants BIO2011-26205 to RG, CSD2007-00008 and BFU2012-36888 to M. C., and ‘Centro de Excelencia Severo Ochoa 2013-2017’, SEV-2012-0208, and of the ERC/European Community PF7 with grant 294653 RNA-MAPS to R. G. E. B. is supported by the European Commission’s 7th Framework Program 4DCellFate grant number 277899. This research reflects only the authors’ views and that the Community is not liable for any use that may be made of the information contained therein. J. C. is supported by grant SFRH/BD/33535/2008 from the Portuguese Foundation to Science and Technology.
Footnotes
Accession codes
ChIPSeq and RNASeq raw data and profiles of read counts were deposited in the NCBI-GEO repository under the accession number GSE56551.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong X, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13:R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107:2926–31. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–31. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodl M, Basler K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol. 2012;22:2253–7. doi: 10.1016/j.cub.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Chen K, et al. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Gao L, Anandhakumar J, Gross DS. Uncoupling transcription from covalent histone modification. PLoS Genet. 2014;10:e1004202. doi: 10.1371/journal.pgen.1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–9. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–41. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Lluch S, et al. Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res. 2011;39:4628–39. doi: 10.1093/nar/gkq1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luque CM, Milan M. Growth control in the proliferative region of the Drosophila eye-head primordium: the elbow-noc gene complex. Dev Biol. 2007;301:327–39. doi: 10.1016/j.ydbio.2006.07.050. [DOI] [PubMed] [Google Scholar]
- 20.Weihe U, Dorfman R, Wernet MF, Cohen SM, Milan M. Proximodistal subdivision of Drosophila legs and wings: the elbow-no ocelli gene complex. Development. 2004;131:767–74. doi: 10.1242/dev.00979. [DOI] [PubMed] [Google Scholar]
- 21.Ng M, Diaz-Benjumea FJ, Cohen SM. Nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development. 1995;121:589–99. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- 22.Beltran S, Angulo M, Pignatelli M, Serras F, Corominas M. Functional dissection of the ash2 and ash1 transcriptomes provides insights into the transcriptional basis of wing phenotypes and reveals conserved protein interactions. Genome Biol. 2007;8:R67. doi: 10.1186/gb-2007-8-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltran S, et al. Transcriptional network controlled by the trithorax-group gene ash2 in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100:3293–8. doi: 10.1073/pnas.0538075100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeitlinger J, Stark A. Developmental gene regulation in the era of genomics. Dev Biol. 2010;339:230–9. doi: 10.1016/j.ydbio.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 25.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–32. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 27.Gaertner B, et al. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2012;2:1670–83. doi: 10.1016/j.celrep.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskins RA, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–92. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rach EA, Yuan HY, Majoros WH, Tomancak P, Ohler U. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 2009;10:R73. doi: 10.1186/gb-2009-10-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni T, et al. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods. 2010;7:521–7. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–45. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 32.Teves SS, Henikoff S. The heat shock response: A case study of chromatin dynamics in gene regulation. Biochem Cell Biol. 2013;91:42–8. doi: 10.1139/bcb-2012-0075. [DOI] [PubMed] [Google Scholar]
- 33.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–9. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farley E, Levine M. HOT DNAs: a novel class of developmental enhancers. Genes Dev. 2012;26:873–6. doi: 10.1101/gad.192583.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–87. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Froldi F, et al. The transcription factor Nerfin-1 prevents reversion of neurons into neural stem cells. Genes Dev. 2015;29:129–43. doi: 10.1101/gad.250282.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georlette D, et al. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21:2880–96. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennington KL, Marr SK, Chirn GW, Marr MT., 2nd Holo-TFIID controls the magnitude of a transcription burst and fine-tuning of transcription. Proc Natl Acad Sci U S A. 2013;110:7678–83. doi: 10.1073/pnas.1221712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pineyro D, Blanch M, Badal M, Kosoy A, Bernues J. GAGA factor repression of transcription is a rare event but the negative regulation of Trl is conserved in Drosophila species. Biochim Biophys Acta. 2013;1829:1056–65. doi: 10.1016/j.bbagrm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Turkel N, et al. The BTB-zinc finger transcription factor abrupt acts as an epithelial oncogene in Drosophila melanogaster through maintaining a progenitor-like cell state. PLoS Genet. 2013;9:e1003627. doi: 10.1371/journal.pgen.1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho JW, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512:449–52. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 46.van Bemmel JG, et al. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–5. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 48.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 49.de Almeida SF, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–83. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 50.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delest A, Sexton T, Cavalli G. Polycomb: a paradigm for genome organization from one to three dimensions. Curr Opin Cell Biol. 2012;24:405–14. doi: 10.1016/j.ceb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Espada J, Esteller M. DNA methylation and the functional organization of the nuclear compartment. Semin Cell Dev Biol. 2010;21:238–46. doi: 10.1016/j.semcdb.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Gan Q, et al. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010;20:763–83. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol. 2015;427:608–25. doi: 10.1016/j.jmb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 56.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
References of online methods
- 57.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 58.Cagan RL, Kramer H, Hart AC, Zipursky SL. The bride of sevenless and sevenless interaction: internalization of a transmembrane ligand. Cell. 1992;69:393–9. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- 59.Derrien T, et al. Fast computation and applications of genome mappability. PLoS One. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–7. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanco E, Messeguer X, Smith TF, Guigo R. Transcription factor map alignment of promoter regions. PLoS Comput Biol. 2006;2:e49. doi: 10.1371/journal.pcbi.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Portales-Casamar E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–10. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–32. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 64.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu LJ, et al. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 2011;39:D111–7. doi: 10.1093/nar/gkq858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cherbas L, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–14. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.