Abstract
Grainyhead transcription factors control epithelial barriers, tissue morphogenesis, and differentiation, but their role in the kidney is poorly understood. Here, we report that nephric duct, ureteric bud, and collecting duct epithelia express high levels of grainyhead-like homolog 2 (Grhl2) and that nephric duct lumen expansion is defective in Grhl2-deficient mice. In collecting duct epithelial cells, Grhl2 inactivation impaired epithelial barrier formation and inhibited lumen expansion. Molecular analyses showed that GRHL2 acts as a transcriptional activator and strongly associates with histone H3 lysine 4 trimethylation. Integrating genome-wide GRHL2 binding as well as H3 lysine 4 trimethylation chromatin immunoprecipitation sequencing and gene expression data allowed us to derive a high-confidence GRHL2 target set. GRHL2 transactivated a group of genes including Ovol2, encoding the ovo-like 2 zinc finger transcription factor, as well as E-cadherin, claudin 4 (Cldn4), and the small GTPase Rab25. Ovol2 induction alone was sufficient to bypass the requirement of Grhl2 for E-cadherin, Cldn4, and Rab25 expression. Re-expression of either Ovol2 or a combination of Cldn4 and Rab25 was sufficient to rescue lumen expansion and barrier formation in Grhl2-deficient collecting duct cells. Hence, we identified a Grhl2/Ovol2 network controlling Cldn4 and Rab25 expression that facilitates lumen expansion and barrier formation in subtypes of renal epithelia.
Keywords: Grainyhead transcription factors, collecting duct, lumen expansion, barrier, formation, nephric duct, Grainyhead-like 2
The renal collecting duct’s vital electrolyte and water-regulatory functions are carried out by highly specialized cell populations.1 The collecting duct itself is composed of a tight epithelium, which separates the urinary compartment from a hypertonic interstitium and maintains a barrier to concentration gradients. The collecting duct derives from the ureteric bud, which emanates from the nephric duct and then undergoes branching morphogenesis in response to signals from the adjacent metanephric mesenchyme.2 Many of the genes that are critical for aspects of nephric duct development continue to be expressed in the ureteric bud and collecting duct, serving roles in development, differentiation, and maintenance of these cells (e.g., Pax2/8, Gata3, Emx2, and Hnf1b).3 Little is known about the molecular pathways governing the specific epithelial properties of these cells, although it is clear that these epithelia share several cell biologic properties such as a uniform tubular appearance characterized by a cuboidal epithelium surrounding a fluid-filled lumen and a molecular composition of the apical junctional complex that includes E-cadherin–based adherence junctions and CLDN4-containing tight junctions, both of which are characteristic of a tight epithelial barrier.
The grainyhead transcription factor family regulates various aspects of epithelial function, including differentiation and morphogenesis.4–6 Grainyhead-like 2 (Grhl2) is one of three mammalian homologs of Drosophila grainyhead. It is expressed in the surface ectoderm, the gut tube, and the otic vesicle, which are among the earliest barrier-forming epithelia of the embryo.7,8 Grhl2 expression remains uniformly high in E-cadherin–positive epithelia throughout development and adulthood.8 Grhl2 is required for important morphogenetic events during embryonic development, with gene deletion in mice leading to defective neural tube closure, split face malformations, exencephaly, and death at embryonic day (E) 11.5.8–10 Grhl2 was reported to participate in epithelial morphogenesis and differentiation in cultured airway,11 in biliary and hepatic epithelia,12,13 and in tumorigenesis of skin and mammary epithelial cells.14,15 However, the underlying genetic networks are incompletely understood.
Here, we show that epithelia of the nephric duct, ureteric bud, and collecting duct express high levels of GRHL2. Grhl2 inactivation results in defects of epithelial barrier formation and lumen expansion. We combine gene expression and genome-wide chromatin immunoprecipitation data to show that GRHL2 strongly associates with H3K4 trimethylation, a chromatin mark important for transcriptional activity. Next, we derive a high-confidence target gene set and show that GRHL2 activates genes encoding the apical junctional complex proteins CDH1, CLDN4, and the small GTPase RAB25, as well as the zinc finger transcription factor ovo-like 2 (OVOL2). In functional studies, we identify a novel Grhl2/Ovol2 pathway that reinforces CDH1, CLDN4, and RAB25, and regulates lumen expansion and barrier formation in collecting duct epithelia.
Results
GRHL2 Localization and DNA Binding Sites in the Kidney
In mice, strong GRHL2 immunoreactivity was detected in nuclei of nephric ducts in E10.0 mouse embryos, of ureteric bud epithelia in E15.5 mouse embryos, and of distal nephron epithelia and collecting ducts in E18 mouse embryonic kidney, adult mouse kidney, and adult human kidney (Figure 1). GRHL2 was not detectable in metanephric mesenchymal cells, in renal stromal cells, in glomeruli, and in proximal segments of the nephron. GRHL2 binds to a DNA consensus sequence highly similar to the DNA matrix for Drosophila grainyhead.11,16 We used chromatin immunoprecipitation with a GRHL2 antibody and sequencing (ChIP-seq) on kidneys from E18 and adult mice, as well as on murine inner medullary collecting duct (IMCD-3) cells. We detected genomic regions bound by GRHL2 (peaks) with model-based analysis for ChIP-seq (MACS) using default parameters17 and found 7366, 7890, and 10,621 peaks in IMCD-3 cells, adult mouse kidney, and E18 mouse kidney, respectively. The three data sets revealed a marked overlap, which became even more pronounced when only the top 25% scoring peaks were considered (Figure 2A, Supplemental Figure 1A). The distance distribution of IMCD-3 cell GRHL2 peaks from adult kidney peaks revealed two distinct large groups with ChIP peaks that either overlapped or occurred at large distances from each other (Supplemental Figure 1B). The overlap of in vivo GRHL2 ChIP peaks with IMCD-3 GRHL2 peaks was highly nonrandom (P<0.001). Peak annotation of all three GRHL2 ChIP-seq experiments showed a statistically significant (P<0.001) clustering near transcriptional start sites, promoters, and intronic regions (Figure 2B). De novo motif discovery with MEME revealed a sequence (Figure 2C) that is identical to previously published GRHL2 binding motifs.18
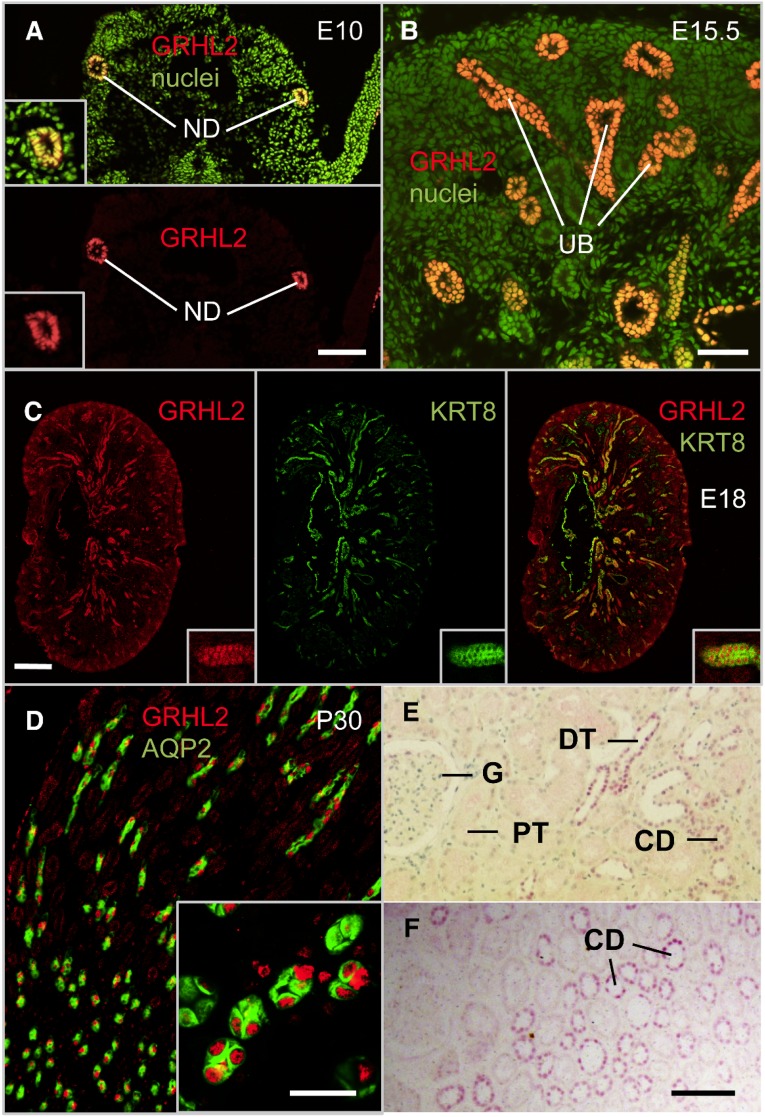
Figure 1.
GRHL2 is localized in epithelial nuclei in the nephric duct, ureteric bud, collecting duct, and distal nephron. (A) Immunofluorescence staining of a cross-section of an E10.0 mouse embryo at the level of the urogenital ridges revealing GRHL2-positive nuclei in the nephric ducts. (B) Immunofluorescence staining of murine embryonic kidney at E15.5 shows GRHL2 expression in the branching ureteric bud. (C and D) Immunofluorescence staining of murine embryonic (E18) (C) and adult (P30) (D) kidney tissue using antibodies against GRHL2 (colors as indicated), the ureteric bud marker keratin 8 (KRT8), and the collecting duct principal cell marker aquaporin 2 (AQP2). (E and F) Immunohistochemical staining of human adult kidney sections using a GRHL2 antibody (purple staining). The section in E is slightly counterstained with hematoxylin and eosin, and the section in F is without counterstaining. ND, nephritic duct; UB, ureteric bud; G, glomerulus; PT, proximal tubule; DT, distal tubule; CD, collecting duct. Bar, 100 µm in A, E, and F; 50 µm in B; 300 µm in C; 20 µm in D.
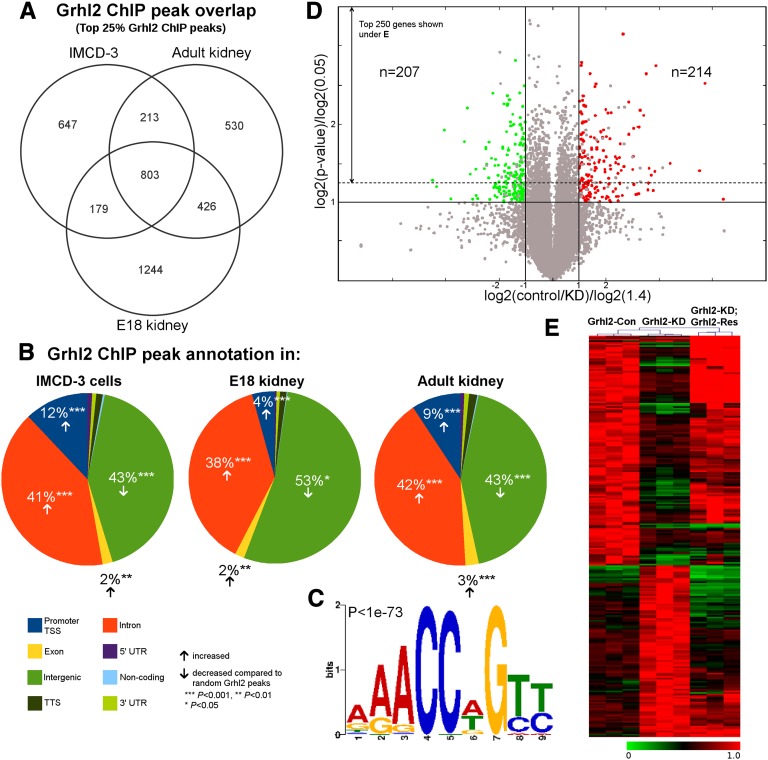
Figure 2.
GRHL2 binding in IMCD-3 cells, adult kidney, and embryonic kidney, and gene expression data. (A) Venn diagram for the top 25% GRHL2 ChIP peaks shows a significant overlap of peaks between IMCD-3 cells, adult, and E18 kidney with a substantial number of peaks present in all three scenarios. (B) Promoters/TSSs (blue), exons (yellow), and introns (orange) show a statistically significant enrichment for GRHL2 ChIP peaks compared with random peaks, whereas intergenic regions (green) display a significant de-enrichment. (C) De novo motif discovery from the top 10% of IMCD-3 GRHL2 peaks yields a consensus sequence identical to previously published grainyhead binding motifs (P value provided by MEME). (D) Volcano plot of the expression data of all genes displaying log-transformed P values and fold changes. The solid lines represent the fold change thresholds of 1.4 (orthogonal) and the P value threshold of 0.05 (horizontal). According to the axes labels, genes colored in red are downregulated in Grhl2-KD (n=214) and genes in green are upregulated in Grhl2-KD (n=207) compared with Grhl2-Con. Gray data points refer to nondifferential genes or differentially expressed genes not rescued in Grhl2-KD; Grhl2-Res. The dotted line marks the border to the top 250 differentially expressed genes depicted in E. (E) Heatmap of the top 250 differentially regulated genes from microarray data for control (n=3), Grhl2 knockdown (n=3), and Grhl2 rescue (n=3) in IMCD-3 cells. Expression values are maximum normalized for each gene. *P<0.05; **P<0.01; ***P<0.001. TSS, transcriptional start site; UTR, untranslated region.
GRHL2 Acts as a Transcriptional Activator and Strongly Associates with Regions of H3K4 Trimethylation
To gain further insight into the transcriptional network downstream of GRHL2, we generated IMCD-3 cells with a Grhl2 knockdown (Grhl2-KD) using lentiviral gene transfer of a short hairpin RNA (shRNA) expression cassette (Supplemental Figure 2, A and B). To control the specificity of the shRNA, we also generated IMCD-3 cells expressing a control shRNA (Grhl2-Con), and IMCD-3 cells expressing the Grhl2 shRNA and a rescue construct consisting of a full-length Grhl2 cDNA with silent mutations at the shRNA target site rendering it insensitive to shRNA-mediated degradation (Grhl2-KD; Grhl2-Res). GRHL2 protein was reduced to 26.3%±4.9% in Grhl2-KD compared with Grhl2-Con and reinduced up to 348.4%±49.5% in Grhl2-KD; Grhl2-Res (Supplemental Figure 2, D and E). We performed gene expression analysis on Grhl2-Con, Grhl2-KD, and Grhl2-KD; Grhl2-Res IMCD-3 cells using microarrays. We found 421 genes that were differentially expressed (P<0.05, ≥1.4-fold change, Grhl2-Con versus Grhl2-KD). Of these genes, 207 were upregulated and 214 were downregulated upon Grhl2 deficiency (Figure 2, D and E, see Supplemental Table 1 for a list of these genes). Expression of 372 (88.4%) of these genes was rescued toward control levels in Grhl2-KD; Grhl2-Res (Supplemental Table 1). Gene ontology analysis of differentially expressed genes in Grhl2-KD, using the HOMER software package,19 revealed a high enrichment in key epithelial cell programs such as differentiation, development, and cell organization (Supplemental Table 2). These genes included previously reported GRHL2 targets such as Rab25 (encoding the GTPase Rab 25), Cdh1 (encoding E-cadherin), and Cldn4 (encoding claudin 4; Supplemental Figure 2, C and D).8,12
Histone H3 lysine 4 trimethylation (H3K4me3) is a chromatin mark associated with actively transcribed genes.20,21 We previously reported that H3K4 trimethylation at the Cdh1 promoter is dependent on GRHL2.8 To better understand the association of GRHL2 and H3K4 trimethylation in a genome-wide context, we utilized H3K4me3 ChIP-seq from Grhl2-Con and Grhl2-KD cells. Using MACS, we found 19,904 absolute H3K4me3 peaks corresponding to regions of strong H3K4 trimethylation in Grhl2-Con cells. Of these regions, 1440 displayed significantly reduced H3K4 trimethylation in Grhl2-KD (MACS on Grhl2-Con using Grhl2-KD as background model) and 417 displayed significantly enhanced H3K4 trimethylation in Grhl2-KD cells (MACS on Grhl2-KD using Grhl2-Con as a background model). This indicated widespread modulation of H3K4 trimethylation by GRHL2, particularly toward a loss of H3K4 trimethylation in the absence of GRHL2. Regions of significantly reduced H3K4 trimethylation in Grhl2-KD were strongly overlapping with GRHL2 peaks, genes, and their transcriptional start sites (Figure 3A). In fact, GRHL2 binding sites were observed in more than one third of the regions that displayed significantly reduced H3K4 trimethylation in Grhl2-KD compared with Grhl2-Con, and among more than half of the top differentially regulated regions (Figure 3B). Conversely, they were rarely found in other regions of H3K4 trimethylation, particularly in regions that displayed increased H3K4 trimethylation in Grhl2-KD cells (Figure 3B). We observed a tight association of GRHL2 binding with genes downregulated in Grhl2-KD compared with Grhl2-Con, but not with genes that were upregulated (Figure 3B). Hence, our data are consistent with a model in which GRHL2 associates with H3K4 trimethylation and acts mainly as a transcriptional activator. Conversely, although several genes were negatively regulated by GRHL2, including epithelial-to-mesenchymal transition (EMT)–associated genes encoding vimentin (Vim), zinc finger E-box binding homeobox 1 (Zeb1), and twist basic helix-loop-helix transcription factor 2 (Twist2), we did not find evidence for direct physical interaction of GRHL2 with these genes (Figure 3B and Supplemental Figure 3 showing GRHL2 ChIP-seq data at the Vim, Zeb1, and Twist2 loci), suggesting that GRHL2 may regulate their expression by indirect effects.
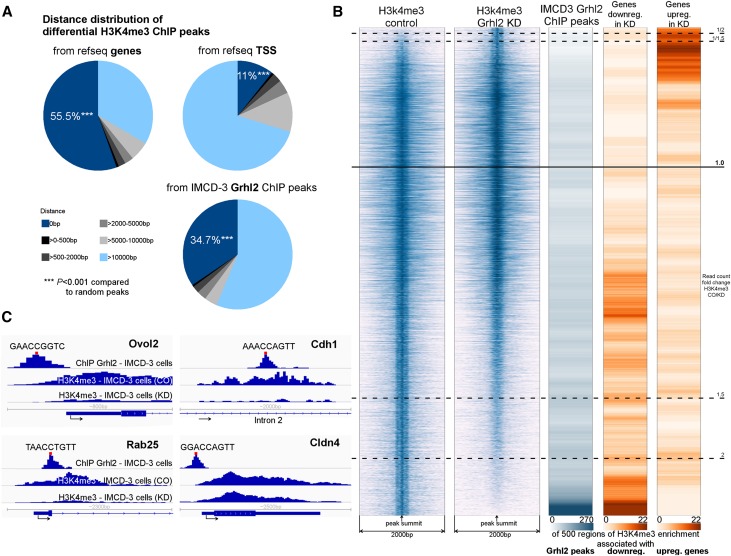
Figure 3.
GRHL2 acts as a transactivator and strongly associates with regions of H3K4 trimethylation. (A) Differential H3K4me3 peaks (significantly more H3K4me3 in Grhl2-Con compared with Grhl2-KD) strongly cluster at genes, TSSs, and GRHL2 ChIP peaks (P<0.001 for 0 bp distance compared with random peaks). (B) Integrated analysis of ChIP-seq and gene expression data. The first two columns display regions of H3K4me3 enrichment in Grhl2-Con and Grhl2-KD cells (26,500 regions corresponding to H3K4me3 peaks detected). Row-wise, a window of 2 kb around the peak summit is shown. Peaks are sorted by read count ratio (Grhl2-Con/Grhl2-KD). The horizontal black lines mark read count fold change thresholds. Note that in response to Grhl2 deficiency, H3K4 trimethylation is more frequently lost than gained. The third column shows a density plot indicating the frequency of GRHL2 ChIP peaks within the respective H3K4me3 regions. The last two columns show density plots representing the frequency of differentially expressed genes downregulated (column 4) or upregulated (column 5) in Grhl2-KD cells adjacent to the respective H3K4me3 regions. A H3K4me3 region is considered associated with a gene if it is ≤500 bp away from the gene’s transcriptional start site or/and intersected with the gene body. In total, 175 downregulated and 170 upregulated genes are associated with some H3K4me3 region shown. Note that a gene can associate with multiple H3K4me3 regions. The density plots are generated by calculating a sliding average over 500 H3K4me3 regions. (C) Detailed view of ChIP-seq peak locations in proximity to the genes encoding Ovol2, Cdh1, Rab25, and Cldn4. The red bar indicates the position of a significant GRHL2 binding motif (P<0.0001, sequence depicted). TSS, transcriptional start site. ***P<0.001.
The Core Target Gene Set of GRHL2 Includes the Gene Encoding Ovol2
To search for relevant GRHL2 targets, we combined the data of our experiments and defined strict criteria for possible target genes, including the following: (1) differential expression between Grhl2-Con and Grhl2-KD cells, (2) successful rescue of expression levels in Grhl2-KD; Grhl2-Res cells, (3) differential GRHL2-dependent H3K4 trimethylation in the gene or ≤500 bp from the transcriptional start site, and (4) GRHL2-binding peaks in IMCD-3 and adult kidney chromatin overlapping with the site of differential H3K4 trimethylation. The rationale for these rigorous criteria was derived from our observation that GRHL2-dependent H3K4me3 peaks either intersect with or appear at a large distance from GRHL2 peaks and GRHL2-dependent genes (Figure 3A). This approach yielded a set of 17 genes. Twelve of these genes exhibited a peak center related (±50 bp from peak summit) significant GRHL2 binding motif (P<0.001) (Supplemental Figure 1, C and D, Supplemental Table 3). Among these core target genes, we found previously reported targets, Cdh1 and Rab25,8,12 and additional novel targets, including the gene encoding the zinc finger transcription factor Ovol2. Claudin 4 is a known GRHL2 target gene,8,22 but was not represented within the core target gene set due to a lack of a differential H3K4me3 peak, although all other criteria were fulfilled. However, visual inspection indicated a reduced H3K4 trimethylation level at the Cldn4 locus in Grhl2-KD cells compared with Grhl2-Con, which did not fulfill our strict statistical criteria (Figure 3C).
GRHL2 is Required for Lumen Expansion and Epithelial Barrier Function
To study the role of GRHL2 in renal epithelial morphogenesis, we analyzed Grhl2-deficient mice8 and Grhl2-deficient IMCD-3 cells. Because Grhl2 mutant mice exhibit embryonic lethality at E11.5, we focused our analysis on the nephric duct epithelium at E10.0, a stage at which Grhl2-deficient embryos and their control littermates are still comparable in size and developmental stage. We analyzed nephric duct morphology in the posterior part of the mesonephros where mesonephric tubules are visible but not connected to the nephric duct. There, the nephric duct epithelium forms a uniform straight tubule with a relatively constant lumen area, which can be quantitated in cross-sections of the embryo. In control embryos, the epithelium was cuboidal and formed a fluid-filled lumen at this stage, expressing the nephric duct marker PAX2 (Figure 4C). In Grhl2 mutants, the nephric duct still revealed a cuboidal epithelium, but the lumen was markedly reduced and occasionally displayed a complete collapse (Figure 4, A and C). Average lumen area was reduced to 27% in these ducts compared with control littermate nephric ducts (Figure 4B). Epithelial polarization of Grhl2 mutant nephric ducts appeared intact based on the localization of β-catenin and PAR3 (Figure 4C).
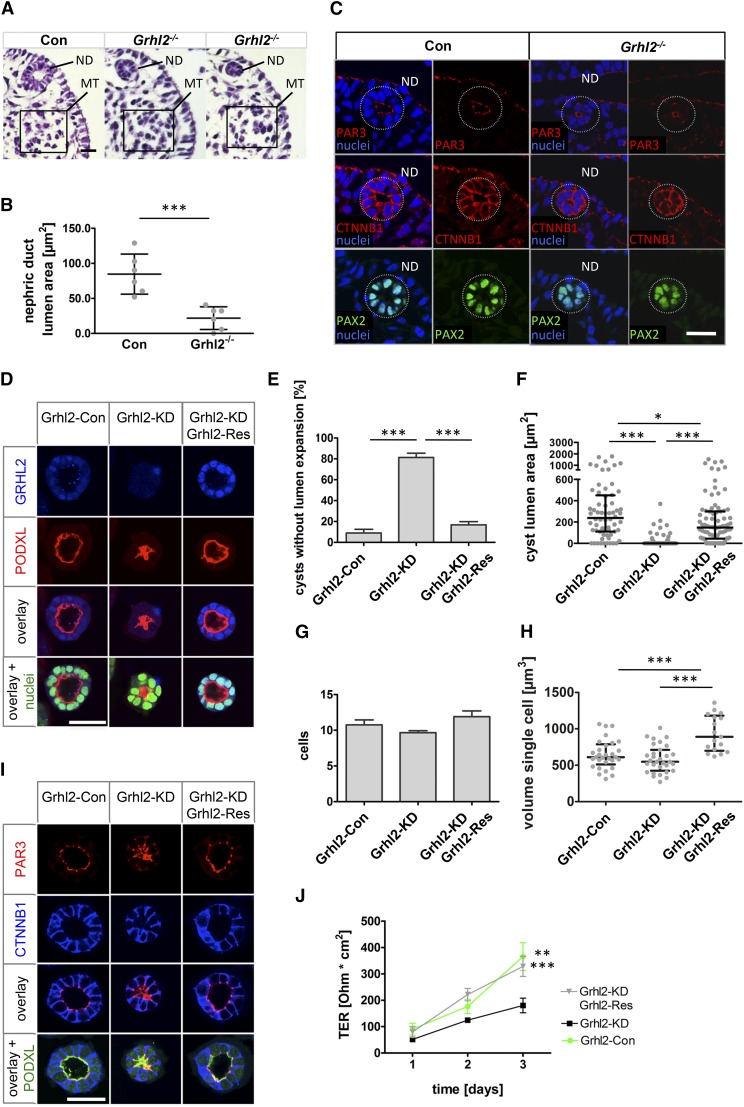
Figure 4.
GRHL2 regulates lumen expansion and barrier formation. (A) Hematoxylin and eosin staining of E10.0 Grhl2-deficient (Grhl2−/−) and control embryo cross-sections at the level of the caudal mesonephros. Grhl2−/− nephric ducts show a reduced (middle) or collapsed (right) lumen compared with control embryos. (B) Statistical analysis of E10.0 nephric duct lumen area with a significantly reduced lumen area in Grhl2−/− embryos is shown (mean±SD). (C) Immunofluorescence staining of E10.0 mouse embryo sections at the same level as in A using antibodies against PAX2, β-catenin (CTNNB1), and PAR3 in control and Grhl2−/− nephric ducts. (D–I) IMCD-3 collecting duct cell cysts formed in Matrigel after 5 days of culture are immunofluorescence stained and analyzed by confocal microscopy. (D) To evaluate lumen expansion cysts are stained with antibodies against GRHL2 and podocalyxin (PODXL). Nuclei are counterstained using SYTOXGreen. (E) Percentages of cysts that did not develop a single open lumen are shown (mean±SEM). (F) The area bounded by the apical, podocalyxin-marked membrane of the cells is determined at the level of the largest lumen diameter of each cyst (median and IQR). Cysts that did not develop an open lumen are assigned an area of 0. (G) Cysts are analyzed for the number of cells surrounding the lumen at the largest diameter (mean±SEM). (H) Volumes of individual cells are determined using Imaris software after three-dimensional reconstruction (median and IQR). (I) Representative cysts are shown after immunofluorescence staining using antibodies against PAR3, β-catenin (CTNNB1), and PODXL. Grhl2-Con, Grhl2-KD, and Grhl2-KD; Grhl2-Res cells are grown on polycarbonate membranes. Measurements of TERs are shown for the indicated days (mean±SEM). *P<0.05; **P<0.01; ***P<0.001. Con, control; ND, nephric duct; MT, mesonephric tubules; IQR, interquartile range. Bar, 25 µm in A and C; 38 µm in D; 35 µm in I.
We next examined IMCD-3 epithelial morphogenesis in three-dimensional cultures and barrier formation through measurements of transepithelial resistance (TER) of IMCD-3 monolayers in transwells. Grhl2-Con, Grhl2-KD, and Grhl2-KD; Grhl2-Res IMCD-3 cells were seeded in Matrigel. During a culture period of 5 days, Grhl2-Con cells formed epithelial cysts, consisting of polarized epithelial cells surrounding a central lumen outlined by podocalyxin, a marker of the apical membrane. Conversely, Grhl2-KD cells displayed a collapse of the cyst lumen. Although the apical plasma membranes of the Grhl2-KD cells were formed and podocalyxin was localized apically, the lumen frequently failed to expand with an undetectable or markedly diminished luminal space (Figure 4, D and E). Expression of Grhl2-Res in Grhl2-KD cells restored the number of open lumen-forming cysts, indicating that GRHL2 specifically mediates this effect (Figure 4, D and E). Extending the culture period to 9 days did not substantially alter these observations, indicating a qualitative defect of lumen expansion rather than a temporal delay (Supplemental Figure 4, A and B). Of note, we observed multiple lumen formation only in rare instances, both in Grhl2-Con cysts and in Grhl2-KD cysts. To quantitate the defect in lumen expansion, the lumen area bounded by the apical, podocalyxin-marked membrane at the center and at maximal lumen diameter of each cyst was analyzed. This revealed profound differences in lumen size between Grhl2-Con and Grhl2-KD cells and their rescue in Grhl2-KD; Grhl2-Res cells (Figure 4F).
To determine whether a reduced number of cells or differences in the cell volume of Grhl2-KD cells relative to controls contributed to the defective lumen expansion in Grhl2-KD cysts, we analyzed the number of cells surrounding the lumen at the largest cyst diameter and the single cell volume within each cyst. Analysis exhibited no significant differences in the number of cyst-lining cells between Grhl2-Con, Grhl2-KD, and Grhl2-KD; Grhl2-Res cells (Figure 4G). Grhl2-KD cells showed no significant reduction in single cell volume compared with control cells, whereas Grhl2-KD; Grhl2-Res cells displayed a significantly increased single cell volume compared with Grhl2-KD and Grhl2-Con cells (Figure 4H). Recent investigations showed that a defective single lumen formation might be explained by a disrupted orientation of the mitotic spindle during cell division.23–25 We found that in Grhl2-KD cells, the mitotic spindle axis was mainly perpendicular to the podocalyxin-marked center of the cysts, comparable to Grhl2-Con and Grhl2-KD; Grhl2-Res cells (Supplemental Figure 4, C and D). We next analyzed markers of apicobasal polarity by staining for the basolateral marker β-catenin, the cell polarity complex component PAR3, and the tight junction protein zonula occludens 1 (Figure 4I and Supplemental Figure 4E), revealing no overt abnormalities in Grhl2-KD cysts compared with Grhl2-Con and Grhl2-KD; Grhl2-Res cysts. Together, these analyses indicate that Grhl2-deficient collecting duct cells display a selective defect of lumen expansion without evidence of mitotic axis deviation, multiple lumen formation, or disruption of cell polarity.
Next, we analyzed the effect of Grhl2 inactivation on barrier formation in IMCD-3 monolayers by measuring TER over a 3-day culture period. Grhl2-KD cells displayed a significantly lower TER than Grhl2-Con cells, which was appropriately rescued in Grhl2-KD; Grhl2-Res cells (Figure 4J). Extending the culture period for up to 7 days did not alter this finding (Supplemental Figure 4F). Together, these data suggest that GRHL2 regulates collecting duct cell lumen expansion independent of cell number or alteration of apicobasal cell polarization, but by a mechanism that may involve enforcement of the epithelial barrier.
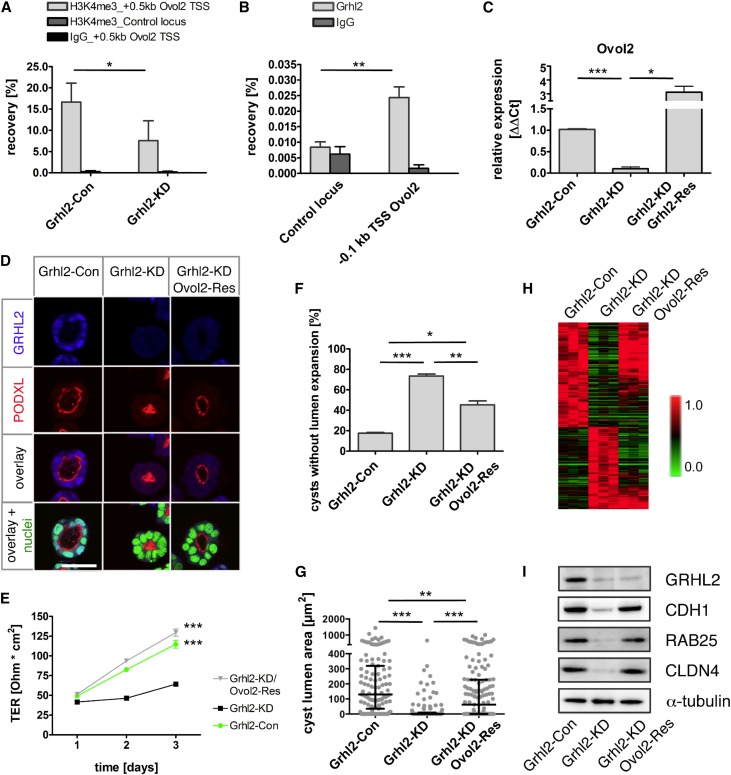
Ovol2 Reinduces Central Components of the GRHL2 Network and Rescues TER and Lumen Expansion in Grhl2-Deficient Cells
The gene encoding OVOL2, a mammalian homolog of Drosophila ovo,26 was among the core targets of GRHL2. Deletion of Ovol2 in mice results in an embryonic phenotype and death by E10.5,27 similar to the phenotype observed in Grhl2 mutant mice. Ovol2 expression correlates with hallmark genes of epithelial differentiation in cancer cell lines.28 Therefore, we decided to further investigate the potential functional role of Ovol2 downstream of GRHL2. Downregulation of Ovol2 and H3K4 trimethylation at the Ovol2 promoter in Grhl2-KD cells and GRHL2-binding to the Ovol2 promoter in IMCD-3 cells were validated by quantitative RT-PCR and ChIP-quantitative PCR, respectively (Figure 5, A–C). We next analyzed the effect of ectopically expressed full-length Ovol2 in Grhl2-KD cells on lumen formation and barrier function by generating Grhl2-KD; Ovol2-Res cells (Supplemental Figure 5A). Remarkably, Ovol2 re-expression significantly increased the percentage of cysts establishing a single open lumen and resulted in a higher TER compared with Grhl2-KD cells (Figure 5, D–G), indicating that reinduction of Ovol2 expression in Grhl2-deficient cells restored the hallmarks of GRHL2 inactivation. To analyze the molecular basis of these effects, we performed gene expression profiling using microarrays in Grhl2-Con, Grhl2-KD, and Grhl2-KD; Ovol2-Res cells. Unexpectedly, this revealed that 75.7% of the genes downregulated in Grhl2 deficiency were reinduced by Ovol2 overexpression (Figure 5H). Among these genes were Cldn4, Cdh1, and Rab25, which were rescued to baseline levels in Grhl2-KD; Ovol2-Res cells based on mRNA and protein levels (Figure 5I, Supplemental Figure 5B). Interestingly, re-expression of Ovol2 also resuppressed the expression of 85.5% of genes that were upregulated in Grhl2-KD cells, including the EMT-associated genes Vim, Zeb1, and Twist2.
Figure 5.
Re-expression of Ovol2 after Grhl2 knockdown rescues lumen formation and reinduces GRHL2-dependent genes. (A) Validation of differential H3K4 trimethylation at the Ovol2 TSS between Grhl2-KD and Grhl2-Con cells using ChIP-quantitative PCR. Shown are mean values of percent recovery ±SD. Primers amplified a region in the H3K4me3 peak area, 0.5 kb downstream of Ovol2 TSS. Chromatin is amplified for a negative control locus. IgG is used as negative antibody control. (B) ChIP-quantitative PCR using an antibody against GRHL2 reveals an enrichment at the Ovol2 promoter (0.1 kb upstream of Ovol2 TSS) compared with a negative control locus in IMCD-3 chromatin. (C) mRNA expression of Ovol2 is analyzed using quantitative RT-PCR in Grhl2-KD, Grhl2-Con, and Grhl2-KD; Grhl2-Res cells (mean±SEM). (D) After 5 days of culture in Matrigel, cysts from Grhl2-Con, Grhl2-KD, and Grhl2-KD; Ovol2-Res cells are immunofluorescence stained and analyzed by confocal microscopy. Representative cysts stained with antibodies against GRHL2 and PODXL are shown. Nuclei are counterstained using SYTOXGreen. (E) Grhl2-Con, Grhl2-KD, and Grhl2-KD; Ovol2-Res cells are grown on polycarbonate membranes. Measurements of TER are shown for the indicated days (mean±SEM). (F and G) Cysts grown in Matrigel for 5 days are analyzed for lumen formation. (F) Percentages of cysts that did not develop a single open lumen are shown (mean±SEM). (G) Areas generated from sections with the largest lumen diameter are calculated for every cyst (median and IQR). (H) Microarray gene expression analysis is performed on Grhl2-Con (n=3), Grhl2-KD (n=3), and Grhl2-KD; Ovol2-Res (n=3) cells. The top 250 differentially expressed genes are hierarchically clustered (Pearson correlation, average linkage clustering). Note substantial reinduction of Grhl2-dependent genes in Grhl2-KD cells after Ovol2 overexpression. (I) Western blots for CDH1, GRHL2, CLDN4, and RAB25 are performed on Grhl2-Con, Grhl2-KD, and Grhl2-KD; Ovol2-Res cells. α-Tubulin is used as the loading control. *P<0.05; **P<0.01; ***P<0.001. TSS, transcriptional start site; IQR, interquartile range; ΔΔCt, difference in cycle threshold. Bar, 38 µm in D.
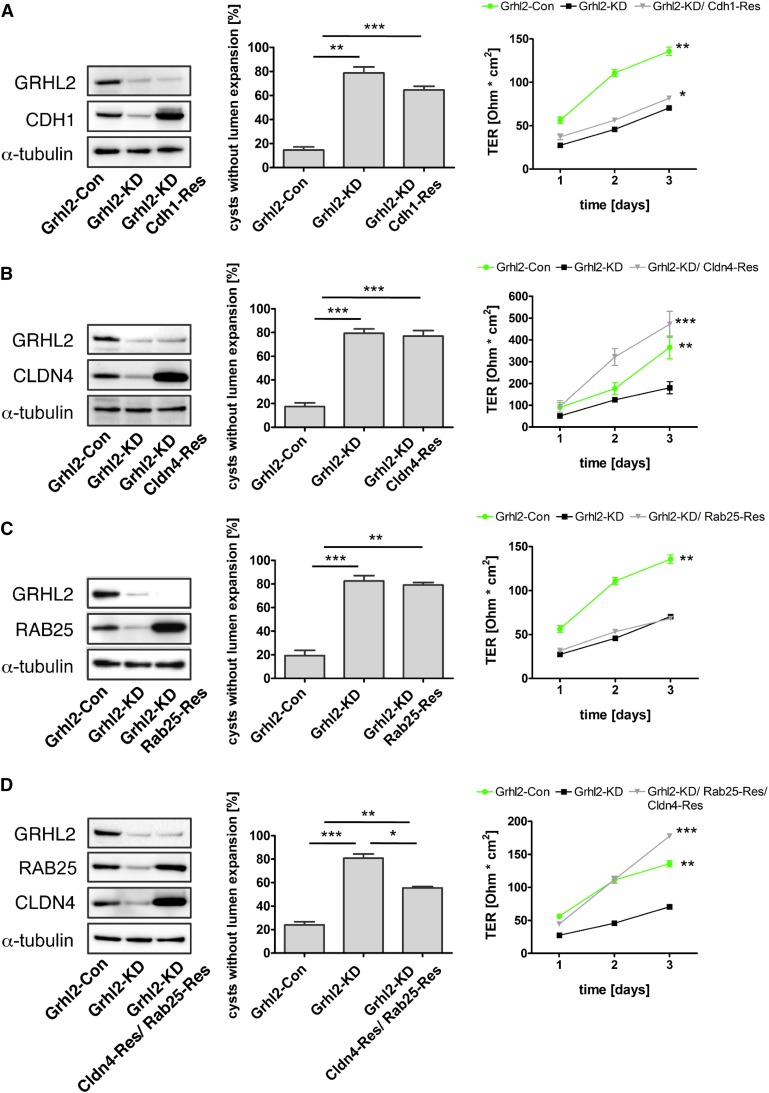
Recent studies implicated CDH1, RAB25, and CLDN4 in epithelial lumen formation.12,29,30 Although CDH1 and CLDN4 are components of the apical junctional complex, RAB25 appears to be important for cargo shuttling to the apical domain. Therefore, we decided to re-express each of these genes in Grhl2-KD cells to assay for their sufficiency to replace GRHL2 function. Re-expression of Cldn4 in Grhl2-KD cells was sufficient to completely restore TER in transwell monolayers, whereas Cdh1 and Rab25 were insufficient to rescue TER (Figure 6, A–C). However, re-expression of neither of these genes alone was sufficient to rescue lumen expansion in Grhl2-KD cells (Figure 6, A–C). This suggested that GRHL2 drove barrier formation via Cldn4 activation but that this alone was insufficient to explain the effect of GRHL2 on lumen expansion. Because Rab25 had previously been implicated in epithelial lumen formation,29 we re-expressed a combination of Cldn4 and Rab25 in Grhl2-KD cells. Notably, this rescued both lumen formation and TER (Figure 6D). Together these data identify a novel Grhl2/Ovol2 pathway that reinforces Cldn4 and Rab25 expression and thereby regulates lumen expansion and barrier formation.
Figure 6.
CLDN4 and RAB25 are critical mediators of GRHL2 function. Cdh1, Cldn4, and Rab25 are re-expressed in Grhl2-KD cells. Protein expression is evaluated using Western blots (left panels). Effects on lumen formation (center panels) and TER (right panels) are quantified as described above. Grhl2-Con, Grhl2-KD, and Grhl2-KD cells overexpressing Cdh1 (Grhl2-KD; Cdh1-Res) (A), Cldn4 (Grhl2-KD; Cldn4-Res) (B), Rab25 (Grhl2-KD; Rab25-Res) (C), or a combination of Cldn4 and Rab25 (Grhl2-KD; Cldn4-Res; Rab25-Res) (D) were analyzed. *P<0.05; **P<0.01; ***P<0.001.
Discussion
We present the first functional and molecular analysis of GRHL2 and its downstream signaling events in renal epithelia. Through an integration of gene expression data, genome-wide GRHL2 binding in vivo and in vitro, and global analysis of H3K4 trimethylation, we were able to derive GRHL2 downstream targets and discovered a Grhl2/Ovol2 network controlling lumen expansion and barrier formation. We show that GRHL2 expression is high in nephric ducts, in ureteric bud epithelia, and in distal nephron epithelia and collecting ducts. A loss of Grhl2 leads to a reduced or even collapsed lumen in E10.0 nephric ducts. Functional analyses using IMCD-3 cells showed that GRHL2 controls epithelial barrier formation (determined by TER) and lumen expansion. Molecular analyses indicate a network involving GRHL2-mediated activation of Ovol2, both of which reinforce expression of important epithelial effectors, including CDH1, CLDN4, and RAB25. In addition, our data suggest that GRHL2-induced CLDN4 expression mediates increased barrier formation, but this is insufficient to completely explain the effect on lumen formation. Only a joint re-expression of CLDN4 and RAB25 was able to rescue lumen expansion in Grhl2-deficient IMCD-3 cells, suggesting that RAB25 mediates important aspects of lumen formation downstream of GRHL2 that go beyond increasing the epithelial barrier.
Rab25, a Rab11 GTPase family member, has been reported to control apical trafficking and transcytosis.31,32 and has previously been associated with defective MDCK cyst lumen formation.29 Inactivation of Rab25 or the related Rab11a resulted in a multiple lumen phenotype in epithelial cysts, which is characterized by failure of lumen expansion and retention of podocalyxin in subapical vesicles.29,33 Multiple lumen formation has been linked with defective CDC42-mediated orientation of the mitotic spindle.23,24 Interestingly, in response to Grhl2 inactivation in IMCD-3 cells, we found no evidence of multiple lumen formation or misorientation of the mitotic spindle. In fact, even extended culture of Grhl2-deficient IMCD-3 cysts for 9 days produced regularly aligned polarized cells surrounding a single large patch of podocalyxin without any visible lumen (Supplemental Figure 4A). This may be an intrinsic property of collecting duct cells and distinct from the MDCK or Caco-2 cell systems. However, it suggests that Grhl2-deficient collecting duct cells exhibit a defect of lumen expansion that occurs subsequent to adequate cell alignment and polarization. Because previous studies indicated that lumen expansion is driven by fluid accumulation34–37 and requires the formation of tight paracellular barriers,36 we propose that GRHL2 regulates lumen expansion by stabilizing the epithelial barrier via CLDN4 and facilitating apical fluid excretion via RAB25. However, this hypothesis will require additional experimental support.
Our study also provides novel insights into the molecular regulation of target genes by GRHL2. We show that GRHL2 acts as a predominant activator of target genes, supporting and expanding our previous finding of GRHL2 transactivation of Cdh1 and Cldn4.8 This conclusion is based on genome-wide evidence of strong association of GRHL2 with genes that are downregulated when GRHL2 function is lost and with regions of decreased H3K4 trimethylation in Grhl2-KD. Conversely, we found little evidence of a direct repressor function of GRHL2, because we saw no evidence of a direct association of GRHL2 with genes that are upregulated in Grhl2-deficient cells or with corresponding regions of upregulated H3K4 trimethylation in Grhl2-KD. This finding is in contrast with previous reports of a direct suppressor effect of GRHL2 at for instance EMT-associated genes, including the mesenchymal transcription factor ZEB1.14,38 Although we observed an activation of EMT-associated genes Zeb1, Vim, and Twist2 in Grhl2-deficient cells, we did not find direct association of GRHL2 with their promoters or other adjacent gene-regulatory regions. These discrepancies may be related to differences in tissues or cell types between different studies.
Our identified core target Ovol2 is a member of an evolutionary conserved family of zinc finger transcription factors. Ovol2 is involved in diverse processes such as keratinocyte differentiation and cell fate decisions, as well as embryonic development.27,39,40 Our current data reveal that most differentially regulated genes in Grhl2-deficient cells were rescued to their original expression levels, when Ovol2 was re-expressed in these cells. Furthermore, we show that Ovol2 is sufficient to functionally replace Grhl2 in lumen expansion and barrier function through induction of Cldn4 and Rab25. Potentially, OVOL2 may mediate the previously identified function of GRHL2 as a suppressor of EMT. In a recent publication, it was shown that OVOL2 is necessary to suppress EMT in mammary cells with OVOL2 ChIP peaks near the Vim, Twist1, and Zeb1 promoters.41 Consistently, our data show that Ovol2 overexpression can abolish the upregulation of EMT genes Vim, Zeb1, and Twist2 in Grhl2-deficient cells. The fact that CLDN4 and RAB25 together were sufficient to functionally replace GRHL2 in lumen formation and barrier function suggests that suppression of EMT genes is not a mandatory prerequisite for these processes.
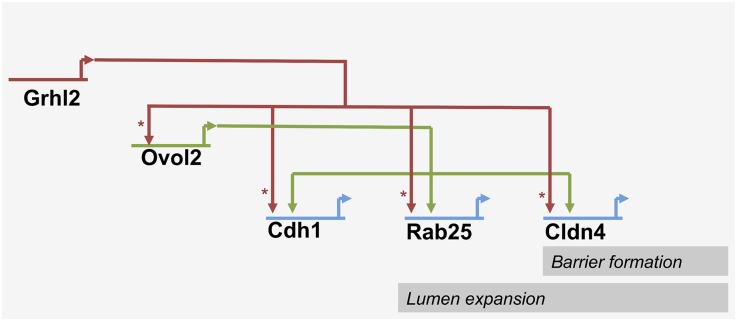
The novel gene network we derived has two input transcription factors, one of which (GRHL2) regulates the other (OVOL2), both jointly regulating effector target genes (e.g., Cdh1, Cldn4, and Rab25; see Figure 7 for a putative transcriptional network). This system bears the characteristics of a feed-forward signaling system.42 Feed-forward loops of this type are widespread regulatory elements and are implicated in sign-sensitive delay and preventing inactivation of the target gene program when input signals are lost (i.e., they serve as stabilizers of target gene expression) and have been shown to be relevant in the context of cellular differentiation.42–45
Figure 7.
A putative transcriptional network downstream of Grhl2 regulating barrier formation and lumen expansion in collecting duct cells. Asterisks indicate direct regulations, whereas interactions indicated by open arrowheads may be direct or indirect.
Our analysis of Grhl2-deficient mice revealed that Grhl2 is required for nephric duct lumen expansion in vivo. Because globally Grhl2-deficient mice exhibit embryonic lethality at E11.5,8 we were unable to analyze the role of Grhl2 in the ureteric bud or the collecting duct of the developing and mature kidney in this model. This will require conditional inactivation of Grhl2, which our laboratory is currently pursuing.
Ablation of Ovol2 in mouse embryos leads to lethality by E10.5 accompanied by severe embryonic defects including abnormal neural tube closure.27,46 This phenotype is highly reminiscent of the Grhl2 knockout phenotype,8,10 suggesting a more widespread functional similarity of GRHL2 and OVOL2 and a potential relevance of the Grhl2/Ovol2 signaling loop in cells and tissues beyond the kidney.
Concise Methods
For this study, we used murine embryonic, murine, and human kidney tissues as well as murine IMCD-3 cells cultured in cell culture plates or in Matrigel and applied cell culture methodology, generation of recombinant lentiviruses, lentiviral gene transfer, PCR, microarray analysis, ChIP-seq technology, immunofluorescence and immunohistochemical staining, Western blot, and measurements of TERs as outlined in detail in the Supplemental Materials and Methods.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Antje Sommer, Tatjana Luganskaja, Gabriel Kirchgraber, Kirstin Rautenberg, and Siegrun Blauhut for their excellent technical support. We also thank Dr. Nora Mecklenburg for help with paraffin embedding of murine embryos, Dr. Friedrich C. Luft for critical comments concerning the manuscript, Dr. Hans-Peter Rahn (Max Delbrück Center Preparative Flow Cytometry Facility), and Dr. Anje Sporbert (Max Delbrück Center Advanced Light Microscopy Facility).
This work was funded by the German Research Foundation (Emmy-Noether grant (to K.S.), Research Unit 667 grant (to K.S.), and Research Unit 1368 grant (to K.S.)) and the Urological Research Foundation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014080759/-/DCSupplemental.
References
- 1.Madsen KM, Clapp WL, Verlander JW: Structure and function of the inner medullary collecting duct. Kidney Int 34: 441–454, 1988 [DOI] [PubMed] [Google Scholar]
- 2.Little M, Georgas K, Pennisi D, Wilkinson L: Kidney development: Two tales of tubulogenesis. Curr Top Dev Biol 90: 193–229, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Costantini F: Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. Wiley Interdiscip Rev Dev Biol 1: 693–713, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narasimha M, Uv A, Krejci A, Brown NH, Bray SJ: Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J Cell Sci 121: 747–752, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, Andersen B: The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol 299: 122–136, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Yu Z, Mannik J, Soto A, Lin KK, Andersen B: The epidermal differentiation-associated Grainyhead gene Get1/Grhl3 also regulates urothelial differentiation. EMBO J 28: 1890–1903, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM: Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns 6: 964–970, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Werth M, Walentin K, Aue A, Schönheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M, Barasch J, Rosenbauer F, Luft FC, Schmidt-Ott KM: The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137: 3835–3845, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Pyrgaki C, Liu A, Niswander L: Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev Biol 353: 38–49, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rifat Y, Parekh V, Wilanowski T, Hislop NR, Auden A, Ting SB, Cunningham JM, Jane SM: Regional neural tube closure defined by the Grainy head-like transcription factors. Dev Biol 345: 237–245, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH, Reddy TE, Hogan BL: Evidence for multiple roles for grainyheadlike 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc Natl Acad Sci U S A 110: 9356–9361, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N: Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell 23: 2845–2855, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanimizu N, Nakamura Y, Ichinohe N, Mizuguchi T, Hirata K, Mitaka T: Hepatic biliary epithelial cells acquire epithelial integrity but lose plasticity to differentiate into hepatocytes in vitro during development. J Cell Sci 126: 5239–5246, 2013 [DOI] [PubMed] [Google Scholar]
- 14.Cieply B, Riley P, 4th, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM: Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res 72: 2440–2453, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang X, Chen W, Kim RH, Kang MK, Park NH: Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene 28: 565–574, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilanowski T, Tuckfield A, Cerruti L, O’Connell S, Saint R, Parekh V, Tao J, Cunningham JM, Jane SM: A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev 114: 37–50, 2002 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C: Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36, 1994 [PubMed] [Google Scholar]
- 19.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK: Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL: Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 99: 8695–8700, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T: Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411, 2002 [DOI] [PubMed] [Google Scholar]
- 22.Varma S, Cao Y, Tagne JB, Lakshminarayanan M, Li J, Friedman TB, Morell RJ, Warburton D, Kotton DN, Ramirez MI: The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J Biol Chem 287: 37282–37295, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe AB, Kaji N, Durgan J, Hall A: Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol 183: 625–633, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Fraticelli AE, Vergarajauregui S, Eastburn DJ, Datta A, Alonso MA, Mostov K, Martín-Belmonte F: The Cdc42 GEF Intersectin 2 controls mitotic spindle orientation to form the lumen during epithelial morphogenesis. J Cell Biol 189: 725–738, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durgan J, Kaji N, Jin D, Hall A: Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J Biol Chem 286: 12461–12474, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dai Q, Li L, Nair M, Mackay DR, Dai X: Ovol2, a mammalian homolog of Drosophila ovo: Gene structure, chromosomal mapping, and aberrant expression in blind-sterile mice. Genomics 80: 319–325, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackay DR, Hu M, Li B, Rhéaume C, Dai X: The mouse Ovol2 gene is required for cranial neural tube development. Dev Biol 291: 38–52, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roca H, Hernandez J, Weidner S, McEachin RC, Fuller D, Sud S, Schumann T, Wilkinson JE, Zaslavsky A, Li H, Maher CA, Daignault-Newton S, Healy PN, Pienta KJ: Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE 8: e76773, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant DM, Datta A, Rodríguez-Fraticelli AE, Peränen J, Martín-Belmonte F, Mostov KE: A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol 12: 1035–1045, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia L, Liu F, Hansen SH, Ter Beest MB, Zegers MM: Distinct roles of cadherin-6 and E-cadherin in tubulogenesis and lumen formation. Mol Biol Cell 22: 2031–2041, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casanova JE, Wang X, Kumar R, Bhartur SG, Navarre J, Woodrum JE, Altschuler Y, Ray GS, Goldenring JR: Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol Biol Cell 10: 47–61, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzaban S, Massol RH, Yen E, Hamman W, Frank SR, Lapierre LA, Hansen SH, Goldenring JR, Blumberg RS, Lencer WI: The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J Cell Biol 185: 673–684, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desclozeaux M, Venturato J, Wylie FG, Kay JG, Joseph SR, Le HT, Stow JL: Active Rab11 and functional recycling endosome are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am J Physiol Cell Physiol 295: C545–C556, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Bagnat M, Cheung ID, Mostov KE, Stainier DY: Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol 9: 954–960, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Barcroft LC, Offenberg H, Thomsen P, Watson AJ: Aquaporin proteins in murine trophectoderm mediate transepithelial water movements during cavitation. Dev Biol 256: 342–354, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Ferrari A, Veligodskiy A, Berge U, Lucas MS, Kroschewski R: ROCK-mediated contractility, tight junctions and channels contribute to the conversion of a preapical patch into apical surface during isochoric lumen initiation. J Cell Sci 121: 3649–3663, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Meder D, Shevchenko A, Simons K, Füllekrug J: Gp135/podocalyxin and NHERF-2 participate in the formation of a preapical domain during polarization of MDCK cells. J Cell Biol 168: 303–313, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM: Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res 73: 6299–6309, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells J, Lee B, Cai AQ, Karapetyan A, Lee WJ, Rugg E, Sinha S, Nie Q, Dai X: Ovol2 suppresses cell cycling and terminal differentiation of keratinocytes by directly repressing c-Myc and Notch1. J Biol Chem 284: 29125–29135, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T, Zhu Q, Xie Z, Chen Y, Qiao Y, Li L, Jing N: The zinc finger transcription factor Ovol2 acts downstream of the bone morphogenetic protein pathway to regulate the cell fate decision between neuroectoderm and mesendoderm. J Biol Chem 288: 6166–6177, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Villarreal-Ponce A, Sun P, Salmans ML, Fallahi M, Andersen B, Dai X: Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev Cell 29: 59–74, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangan S, Alon U: Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A 100: 11980–11985, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murugan R: Theory on the dynamics of feedforward loops in the transcription factor networks. PLoS ONE 7: e41027, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R: The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2: e328, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojas-Sutterlin S, Lecuyer E, Hoang T: Kit and Scl regulation of hematopoietic stem cells. Curr Opin Hematol 21: 256–264, 2014 [DOI] [PubMed] [Google Scholar]
- 46.Unezaki S, Horai R, Sudo K, Iwakura Y, Ito S: Ovol2/Movo, a homologue of Drosophila ovo, is required for angiogenesis, heart formation and placental development in mice. Genes Cells 12: 773–785, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.