Abstract
Peptide and protein sequence analysis using a combination of gas-phase ion–ion chemistry and tandem MS is described. Samples are converted to multiply charged ions by ESI and then allowed to react with fluoranthene radical anions in a quadrupole linear ion trap mass spectrometer. Electron transfer from the radical anion to the multiply charged peptide or protein promotes random fragmentation along the amide backbone that is independent of peptide or protein size, sequence, or the presence of post-translational modifications. Examples are provided that demonstrate the utility of electron-transfer dissociation for characterizing post-translational modifications and for identifying proteins in mixtures on a chromatographic timescale (500 ms/protein).
Keywords: cell migration, chromatin, HIV regulator of expression of virion products, mass spectrometry, O-GlcNAcylation, phosphorylation, post-translational modifications, protein identification
Introduction
The traditional method of identifying proteins in complex mixtures by tandem MS involves the following steps: (a) enzymatic digestion with trypsin; (b) fractionation of the resulting tryptic peptides (usually 10–30 residues in length) by nanoflow HPLC interfaced to a mass spectrometer equipped for ESI; (c) fragmentation of individual peptides by collision-activated dissociation (CAD); and (d) a search of the resulting tandem mass spectra against a database of spectra predicted for tryptic peptides of all known proteins. Thousands of proteins in cultured cells, tissues and biological fluids have been identified by this approach. Unfortunately, the CAD step in the above protocol often promotes the loss of labile post-translational modifications (PTMs) (i.e. phosphate [1–3] and carbohydrate [4] modifications) and provides only limited sequence information from large peptides and intact proteins.
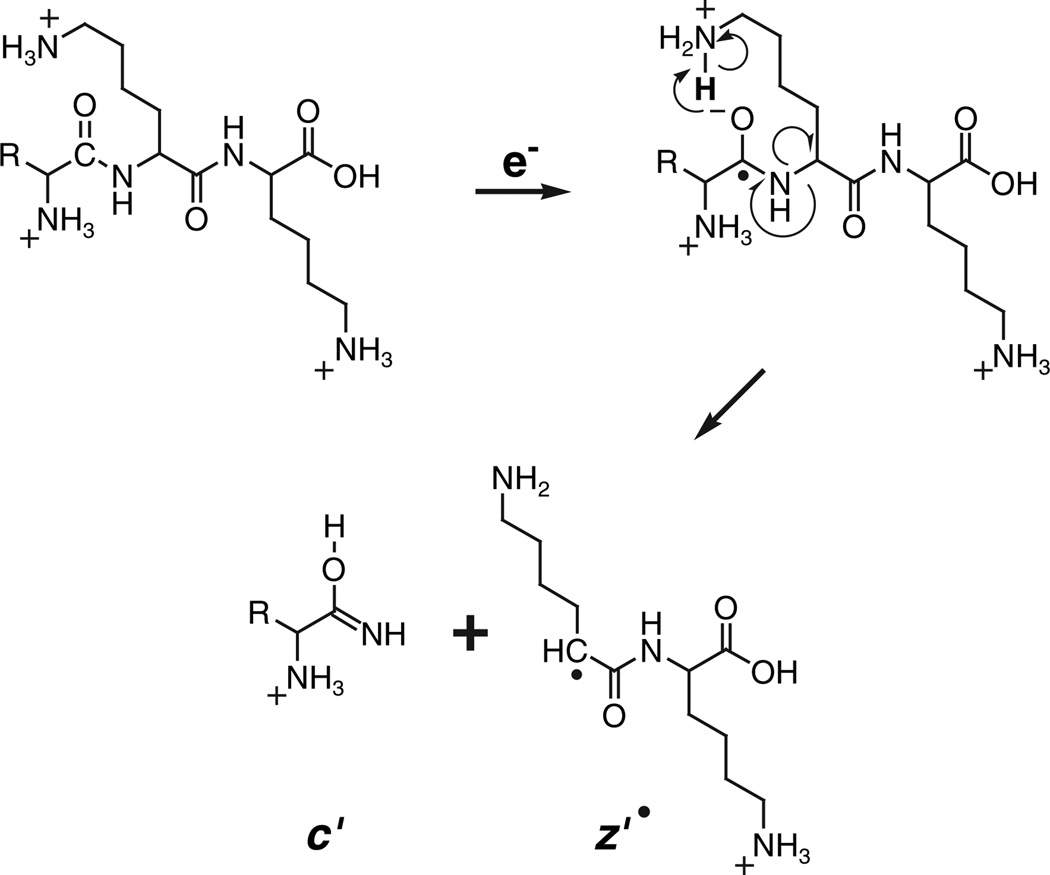
Electron-capture dissociation (ECD), a technique introduced by the McLafferty laboratory in 1998, overcomes the above limitations [5]. In this method, multiply protonated peptides or proteins are confined in the Penning trap of an FT ion cyclotron resonance mass spectrometer and allowed to interact with a beam of electrons having thermal or near- thermal energies. Capture of a thermal electron by a protonated peptide or protein is exothermic by ~6 eV and causes the peptide backbone to fragment by a nonergodic process, e.g. one that does not involve intramolecular vibrational energy redistribution. One possible pathway for this process (Fig. 1) involves capture of the electron into an amide carbonyl group that is hydrogen bonded to the protonated side chain of a basic amino acid. The resulting radical anion abstracts a proton and generates a radical site that triggers dissociation to produce a complementary pair of fragment ions of type c′ and z′˙. Subtraction of the m/z values for the fragments within a given ion series that differ by a single amino acid affords the mass and thus the identity of the extra residue in the larger of the two fragments. The complete amino acid sequence of a peptide is deduced by extending this process to all homologous pairs of fragments within a particular ion series.
Fig. 1.
Fragmentation scheme for production of ions of type c′ and z′˙ by reaction of a low-energy electron with a multiply protonated peptide.
Because ECD occurs along the peptide backbone in a size- and sequence-independent manner and preserves PTMs, it has become the method of choice for analysis of intact large proteins on FT ion cyclotron resonance mass spectrometers [6,7]. Unfortunately, ECD in its most efficient form requires that sample ions be immersed in a dense population of near-thermal electrons. This requirement makes it technically challenging to implement ECD on the instruments used most commonly for peptide and protein analysis, those that trap ions by radiofrequency electrostatic fields rather than with static magnetic and electric fields. High-quality ECD spectra often require the averaging of data from large numbers of scans acquired over a period of minutes. This latter requirement precludes the widespread use of ECD for the analysis of peptides and proteins in complex mixtures by mass spectrometers interfaced to a chromatographic technique such as HPLC.
Electron-transfer dissociation (ETD), a technique introduced by the Hunt laboratory in 2004, overcomes both of the above limitations [3]. For ETD, radical anions of polyaromatic hydrocarbons, such as fluoranthene, are formed under chemical ionization conditions, stored in a quadrupole linear ion trap (QLT) mass spectrometer, and then allowed to react with multiply protonated peptides and proteins in the gas phase.
In the above ion–ion reaction, the fluoranthene radical anion transfers an electron to the [M + 3H]+3 species and the charge-reduced peptide ion then fragments by the same mechanism believed to be responsible for ECD [3,8]. As with ECD, the observed fragmentation along the peptide backbone is independent of peptide or protein size, sequence, or the presence of PTMs. Because the ion–ion chemistry is highly efficient and requires only milliseconds to complete, ETD can easily be performed with femtomole quantities of sample on a timescale that is compatible with LC-MS [3].
Discussed below are examples that illustrate the utility of ETD for assigning sites of PTMs and for both identifying and characterizing intact proteins in mixtures on a chromatographic timescale.
Identification of PTMs
One of the first research applications demonstrating the value of ETD involves strategies employed to analyze the yeast phosphoproteome. Prior to the development of ETD, strategies to identify phosphophorylation sites were limited to analysis of tryptic peptides by low-energy CAD. In an earlier study, a yeast whole cell lysate was proteolytically digested with trypsin, and phosphopeptides were enriched from the sample using immobilized metal affinity chromatography and analyzed using nanoflow HPLC interfaced to a QLT mass spectrometer (Thermo Electron LTQ, Thermo Scientific, San Jose, CA) [2]. The eluting peptides were introduced into the LTQ mass spectrometer via ESI, and CAD was utilitzed for tandem MS to assign sites of phosphorylation. We detected > 1000 phosphopeptides but defined only 383 sites, largely because the CAD process often promoted elimination of phosphoric acid from Ser and Thr residues without breaking amide bonds along the peptide backbone. The resulting MS/MS spectra were deficient in the necessary sequence information.
In a more recent study, we used endoproteinase LysC as the proteolytic enzyme to produce more highly charged, longer peptides, utilized immobilized metal affinity chromatography for phosphopeptide enrichment, and employed ETD for peptide fragmentation on an LTQ mass spectrometer modified for ETD. From a single nanoflow LC-MS/MS experiment on 20 µg of cell lysate, we identified 1252 phosphorylation sites on 629 proteins that were expressed at levels from < 50 copies per cell to 1.2 × 106 copies per cell [1]. A single peptide phosphorylated on His was also sequenced. We concluded that ETD is ideally suited for determining sites of phosphorylation.
A second example involves the application of ETD for the identification of PTMs that regulate the formation and disassembly of focal adhesions, protein complexes that allow the cell to migrate through the extracellular matrix [9]. Proteomics methodologies utilizing ETD have been developed for the purpose of mapping phosphorylation sites on migration-related proteins. A summary of our findings to date can be found on the website http://www.cellmigration.org (first select the CMC Activity Center subheading and then select the Proteomics link).
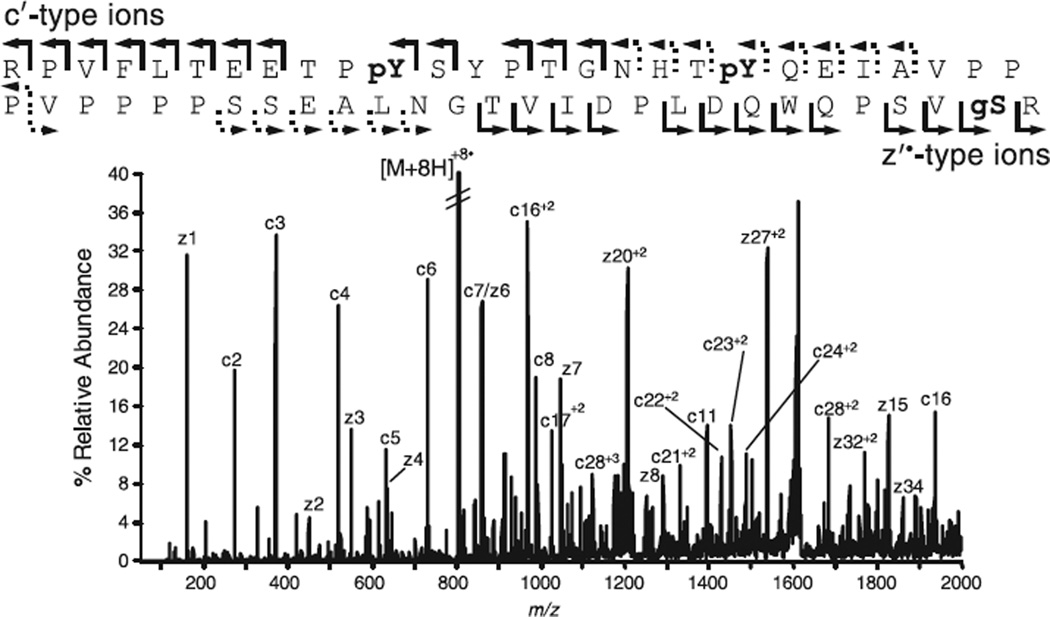
One of the proteins of interest to the Cell Migration Consortium is paxillin, a scaffolding protein involved in focal adhesions [9]. Shown in Fig. 2 is the ETD spectrum recorded on a [M + 8H]+8 ion (m/z 805.5) from a 55 residue tryptic peptide within paxillin [10]. Ions of type c′ define the first eight amino acids of the peptide as residues 21–28 in the paxillin sequence. The next tryptic cleavage site occurs at Arg75, and the predicted mass of this peptide is thus 6073 Da. The measured mass is 363 mass units higher than predicted. Further analysis of c′-type ions (singly and doubly charged) indicates that the Tyr residues at positions 31 and 40 are both 80 mass units higher in mass than expected and are therefore phosphorylated. Analysis of the z′˙-type ions indicates that the Ser residue at position 74 carries an extra 203 mass units and is thus modified with an N-acetylglucosamine (O-GlcNAc) moiety. It should be noted that eight different forms of the 55 residue tryptic peptide (unmodified and all possible forms with the above three PTMs) were observed during a nanoflow LC-MS/MS analysis of tryptic peptides from paxillin. Forty-five different sites of phosphorylation were also identified on this protein.
Fig. 2.
ETD mass spectrum recorded on the [M + 8H]+8 ion (m/z 805.5) from a 55 residue, paxillin, tryptic peptide (residues 21–75). Observed fragment ions of type c′ and z′˙ are shown with right-angle arrows above and below the sequence, respectively. Singly and multiply charged ions are indicated by solid and dashed lines, respectively.
The detection and location of Ser and Thr residues modified with the O-GlcNAc moiety represent a breakthrough in technology, as this modification fails to survive sequence analysis by other MS techniques, including low-energy CAD. Discovered by Hart [11], this modification is found on proteins in both the nucleus and cytoplasm, is added and removed by transferase and O-GlcNAcase enzymes, respectively, and often occurs at sites that are known to be phosphorylated [12]. Western blots of cell lysates with O-GlcNAc-specific antibodies indicate that several hundred if not thousands of proteins carry this modification [13].
The third example is taken from the field of chromatin biology. In this area, ETD-based MS has enabled detection and site-mapping of combinatorial modifications on highly basic histone proteins [14]. Two copies of each of the four core histones, H2A, H2B, H3, and H4, are assembled with DNA to form nucleosomes, which are the building blocks of eukaryotic chromatin [15]. A host of PTMs reside on the N-terminal tails of the histones, and this array of PTMs has been well documented and includes monomethylation and dimethylation of Arg, monomethylation, dimethylation and trimethylation of Lys, acetylation and ubiquitination of Lys, and phosphorylation of Ser and Thr [16–18]. Combinations of these modifications are suggested to constitute a ‘histone code’ that regulates the binding of protein complexes involved in transcription, replication, recombination, DNA damage repair, and gene silencing [18,19]. In a recent study, Taverna et al. used ETD to map long-distance combinations of transcription-associated PTMs on the N-terminus of histone H3 purified from Tetrahymena thermophila [14]. H3 species containing monomethylation, dimethylation and trimethylation of H3K4, marks associated with transcription, were found to be acetylated in hierarchical order, with up to five acetyl groups added to K9, K14, K18, K23, and K27.
Protein identification on a chromatographic timescale using sequential ion–ion reactions and tandem MS
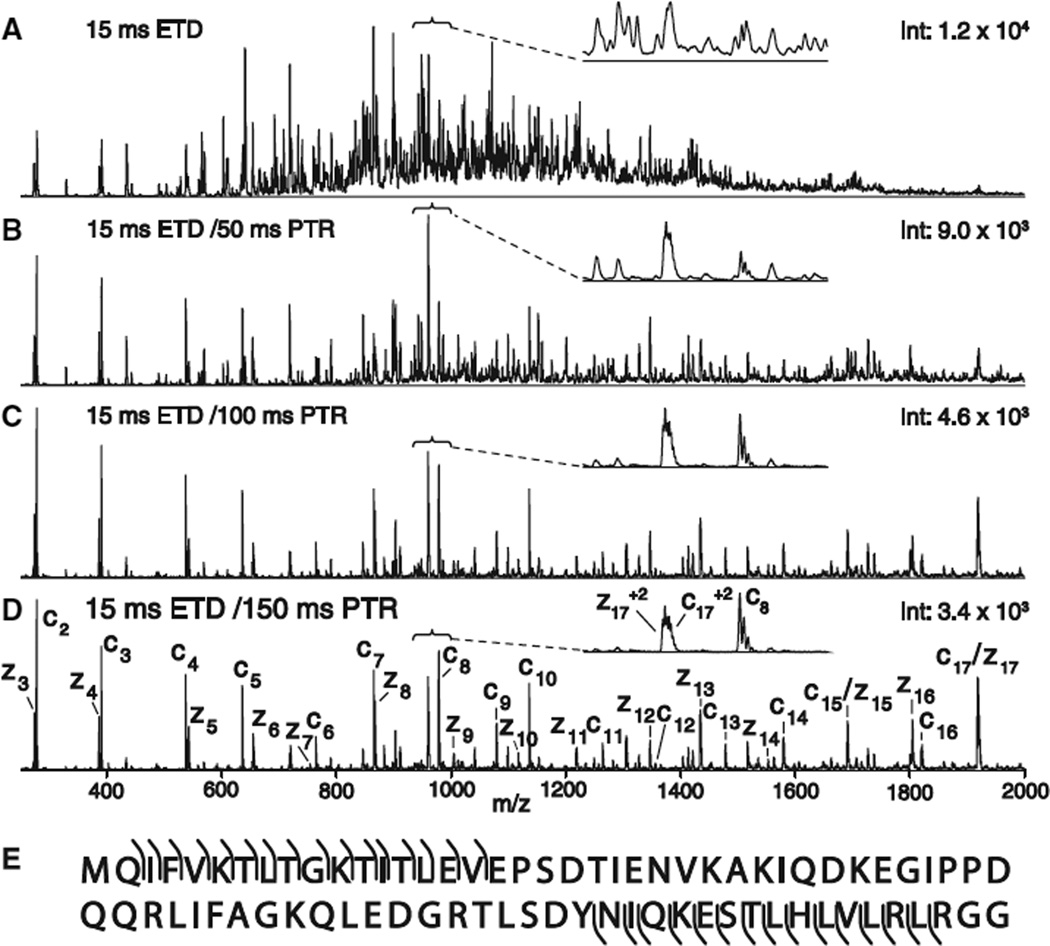
ETD has most recently been utilized for the direct analysis of intact proteins, and ubiquitin is a model protein that was initially interrogated via ETD for tandem MS. Shown in Fig. 3A is the ETD spectrum that results when the [M + 13H]+13 ion (m/z 659) of the 76 residue protein ubiquitin is allowed to react with fluoranthene radical anions for 15 ms [8]. In theory, the spectrum contains the 144 predicted fragment ions of type c′ and z′˙ in a variety of different charge states ranging from +1 to +12. The result is a mixture of ions that cannot be resolved on an LTQ mass spectrometer. Fortunately, the initial ETD spectrum can be simplified by sequestering the entire mixture of highly charged c′- and z′˙-type fragment ions, and then reacting them with a second anion that functions as a base rather than an electron donor. The carboxylate anion of benzoic acid satisfies this requirement and deprotonates the multiply charged fragments. This proton transfer charge reduction (PTR) reaction removes excess charge from the diverse population of multiply charged fragment ions. As the ion–ion reaction rates increase proportionally with the square of the charge [20] (+10 ions react 100 times faster than +1 ions), adjustment of the PTR reaction duration allows one to control the charge state of the resulting products. For the spectra in Fig. 3, multiple PTR reaction times were employed (50, 100 and 150 ms; Fig. 1B–D). As the reaction period is extended, the higher-charged fragments are preferentially concentrated to lower charge states. After a reaction time of 150 ms, singly charged products predominate. By subtracting m/z values for consecutive c′- and z′˙-type ions within a series, it is usually possible to read the amino acid sequence at the N-terminus and C-terminus, respectively, for about 17 residues before the observed m/z values exceed the mass range of the benchtop LTQ mass spectrometer (2000 Da).
Fig. 3.
Tandem mass spectrum of the protein, ubiquitin, generated by sequential ion–ion reactions. (A) ETD spectrum of the [M + 13H]+13 ion (m/z 659) after a 15 ms reaction with the radical anions of fluoranthene. Note that the spectrum contains several hundred, unresolved signals for highly charged fragment ions of type c′ and z′˙. (B–D) Spectra recorded following reactions of these ions with even electron benzoate anions for 50 ms (B), 100 ms (C), and 150 ms (D). Note that this PTR reaction converts the multiply charged ions to a mixture that is dominated by singly and doubly charged species after 150 ms. (E) Ubiquitin sequence with the observed singly charged ions of type c′ and z′˙ are indicated by angled lines above and below the sequence, respectively.
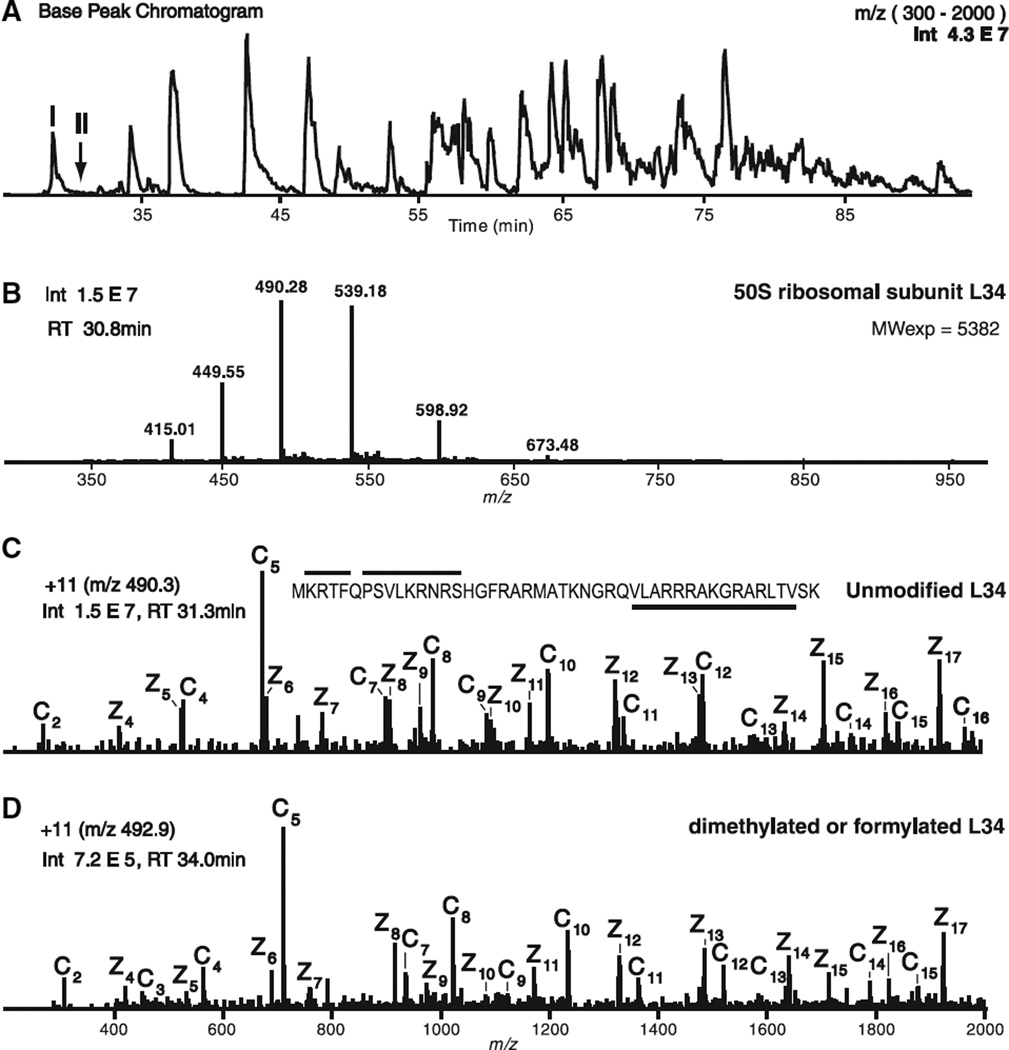
Shown in Fig. 4 are data taken from an experiment that uses the above technology to analyze proteins that constitute the Escherichia coli ribosome [21]. Two subunits make up this 2.3 ×106 Da particle [22]. The small 30S, or S, subunit contains 21 proteins involved in mRNA binding. The 50S, or L, subunit contains 34 proteins, binds to tRNA, and mediates peptidyl transfer. Shown in Fig. 4A is the base peak chromatogram from a 90 min, automated, on-line LC-MS/MS experiment performed on a benchtop LTQ instrument modified for ETD. This instrument was operated in the data-dependent mode and cycled through acquisition of a full mass spectrum and ETD/PTR MS/MS spectra on the six most abundant ions every 3 s (400–500 ms per spectrum). Throughout the automated LC-MS/MS experiment, the ion–ion reaction times for ETD and PTR were set for 35 and 135 ms, respectively. Under these conditions, the resulting spectra are dominated by singly charged ions.
Fig. 4.
Analysis of E. coli ribosomal proteins by LC-MS, tandem (ETD/PTR) MS. (A) Base peak ion chromatogram observed for gradient eluted ribosomal proteins. (B) Single-scan, full (ESI) mass spectrum acquired on the protein eluting under peak I [labeled in (A)] at a retention time of 30.8 min. (C) Single-scan, ETD/PTR tandem mass spectrum of protein [M + 11H]+11 ions at m/z 490.3 in (B). The observed ions of type c′ and z′˙ define sequences at the N-terminus and C-terminus of the protein, respectively. These sequences match to the 50S ribosomal protein, L34. Lines above and below the protein sequence indicate the amino acid sequence coverage defined by ions in the spectrum. (D) Single-scan, ETD/PTR tandem mass spectrum of [M + 11H]+11 ions (m/z 492.9) from a protein that elutes at 34 min [peak II in (A)], ~3 min after peak I. Spectra in (C) and (D) contain an identical set of type z′˙ ions. Ions of type c′ in the two spectra differ in mass by 28 Da. These data suggest that the L34 species in (D) is either monomethylated on the N-terminus and the side chain of Lys2 or is dimethylated/formylated on the N-terminus or side chain of Lys2.
Displayed in Fig. 4B is the full MS spectrum recorded on peak I in the base peak ion chromatogram. Signals in the observed charge envelope carry 8–13 positive charges and correspond to a protein having an average molecular mass of 5382 Da. The ETD/PTR MS/MS spectrum recorded on the [M + 11H]+11 ion (m/z 490.3) in this cluster is shown in Fig. 4C. Ions of type c′ and z′˙ in the spectrum are labeled as such and define the first 15 and last 17 amino acids in the 50S ribosomal protein, L34. The observed sequence coverage for this protein is shown above and below the sequence in Fig. 4C. Because the experimental and calculated average molecular masses for this protein are in agreement (5382 and 5381 Da, respectively), we conclude that protein L34 in peak I is not post-translationally modified.
Three minutes later in the chromatogram (peak II), the instrument records an ETD/PTR MS/MS spectrum (Fig. 4D) on another [M + 11H]+11 ion (m/z 492.9). Ions of type z′˙ in this spectrum occur at m/z values that are identical to those in Fig. 4C. This result suggests that the protein in peak II is a modified form of the 50S ribosomal protein, L34. All ions of type c′ (c′2–c′15) in Fig. 4D occur at m/z values that are 28 Da higher than those in Fig. 4C. We conclude that this version of the L34 protein contains either an N-terminus and Lys2 side chain that are both monomethylated, or an N-terminus or Lys2 side chain that is either formylated or dimethylated. From the observed ion currents, we estimate that modified and unmodified versions of the L34 protein are present in a ratio of 1 : 20.
Analysis of the spectra recorded on the other peaks in the base peak ion chromatogram allowed us to detect and identify 46 of the 55 proteins known to make up the E. coli ribosome. Under our experimental conditions, the other nine proteins were probably retained on the HPLC column. It is of note that the calculated and experimental average molecular masses for 42 of the proteins disagreed. The observed differences could be assigned to: deletion of the N-terminal Met, truncations at either the N-terminus or C-terminus of the protein, and the presence of PTMs such as methylation (14 Da), dimethylation or formylation (28 Da), trimethylation or acetylation (42 Da), and glutamylation (multiples of 129 Da).
Current and future work
Future efforts will involve characterizing the PTMs that regulate the function of biologically important proteins. Rev (regulator of expression of virion products) is one such protein. Rev is expressed by HIV-1 and mediates the export of unspliced viral RNA from the host cell nucleus [23,24]. This is an essential step for the late-stage translation of viral proteins that are necessary for viral replication.
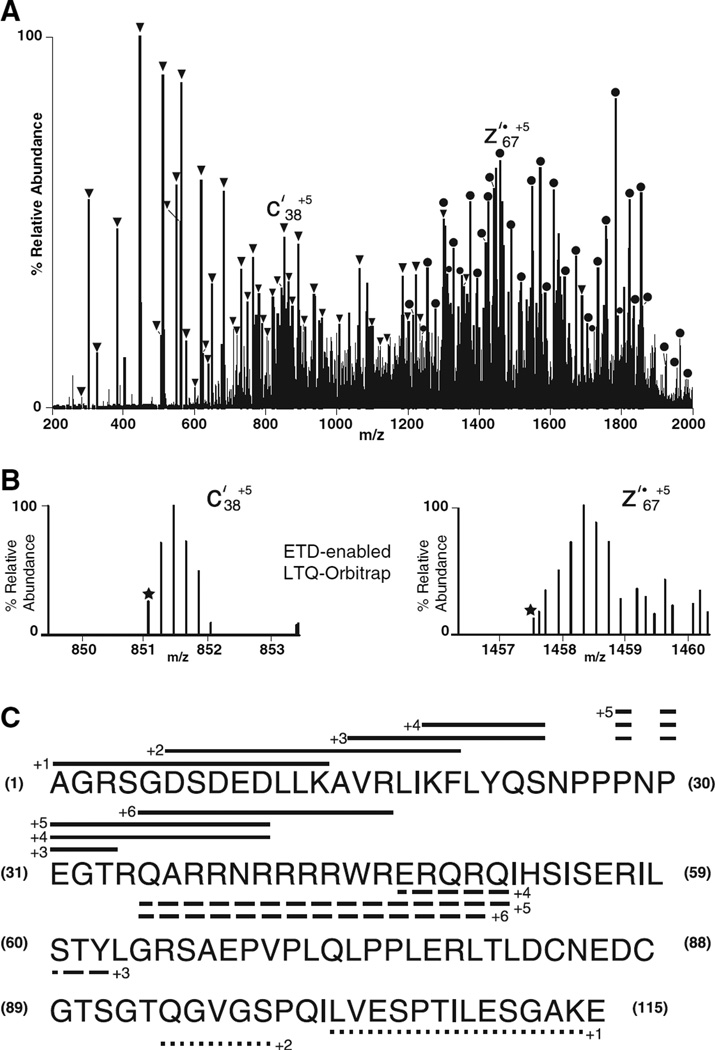
Results from a preliminary experiment on recombinant Rev are shown in Fig. 5. In this experiment, a 500 fmol sample of Rev was analyzed by on-line LC-MS/MS. The ETD-enabled LTQ instrument was operated in the data-dependent mode and cycled through acquisition of a full mass spectrum and ETD MS/MS spectra on the six most abundant ions every 2 s. The reaction time with fluoranthene radical anions was set to 30 ms, and PTR was not implemented. Three ETD scans (300 ms each) recorded on the [M + 15H]+15 ion of Rev were averaged to produce the spectrum in Fig. 5A. As the PTR reaction was not implemented, the observed spectrum contains a large number of multiply charged ions of type c′ and z′˙. These are labeled on the spectrum with solid triangles and circles, respectively. Charge states for many of these ions were assigned by recording the Rev ETD spectra on a high-resolution hybrid LTQ-Orbitrap mass spectrometer (Thermo Electron LTQ-Orbitrap, Thermo Scientific, Bremen, Germany). For this experiment, a prototype atmospheric pressure chemical ionization source [25] was installed on the front end of the LTQ instrument and employed to generate azobenzene radical anions as the electron-transfer reagent. As shown in Fig. 5A, the Orbitrap is capable of resolving signals for multiply charged ions that differ in mass by 1 Da (replacement of one 12C atom by a single 13C atom). For singly charged ions, the separation should be 1 Da. As the separation between isotopes is 1/5 Da in Fig. 5B, the charge on both ion types must be +5. Accordingly, the two ions are assigned as c′38+5 and z′˙67+5, respectively.
Fig. 5.
Analysis of recombinant Rev protein by LC-MS, tandem (ETD) MS. (A) Average of three 300 ms ETD scans recorded on [M + 15H]+15 ions from Rev. For each scan, the reaction time with fluoranthene radical anions was 30 ms. Fragment ions of type c′ and z′˙ are denoted by solid triangles and circles, respectively. (B) Signals observed in two different mass windows from ETD spectra recorded on Rev [M + 15H]+15 ions with the high-resolution LTQ-Orbitrap instrument. In the spectra recorded with the Orbitrap instrument, ion signals are resolved into 12C and 13C isotope peaks separated by 1/5 Da. This indicates that the charge on the ions in both mass windows must be +5 and allows them to be assigned to c′38+5 and z′˙67+5 ions, respectively. (C) Sequence coverage for Rev assigned from ions of type c′ and z′˙ are shown by solid and dashed lines, respectively. Charge states for the observed ions are specified at the beginning of each line. Coverage from ions of type y″ obtained from a CAD spectrum recorded on the same [M + 15H]+15 ions is shown by dotted lines below the Rev sequence.
Sequence coverage of Rev provided by ions of type c′ and z′˙ is indicated by solid and dashed lines, respectively, above and below the sequence in Fig. 5C. Sequence information on the N-terminal 45 amino acids is provided by ions of type c′ having charge states up to +6. Ions of type z′˙ with charge states of +4 to +6 overlap with this region and extend the sequence to residue 50. Unfortunately, Rev lacks multiple basic residues near the C-terminus of the protein. As a result, the ETD spectrum is devoid of low mass fragments of type z′˙ that would provide sequence information at the C-terminus of the protein. Although this region is accessed by recording a CAD spectrum on the same precursor (see coverage indicated by dotted lines below the sequence), we are presently exploring chemistry that will allow us to introduce charge in regions of the protein that are devoid of basic residues. Recording ETD spectra on samples that are both protonated and metalated is one possible strategy to accomplish this goal [26]. Still another objective is to extend the sequence coverage of intact proteins by implementing ETD on high-resolution instrumentation (Orbitrap and FT mass spectrometers) capable of resolving highly charged fragment ions and measuring their masses accurately to three or four decimal places. Preliminary data from several different approaches to this problem have already been reported [27–29].
ETD is a major breakthrough in the field of proteomics and enables rapid sequencing of large peptides and intact proteins on a QLT mass spectrometer. The scan times required for direct analysis of proteins using ETD are comparable to the times required for analysis of peptides using CAD. In addition, coupling nanoflow LC with our ETD technology increases the sensitivity of intact protein analyses, and with ongoing advancements in protein chromatographic fractionation, ETD can be extended to larger proteins and protein complexes. ETD is a powerful dissociation technique that enables the correlation of protein molecular mass with extensive sequence information, allowing identification of PTMs, truncations and splice variants of a protein every ~500 ms. For recent publications on the topic of ETD, see [30–34].
Acknowledgements
This work was supported by grants from the National Institutes of Health (GM37537 and U54 G64346 to D. F. Hunt) and the National Science Foundation (MCB-0209793 to D. F. Hunt).
Abbreviations
- CAD
collision-activated dissociation
- ECD
electron-capture dissociation
- ESI
electrospray ionization
- ETD
electron-transfer dissociation
- FT
Fourier transform
- O-GlcNAc
N-acetylglucosamine
- PTM
post-translational modification
- PTR
proton transfer charge reduction
- QLT
quadrupole linear ion trap
- Rev
regulator of expression of virion products
References
- 1.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 3.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 6.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 7.McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley T. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. FEBS J. 2007;274 doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 8.Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol. 2000;2:E231–E236. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder MJ, Webb DJ, Shabanowitz J, Horwitz AF, Hunt DF. Methods for the detection of paxillin post-translational modifications and interacting proteins by mass spectrometry. J Proteome Res. 2005;4:1832–1841. doi: 10.1021/pr0502020. [DOI] [PubMed] [Google Scholar]
- 11.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 12.Hart GW, Housley MP, Slawson C. Cycling of O-linked [beta]-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 13.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 14.Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci USA. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 16.Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, Hunt DF. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 18.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 20.Stephenson JL, McLuckey SA. Ion/ion reactions in the gas phase. Proton transfer reactions involving multiply-charged proteins. J Am Chem Soc. 1996;118:7390–7397. [Google Scholar]
- 21.Chi A, Bai DL, Geer LY, Shabanowitz J, Hunt DF. Analysis of intact proteins on a chromatographic time scale by electron transfer dissociation tandem mass spectrometry. Int J Mass Spectrom. 2007;259:197–203. doi: 10.1016/j.ijms.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramakrishnan V, White SW. Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem Sci. 1998;23:208–212. doi: 10.1016/s0968-0004(98)01214-6. [DOI] [PubMed] [Google Scholar]
- 23.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 24.Hammarskjold ML. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 25.Liang X, Xia Y, McLuckey SA. Alternately pulsed nanoelectrospray ionization/atmospheric pressure chemical ionization for ion/ion reactions in an electrodynamic ion trap. Anal Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery HA, Blakeslee J, DeLong A, Shabanowitz J, Hunt DF. Arabidopsis thaliana phosphopeptide identification by electron transfer dissociation mass spectrometry. Proteomics: Phosphorylation; Proceedings of the 55th ASMS Conference on Mass Spectrometry and Allied Topics; Indianapolis, IN. 2007. [Google Scholar]
- 27.McAlister G, Makarov A, Horning S, Schwartz JC, Phanstiel D, Good D, Berggren WT, Coon JJ. Implementation of electron transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Developments in Ion Trap MS; Proceedings of the 55th ASMS Conference on Mass Spectrometry and Allied Topics; Indianapolis, IN. 2007. [Google Scholar]
- 28.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Udeshi ND, Misawa Y, Lindsey Rose KM, Chi A, Rekosh D, Shabanowitz J, Hammarskjold M, Hunt DF. A sensitive top down and middle down strategy for analyzing proteins using electron transfer dissociation. Top Down 2007: Tandem MS Above 5 kDa; Proceedings of the 55th ASMS Conference on Mass Spectrometry and Allied Topics; Indianapolis, IN. 2007. [Google Scholar]
- 30.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M700073-MCP200. in press. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Frolov A, Tang N, Hoffmann R, van de Goor T, Metz TO, Smith RD. Application of electron transfer dissociation mass spectrometry in analyses of non-enzymatically glycated peptides. Rapid Commun Mass Spectrom. 2007;21:661–666. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catalina MI, Koeleman CA, Deelder AM, Wuhrer M. Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun Mass Spectrom. 2007;21:1053–1061. doi: 10.1002/rcm.2929. [DOI] [PubMed] [Google Scholar]