Abstract
Purpose.
To compare fluorescence in situ hybridization (FISH) using a centromeric probe for chromosome 3 (CEP3) and 3p26 locus-specific probe with single-nucleotide polymorphism array (SNP-A) analysis in the detection of high-risk uveal melanoma.
Methods.
Fifty cases of uveal melanoma (28 males, 22 females) treated by enucleation between 2004 and 2010 were analyzed. Fresh tissue was used for FISH and SNP-A analysis. FISH was performed using a CEP3 and a 3p26 locus-specific probe. Tumor size, location, and clinical outcome were recorded during the 7-year study period (median follow-up: 35.5 months; mean: 38.5 months). The sensitivity, specificity, positive predictive value, and negative predictive value were calculated.
Results.
Monosomy 3 was detected by FISH-CEP3 in 27 tumors (54%), FISH-3p26 deletion was found in 30 (60%), and SNP-A analysis identified 31 (62%) of the tumors with monosomy 3. Due to technical failures, FISH and SNP-A were noninterpretable in one case (2%) and two cases (4%), respectively. In both cases of SNP-A failure, tumors were positive for FISH 3p26 deletion and in a single case of FISH failure, monosomy 3 was found using SNP-A. No statistically significant differences were observed in any of the sensitivity or specificity measures.
Conclusions.
For prediction of survival at 36 months, FISH CEP3, FISH 3p26, and SNP-A were comparable. A combination of prognostication techniques should be used in an unlikely event of technical failure (2%–4%).
For prediction of survival at 36 months in patients with uveal melanoma, FISH CEP3, FISH 3p26, and SNP-A are comparable. Use of a combination of prognostication techniques is preferable.
Introduction
In the early 1990s, nonrandom genetic abnormalities involving chromosomes 1, 3, 6, and 8 were identified in uveal melanoma tumor samples.1,2 These aberrations were later shown to correlate with poor prognosis.3–5 Of various cytogenetic abnormalities observed, monosomy 3 is the strongest predictor of metastatic risk.5–14 Several techniques are currently being used to detect monosomy 3 and other chromosomal changes associated with the development of metastatic disease. Gene-expression profiling is also being used in prognostication.15,16 Fluorescence in situ hybridization (FISH) is a rapid and economical assay commonly used in the molecular prognostication of cancer that uses fluorochromes linked to DNA probes, enabling determination of chromosome copy number and location of specific DNA sequences. It is primarily a visual technique that requires the use of a fluorescence microscope and readily allows for coincident cytologic confirmation of malignancy.
In general, three basic types of DNA probes are used: centromeric (chromosome enumeration probes [CEPs]), whole chromosome probes (whole chromosome paints), and locus-specific probes.17 Single-nucleotide polymorphism array (SNP-A) analysis is an automated DNA microarray. Whereas SNP-A analysis requires specialized instrumentation, it does offer several advantages to FISH. SNP-A analysis detects loss of heterozygosity of large numbers of moderately polymorphic DNA segments, providing more comprehensive characterization of genomic data. SNP-A analysis is also useful in identifying uniparental disomy and deletions that may be functionally equivalent to monosomy 3.18
Rapid development and adoption of prognostication assays has led to wide variation in practice patterns. To date, there have been few direct comparisons between available techniques. In the majority of uveal melanoma cytogenetic studies using FISH, CEP3 probes have been used.1,7–11,18–41 The prognostic accuracy of SNP-A has been reported to be superior to that of FISH with a CEP3 probe.18,38 Deletion-mapping studies have identified 3p24–26 as a commonly affected region in patients with metastatic uveal melanoma. These loci can be detected by FISH using 3p24 and 3p26 probes.41,42 The manner in which locus-specific FISH analysis of chromosome 3 in patients with uveal melanoma compares with CEP3 and with SNP-A status has not been reported.
The overall purpose of prognostication is to enter high-risk patients into an adjuvant treatment trial aimed at reducing tumor-specific mortality.43 The purpose of this study was to compare techniques of FISH using CEP3, FISH using 3p26 locus-specific probe, and SNP-A in assessing chromosome 3 status within the tumor. Additionally, we wanted to compare predictive values for survival using these techniques.
Methods
Patients
Fifty consecutive patients with uveal melanoma treated by primary enucleation at the Cleveland Clinic Cole Eye Institute were enrolled between 2004 and 2010. The study was approved by the Institutional Review Board, and this research adhered to the tenets of the Declaration of Helsinki. Patients were followed over the study period ending in October 2011 (median follow-up: 35.5 months; mean: 38.5 months). At the time of diagnosis, each patient underwent comprehensive ophthalmic examination with supporting diagnostic studies including fundus photography, ultrasonography, and in some cases optical coherence tomography or indocyanine green angiography. Computed tomography (CT) scans of the chest, abdomen, and pelvis were initially performed to rule out metastatic disease. Following enucleation, all patients underwent scheduled surveillance for the development of metastases every 6 months, with clinical evaluation, hepatic ultrasound, and liver function testing. The cause of death was established (metastatic or nonmetastatic) by evaluation of medical records, imaging studies, and biopsy results. Where necessary, the patient's family or primary care practitioners were contacted as part of ongoing data collection efforts.
Tumor Sampling
Immediately following enucleation, transillumination was used to mark the tumor margins. Dissection was carried out through a scleral flap overlying the tumor base. In all cases, impression smears were made from fresh (or previously frozen) tumor tissue for FISH analysis. Fresh tumor tissue was also further processed for SNP-A analysis.
FISH
Chromosome 3 status was assessed by FISH using both directly labeled enumeration probes (X SpectrumGreen/Y SpectrumOrange Direct Labeled Fluorescent DNA Probe Kit; Abbott Molecular Diagnostics, Des Plaines, IL) for the alphacentromeric locus of chromosome 3 (CEP3) and a locus-specific probe. The locus-specific probe, 3p26 (TelVysion 3p; Abbott Molecular Diagnostics) used for this study is commercially available (Vysis FISH Chromosome Search Tool, Abbott Molecular Diagnostics), having specificity for locus D3S4559 spanning a large number of genes.44
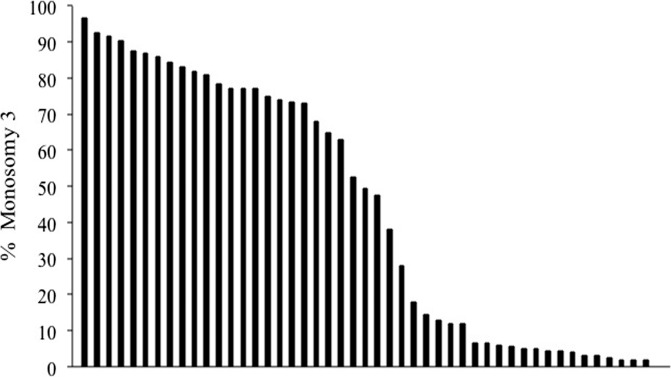
We excluded triploidy/tetraploidy/polyploidy through the use of CEP8 performed using a previously described interphase FISH method for touch preparations.45 Probes were hybridized to fixed fresh cells (Carnoy's solution for cytogenetic investigation) affixed to touch preparations from frozen tissue. A total of 200 interphase cells were scored using a FISH workstation (Carl Zeiss Workstation) to determine the percentage of signals for each locus. Based on frequency distribution of the percentage of cells demonstrating monosomy in each tumor by FISH-CEP3 (Fig. 1) and the fact that many prior studies in the literature have used such a cutoff value,18,30,34,35,37,40 we chose to use a cutoff value of 20% to define monosomy for chromosome 3. Nevertheless, we explored alternative cutoff points using the sum of sensitivity and specificity values. Bootstrap methods were used to evaluate the robustness of the best cutoff point. Statistical analyses were performed using R software (version 2.8; R Foundation for Statistical Computing, Vienna, Austria; available at http://www.r-project.org/). A 0.05 significance level was assumed for all tests.
Figure 1.
Frequency distribution of the percentage of cells demonstrating monosomy 3 in each tumor by FISH-CEP3. Each vertical bar represents a single case. Data are shown from 49 cases because FISH was not informative in a single case.
SNP-A
SNP-A analysis was performed on fresh-frozen tumor tissue that was permeabilized in RNA protective reagent (RNAlater-ICE; Ambion, Austin, TX) in accordance with the manufacturer's protocol. Permeabilized tissue was minced, equilibrated in cold phosphate-buffered saline (PBS), then digested with proteinase-K prior to DNA extraction (DNeasy Kit; Qiagen, Germantown, MD). Using approximately 200 ng of DNA per sample, a labeling and detecting whole-genome genotyping sample kit (Illumina Human 660W-Quad v1; Illumina, San Diego, CA) was used to analyze >660,000 SNP and copy number variation loci to provide high-resolution analysis of chromosome structure. Probe preparation and hybridization were performed by the Cleveland Clinic Genomics Core. Chromosomal aberrations were identified using commercial data analysis software (GenomeStudio with KaryoStudio module; Illumina). Contiguous regions (gain or loss of heterozygosity) of a minimum group size (default 20 adjacent SNPs of the same polarity) and a minimum sequence length of 200 kb were designated as abnormality and quantified by 20-SNP blocks (this is a size of abnormality that can be validated using FISH analysis). A tumor was considered to manifest monosomy 3 if there was contiguous deletion of an entire copy of chromosome 3.
Statistical Analysis
Kaplan–Meier curves were used to assess the degree of correlation between assay results (monosomy 3 determined separately by FISH-CEP3 and SNP-A, as well as 3p26 deletion detected by FISH) and patient survival. Within each method, the statistical significance of differences in disease-free survival was determined using the log-rank test. In these analyses, patients who died without metastases were censored at the time of death, whereas those alive and dead with metastases were considered to have an event at the time of the diagnosis of metastases. Patients who remained alive without metastases were censored at the date of last follow-up.
For both FISH-CEP3 and FISH-3p26, the sensitivity, specificity, positive predictive value, and negative predictive value were calculated using SNP as the gold standard. Then FISH CEP3, FISH 3p26, and SNP were evaluated as predictors of survival at 36 months. McNemar's test was performed to compare sensitivities and specificities.
Results
Patient Characteristics
Tumors from the primary enucleation specimens of 50 white patients, of northern and western European origin, with uveal melanoma were analyzed. This series included 28 males (56%) and 22 females (44%). The median age at diagnosis was 65 years (range: 24–91 years). Using the Collaborative Ocular Melanoma Study (COMS) size criteria, tumors were classified as large in 44 cases (88%), medium in 6 cases (12%), and small in 0 cases.46 The median largest basal diameter was 17.7 mm (range: 11.0–24.0 mm). The median tumor height was 10.1 mm (range: 2.7–14.8 mm). Tumor location was choroidal in 24 (48%), ciliochoroidal in 18 (36%), and iridociliochoroidal in 8 (16%) patients. Patient and tumor characteristics are summarized in Table 1.
Table 1.
Patient and Tumor Characteristics
|
Patient ID |
Sex |
Age (y) |
Tumor |
COMS |
AJCC |
Location |
|
|
LBD (mm) |
HGT (mm) |
||||||
| 1 | F | 86 | 18.0 | 11.0 | Large | 3b | Ciliochoroidal |
| 2 | M | 44 | 20.0 | 10.1 | Large | 4a | Choroidal |
| 3 | F | 74 | 10.0 | 2.7 | Medium | 1a | Choroidal |
| 4 | F | 75 | 18.0 | 9.8 | Large | 3b | Ciliochoroidal |
| 5 | F | 83 | 16.5 | 12.5 | Large | 3b | Iridociliochoroidal |
| 6 | M | 49 | 22.0 | 11.0 | Large | 4b | Ciliochoroidal |
| 7 | M | 79 | 15.0 | 7.2 | Medium | 2b | Ciliochoroidal |
| 8 | F | 79 | 17.7 | 8.1 | Large | 2a | Choroidal |
| 9 | F | 76 | 18.0 | 5.6 | Large | 2b | Iridociliochoroidal |
| 10 | F | 69 | 15.6 | 12.2 | Large | 3b | Ciliochoroidal |
| 11 | M | 58 | 14.0 | 12.0 | Large | 3a | Choroidal |
| 12 | F | 70 | 17.7 | 6.7 | Large | 3a | Choroidal |
| 13 | F | 79 | 18.5 | 14.6 | Large | 4b | Iridociliochoroidal |
| 14 | M | 90 | 21.5 | 7.6 | Large | 4b | Ciliochoroidal |
| 15 | M | 90 | 12.3 | 11.8 | Large | 3b | Ciliochoroidal |
| 16 | M | 59 | 20.0 | 10.0 | Large | 4b | Ciliochoroidal |
| 17 | M | 75 | 17.1 | 8.4 | Large | 2b | Iridociliochoroidal |
| 18 | M | 51 | 18.0 | 7.3 | Large | 2a | Choroidal |
| 19 | M | 60 | 19.0 | 11.2 | Large | 4b | Ciliochoroidal |
| 20 | M | 62 | 19.0 | 8.4 | Large | 4b | Ciliochoroidal |
| 21 | M | 53 | 18.5 | 10.1 | Large | 4a | Choroidal |
| 22 | F | 80 | 15.0 | 9.3 | Medium | 3a | Choroidal |
| 23 | F | 43 | 18.5 | 10.5 | Large | 4b | Iridociliochoroidal |
| 24 | M | 66 | 15.0 | 13.1 | Large | 3b | Iridociliochoroidal |
| 25 | F | 24 | 11.7 | 11.1 | Large | 3a | Choroidal |
| 26 | F | 78 | 19.4 | 3.9 | Large | 4b | Ciliochoroidal |
| 27 | F | 91 | 16.3 | 11.0 | Large | 3a | Choroidal |
| 28 | F | 64 | 16.6 | 11.1 | Large | 3a | Choroidal |
| 29 | M | 51 | 11.0 | 4.0 | Medium | 1a | Choroidal |
| 30 | F | 67 | 15.5 | 12.2 | Large | 3b | Ciliochoroidal |
| 31 | F | 50 | 19.6 | 7.0 | Large | 4a | Choroidal |
| 32 | M | 26 | 17.7 | 13.0 | Large | 3b | Iridociliochoroidal |
| 33 | M | 75 | 18.0 | 14.8 | Large | 3b | Ciliochoroidal |
| 34 | M | 52 | 19.0 | 6.5 | Large | 4b | Ciliochoroidal |
| 35 | F | 68 | 15.0 | 12.2 | Large | 3a | Choroidal |
| 36 | F | 76 | 17.6 | 10.4 | Large | 3a | Choroidal |
| 37 | F | 67 | 15.0 | 7.6 | Medium | 2a | Choroidal |
| 38 | M | 57 | 15.0 | 12.3 | Large | 3a | Choroidal |
| 39 | M | 54 | 14.3 | 12.0 | Large | 3a | Choroidal |
| 40 | M | 56 | 18.0 | 7.8 | Large | 2a | Choroidal |
| 41 | F | 50 | 16.0 | 8.0 | Medium | 2a | Choroidal |
| 42 | M | 58 | 20.0 | 11.9 | Large | 4b | Ciliochoroidal |
| 43 | F | 82 | 17.5 | 12.2 | Large | 3a | Choroidal |
| 44 | M | 73 | 17.3 | 9.0 | Large | 2a | Choroidal |
| 45 | M | 61 | 12.7 | 11.6 | Large | 3b | Ciliochoroidal |
| 46 | M | 50 | 15.4 | 10.6 | Large | 3b | Ciliochoroidal |
| 47 | M | 57 | 24.0 | 4.2 | Large | 4b | Iridociliochoroidal |
| 48 | M | 53 | 19.0 | 8.0 | Large | 4a | Choroidal |
| 49 | M | 43 | 20.0 | 8.2 | Large | 4a | Choroidal |
| 50 | M | 77 | 22.5 | 9.7 | Large | 4b | Ciliochoroidal |
ID, study identification number; F, female; M, male; LBD, largest basal diameter; HGT, height; COMS, Collaborative Ocular Melanoma Study; AJCC, American Joint Commission Classification, 7th edition.
Cutoff Point
The highest combined sensitivity and specificity value was observed with cutoff point at 8%, although several other cutoff points (as high as 49%) provided combined values that were within 0.1 of those observed with a cutoff point of 8% (Table 2). When compared against the traditionally used cutoff point (20%), neither the sensitivity (P = 0.13) nor specificity (P = 0.99) significantly differed between the two cutoff points.
Table 2.
The Sensitivity, Specificity, and Sum of the Two Measures for Various Levels of FISH-CEP3-Positive Cells
|
Positive Cells |
Sensitivity |
Specificity |
Sum |
| 2 | 94.6% | 0.0% | 0.946 |
| 2.5 | 89.2% | 8.3% | 0.975 |
| 3 | 89.2% | 16.7% | 1.059 |
| 4 | 89.2% | 33.3% | 1.225 |
| 4.5 | 86.5% | 33.3% | 1.198 |
| 5 | 81.1% | 41.7% | 1.227 |
| 5.5 | 75.7% | 50.0% | 1.257 |
| 6 | 75.7% | 58.3% | 1.340 |
| 6.5 | 75.7% | 66.7% | 1.423 |
| 8 | 75.7% | 83.3% | 1.590 |
| 9 | 73.0% | 83.3% | 1.563 |
| 12 | 70.3% | 83.3% | 1.536 |
| 13 | 67.6% | 83.3% | 1.509 |
| 14.5 | 64.9% | 83.3% | 1.482 |
| 43.5 | 64.9% | 91.7% | 1.565 |
| 47.5 | 62.2% | 91.7% | 1.538 |
| 49.5 | 59.5% | 91.7% | 1.511 |
| 52.5 | 56.8% | 91.7% | 1.484 |
| 63 | 54.1% | 91.7% | 1.457 |
| 68 | 51.4% | 91.7% | 1.430 |
| 73 | 48.6% | 91.7% | 1.403 |
| 73.5 | 45.9% | 91.7% | 1.376 |
| 74 | 43.2% | 91.7% | 1.349 |
| 75 | 40.5% | 91.7% | 1.322 |
| 77 | 37.8% | 91.7% | 1.295 |
| 78.5 | 29.7% | 91.7% | 1.214 |
| 81 | 27.0% | 91.7% | 1.187 |
| 82 | 24.3% | 91.7% | 1.160 |
| 83 | 21.6% | 91.7% | 1.133 |
| 84.5 | 18.9% | 91.7% | 1.106 |
| 86 | 16.2% | 91.7% | 1.079 |
| 87 | 16.2% | 100.0% | 1.162 |
| 87.5 | 13.5% | 100.0% | 1.135 |
| 90.5 | 10.8% | 100.0% | 1.108 |
| 91.5 | 8.1% | 100.0% | 1.081 |
| 92.5 | 5.4% | 100.0% | 1.054 |
| 96.5 | 2.7% | 100.0% | 1.027 |
Data of 37 cases that were positive by SNP-A are included in the analysis.
Bootstrap methods were also used to evaluate the robustness of the best cutoff point. Two sensitivity analyses were performed. First, the choice of best cutoff point was evaluated. Across 2000 samples, 8% was the best cutoff point in 45% of the bootstrap samples, whereas 9%, 43.5%, and 47% were the best in at least 10% of the bootstrap samples. Again, although 8% provides the best combination of sensitivity and specificity in a given sample, it did not perform significantly better than the currently used cutoff point (20%).
Chromosome 3 Status
FISH-CEP3 identified 27 tumors (54%) with monosomy 3. FISH-3p26 revealed a deletion in 30 cases (60%), including every case detected by FISH-CEP3. SNP-A analysis detected 31 cases (62%) with monosomy 3. Triploidy/tetraploidy/polyploidy was not observed. In two cases (tumors 24 and 25), SNP-A analysis was not interpretable due to a chip failure in one case and sampling of nontumor DNA in the second. In a single case (tumor 29), FISH was not interpretable secondary to sample debris. In both cases of SNP-A failure, tumors were positive for FISH-3p26 deletion and in a single case FISH failure, monosomy 3 was detected using SNP-A. Chromosome 3 status determined by each technique (FISH-CEP3, FISH-3p26, and SNP-A) is listed in Table 3.
Table 3.
Chromosome 3 Copy Number by FISH (using CEP3 and 3p26 probes) and by SNP-A (overall chromosome 3 status, centromeric, and 3p26 regions)
|
ID |
FISH |
SNP-A |
Clinical Data |
|||||||
|
Copy # |
CEP3 |
%M |
3p26 |
Copy # |
Centromere |
3p26 |
F/U |
Status |
Mets |
|
| 1 | Monosomy 3 | Del | 73.0 | Del | Monosomy 3 | Del | Del | 43 | Dead | No |
| 2 | Monosomy 3 | Del | 75.0 | Del | Monosomy 3 | Del | Del | 59 | Alive | No |
| 3 | Monosomy 3 | Del | 49.5 | Del | Monosomy 3 | Del | Del | 49 | Alive | No |
| 4 | Monosomy 3 | Del | 73.5 | Del | Monosomy 3 | Del | Del | 67 | Alive | No |
| 5 | Monosomy 3 | Del | 84.5 | Del | Monosomy 3 | Del | Del | 83 | Dead | No |
| 6 | Monosomy 3 | Del | 47.5 | Del | Monosomy 3 | Del | Del | 37 | Dead | No |
| 7 | Monosomy 3 | Del | 92.5 | Del | Monosomy 3 | Del | Del | 36 | Alive | No |
| 8 | Monosomy 3 | Del | 87.0 | Del | Monosomy 3 | Del | Del | 28 | Dead | Yes |
| 9 | Monosomy 3 | Del | 77.0 | Del | Monosomy 3 | Del | Del | 35 | Alive | No |
| 10 | Monosomy 3 | Del | 63.0 | Del | Monosomy 3 | Del | Del | 21 | Dead | Yes |
| 11 | Monosomy 3 | Del | 65.0 | Del | Monosomy 3 | Del | Del | 28 | Alive | No |
| 12 | Monosomy 3 | Del | 81.0 | Del | Monosomy 3 | Del | Del | 26 | Dead | Yes |
| 13 | Monosomy 3 | Del | 91.5 | Del | Monosomy 3 | Del | Del | 16 | Dead | No |
| 14 | Monosomy 3 | Del | 82.0 | Del | Monosomy 3 | Del | Del | 37 | Dead | No |
| 15 | Monosomy 3 | Del | 74.0 | Del | Monosomy 3 | Del | Del | 18 | Dead | Yes |
| 16 | Monosomy 3 | Del | 68.0 | Del | Monosomy 3 | Del | Del | 24 | Dead | Yes |
| 17 | Monosomy 3 | Del | 83.0 | Del | Monosomy 3 | Del | Del | 2 | Dead | Yes |
| 18 | Monosomy 3 | Del | 52.5 | Del | Monosomy 3 | Del | Del | 33 | Dead | Yes |
| 19 | Monosomy 3 | Del | 96.5 | Del | Monosomy 3 | Del | Del | 12 | Dead | Yes |
| 20 | Monosomy 3 | Del | 78.5 | Del | Monosomy 3 | Del | Del | 31 | Dead | Yes |
| 21 | Monosomy 3 | Del | 90.5 | Del | Monosomy 3 | Del | Del | 33 | Dead | Yes |
| 22 | Monosomy 3 | Del | 77.0 | Del | Monosomy 3 | Del | Del | 39 | Dead | Yes |
| 23 | Monosomy 3 | Del | 87.5 | Del | Monosomy 3 | Del | Del | 26 | Dead | Yes |
| 24 | Monosomy 3 | Del | 77.0 | Del | Chip Failure | NA | NA | 27 | Dead | Yes |
| 25 | Monosomy 3 | Del | 86.0 | Del | No Tumor DNA | NA | NA | 17 | Dead | No |
| 26 | Monosomy 3 | Del | 28.0 | Del | Monosomy 3 | Del | Del | 15 | Dead | Yes |
| 27 | Disomy 3 | Normal | 0.0 | Normal | Monosomy 3 | Del | Del | 28 | Dead | No |
| 28 | Disomy 3 | Normal | 13.0 | Del | Monosomy 3 | Del | Del | 38 | Alive | No |
| 29 | Sample Debris | NA | NA | NA | Monosomy 3 | Del | Del | 82 | Alive | No |
| 30 | Disomy 3 | Normal | 2.0 | Del | Monosomy 3 | Del | Del | 26 | Alive | No |
| 31 | Disomy 3 | Normal | 18.0 | Del | Monosomy 3 | Del | Del | 11 | Dead | Yes |
| 32 | Disomy 3 | Normal | 12.0 | Normal | Monosomy 3 | Del | Del | 14 | Dead | Yes |
| 33 | Monosomy 3 | Del | 38.0 | Del | Monosomy 3 | Del | Del | 18 | Dead | Yes |
| 34 | Disomy 3 | Normal | 2.5 | Normal | Disomy 3 | Normal | Normal | 69 | Alive | No |
| 35 | Disomy 3 | Normal | 4.5 | Normal | Disomy 3 | Normal | Normal | 50 | Alive | No |
| 36 | Disomy 3 | Normal | 3.0 | Normal | Disomy 3 | Normal | Normal | 46 | Alive | No |
| 37 | Disomy 3 | Normal | 14.5 | Normal | Disomy 3 | Normal | Normal | 44 | Alive | No |
| 38 | Disomy 3 | Normal | 6.5 | Normal | Disomy 3 | Normal | Normal | 90 | Alive | No |
| 39 | Disomy 3 | Normal | 5.0 | Normal | Disomy 3 | Normal | Normal | 95 | Alive | No |
| 40 | Disomy 3 | Normal | 6.0 | Normal | Disomy 3 | Normal | Normal | 41 | Alive | No |
| 41 | Disomy 3 | Normal | 6.5 | Normal | Disomy 3 | Normal | Normal | 33 | Alive | No |
| 42 | Disomy 3 | Normal | 3.0 | Normal | Disomy 3 | Normal | Normal | 67 | Alive | No |
| 43 | Disomy 3 | Normal | 4.0 | Normal | Disomy 3 | Normal | Normal | 70 | Dead | No |
| 44 | Disomy 3 | Normal | 5.5 | Normal | Disomy 3 | Normal | Normal | 48 | Dead | No |
| 45 | Disomy 3 | Normal | 2.0 | Normal | Disomy 3 | Normal | Normal | 40 | Dead | Yes |
| 46 | Disomy 3 | Normal | 4.5 | Normal | 100% Del 3q | Normal | Normal | 43 | Alive | No |
| 47 | Disomy 3 | Normal | 5.0 | Del | 100% Del 3q | Del | Del | 51 | Alive | No |
| 66.5% Gain 3p | ||||||||||
| 33.5% Del 3p | ||||||||||
| 48 | Disomy 3 | Normal | 2.0 | Del | 10% Del 3p | Normal | Del | 36 | Alive | No |
| 49 | Disomy 3 | Normal | 0.0 | Normal | 50% Gain 3q | Normal | Normal | 18 | Dead | Yes |
| 50 | Disomy 3 | Normal | 12.0 | Del | 75% Gain 3p | Normal | Gain | 23 | Dead | No |
ID, identification; %M, percentage of monosomy 3 cells by FISH-CEP3; F/U, follow-up (months); Mets, metastases; Del, deletion; NA, not applicable.
The sensitivity, specificity, positive predictive value, and negative predictive value for FISH-CEP3 and FISH-3p26, calculated using SNP-A as the gold standard (Table 4). For prediction of survival at 36 months, the sensitivity and specificity for FISH-CEP3, FISH-3p26, and SNP-A were also calculated (Table 5).
Table 4.
Predictors of Chromosome 3 Status
|
Variable |
Levels |
SNP Disomy 3 |
SNP Monosomy 3 |
Sensitivity |
Specificity |
PPV |
NPV |
| FISH- CEP3 | Disomy 3 | 15 | 7 | 0.78 (0.61, 0.89) | 1.00 (0.80, 1.00) | 1.00 (0.87, 1.00) | 0.68 (0.47, 0.84) |
| Monosomy 3 | 0 | 25 | |||||
| FISH- 3p26 | Normal | 15 | 2 | 0.94 (0.80, 0.98) | 1.00 (0.80, 1.00) | 1.00 (0.89, 1.00) | 0.88 (0.66, 0.97) |
| Deleted | 0 | 30 |
For both FISH CEP3 and FISH 3p26, the sensitivity, specificity, positive predictive value, and negative predictive value were calculated using SNP-A as the gold standard. PPV, positive predictive value; NNP, negative predictive value. Values within parentheses indicate 95% confidence limits.
Table 5.
Predictors of Survival at 36 Months
|
Variable |
Levels |
Alive |
Dead |
Sensitivity |
Specificity |
PPV |
NPV |
| FISH- CEP3 | Disomy 3 | 16 | 4 | 0.78 (0.55, 0.91) | 0.64 (0.45, 0.80) | 0.61 (0.41, 0.78) | 0.80 (0.58, 0.92) |
| Monosomy 3 | 9 | 14 | |||||
| FISH- 3p26 | Normal | 13 | 3 | 0.83 (0.61, 0.94) | 0.52 (0.33, 0.70) | 0.56 (0.37, 0.72) | 0.81 (0.57, 0.93) |
| Deleted | 12 | 15 | |||||
| SNP-A | Disomy 3 | 12 | 2 | 0.89 (0.67, 0.97) | 0.48 (0.30, 0.67) | 0.55 (0.38, 0.72) | 0.86 (0.60, 0.96) |
| Monosomy 3 | 13 | 16 |
FISH-CEP3, FISH-3p26, and SNP-A were evaluated as predictors of survival. This analysis was performed on 43 patients, since 4 patients were alive but had not yet reached 36 months of follow-up. Values within parentheses indicate 95% confidence limits.
Correlation with Clinical Outcome
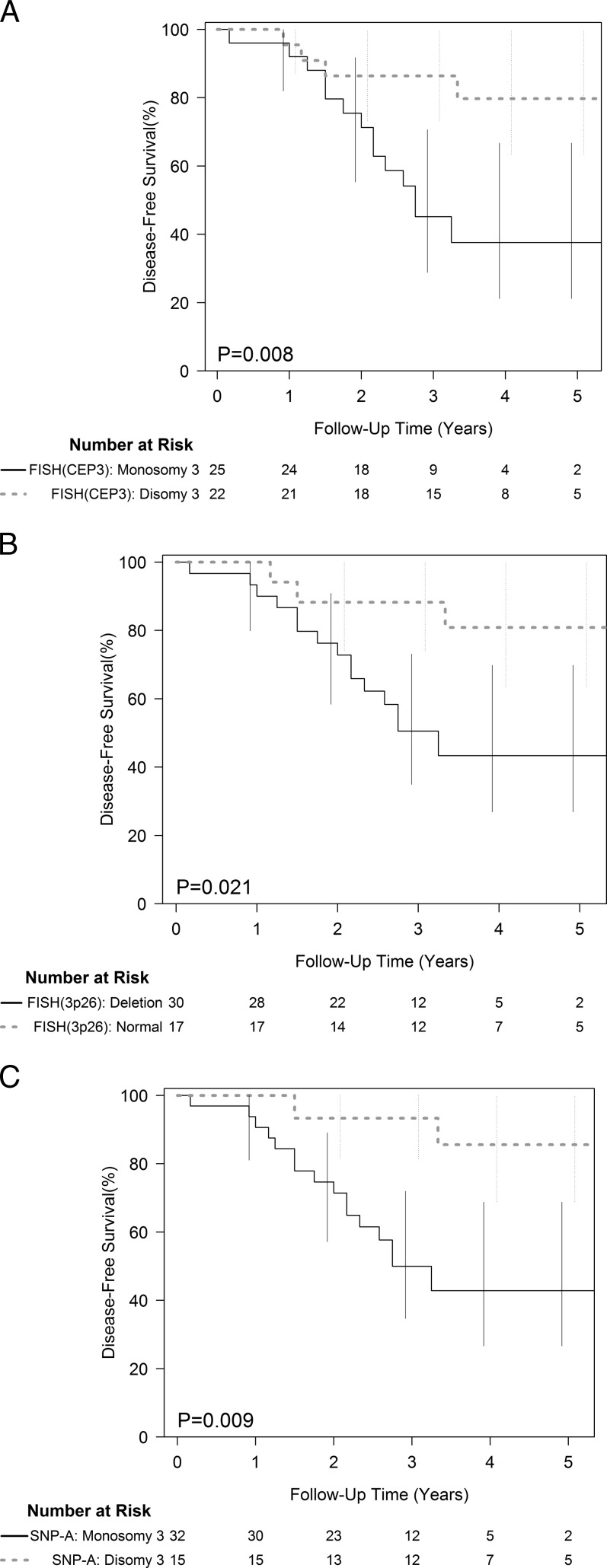
Of 33 patients with monosomy 3 tumors, there were 17 (52%) who developed clinically detectable metastases. All of these patients were deceased (median time from initial diagnosis to death of 21 months). Of the 17 patients with disomy 3 tumors, 12 were alive without metastatic disease at a median follow-up of 47 months. Three patients with disomy-3 tumors died from causes unrelated to uveal melanoma. In two patients (ID 27 and ID 32), FISH-CEP3 and FISH-3p26 indicated normal chromosomal status, whereas SNP-A revealed monosomy 3. Both patients died, one with confirmed metastases (ID 32). Two additional patients (ID 45 and ID 49) with tumor characterized as disomy 3 by FISH-CEP3, FISH-3p26, and SNP-A developed metastases. These patients died from metastases 18 and 40 months following initial diagnosis. Kaplan–Meier survival curves for patients with and without chromosome 3 abnormalities detected by each technique (FISH-CEP3, FISH-3p26, and SNP-A) are shown (Fig. 2).
Figure 2.
Disease-free survival and chromosome 3 status defined by the FISH-CEP3 (A), FISH-3p26 (B), and by SNP-A (C).
For prediction of survival at 36 months, the sensitivity and specificity for FISH-CEP3, FISH-3p26, and SNP-A were 0.78 (0.55, 0.91), 0.64 (0.45, 0.80); 0.83 (0.61, 0.94), 0.52 (0.33, 0.70); 0.89 (0.67, 0.97), 0.48 (0.30, 0.67), respectively (Table 5). No significant differences between measures were observed in any of the sensitivity or specificity measures.
Discussion
For the past decade, prognostication techniques have been a major focus of ophthalmic oncology research. As a result, technologies for identifying high-risk tumors have evolved rapidly. Each treatment center has adopted preferred methodologies and, at times, there has been a lack of consensus regarding standards for performing prognostication studies. FISH in particular is a flexible technology with many adjustable parameters including: type of tissue analyzed (fresh or frozen versus paraffin-embedded), number and type of probes used, cutoff point value used to determine the presence of monosomy 3, and the number of cells scored. We chose a cutoff value of 20% to define monosomy for chromosome 3 based on frequency distribution of the percentage of cells demonstrating monosomy in each tumor (Fig. 1) and because of the fact that many prior studies in the literature have used such a cutoff value.18,30,34,35,37,40 Additionally, although 8% cutoff provided the best combination of sensitivity and specificity, it did not perform significantly better than the currently used cutoff point (20%). Of note, the cutoff point of 8% to define monosomy 3 observed in our study is similar to the cutoff value of 5% (using a nuclear enrichment technique) that has been reported to correlate significantly with the risk of metastases at 5 years.47
There is a paucity of studies that directly compare prognostic techniques in uveal melanoma. A study based on microsatellite analysis (MSA) done on a large number of cases reported a 3-year metastasis rate of 24% in the complete monosomy group and 3% in the disomy group.48 A more recent MSA-based study reported disease-specific mortality rates for tumors with disomy 3 of 13.2% compared with 75.1% in the monosomy 3 group.49 These studies did not include other techniques for comparison.48 In a study using multiplex ligation-dependent probe amplification (MLPA) to detect chromosome 3 loss, 10-year disease-specific mortality was 0% in 133 tumors without chromosome 3 loss.50 Although these authors did not directly compare MLPA with other techniques, Vaarater et al.51 demonstrated that MLPA was equivalent to FISH for the identification of patients at risk for metastatic disease in uveal melanoma. The sensitivity of MLPA to detect patients at risk for metastatic disease was higher than that of FISH (0.795 vs. 0.692) but the specificity was equal for both techniques (0.840).51
Onken et al.18 reported that loss of heterozygosity of chromosome 3 detected by SNP-A was superior to that of FISH in predicting metastatic outcome. However, FISH analysis was suboptimal in that study because investigators used paraffin-embedded tissue (replete with truncation artifacts) and counted only 100 cells. A threshold of 30% (higher than the conventional cutoff of 20%) was used to define monosomy 3 status.18 Young and colleagues reported that monosomy 3 status could be successfully determined in choroidal melanoma in only 64% of cases analyzed by FISH, compared with 73% of cases evaluated by SNP-A. In their series, fresh tissue was acquired by fine-needle aspiration biopsy (FNAB) using a 30-gauge needle via a transscleral approach. For FISH analysis, a CEP3 probe was used, although the threshold for determining monosomy 3 status and the exact number of cells counted were not specified.38
The manner in which FISH using a locus-specific probe compares with SNP-A analysis has not been previously reported. Monosomy 3 was detected in 33 cases (66%) by either FISH or SNP-A status, which is commensurate with the previous reports involving large tumors or those treated by enucleation.11,19,21,22,26,32 SNP-A analysis was the most sensitive technique in identifying 31 of these cases, yielding a detection rate of 62%. When the locus-specific FISH probe was used, 3p26 deletion was found in 60% of monosomy 3 tumors. Of particular importance, FISH-3p26 deletion was present in every case of monosomy 3 detected by FISH-CEP3 (54%). There was a very high correlation between cases of monosomy 3 identified using SNP-A and those with FISH-3p26 deletion. The sensitivity of FISH-3p26 (0.78 [0.61, 0.89]) was higher than that of FISH-CEP3 (0.94 [0.80, 0.98]) using SNP-A as the gold standard.
The detection of 3p26 deletion signifies only that a particular locus is absent, and therefore does not definitively indicate the absence of whole chromosome 3 (monosomy). FISH-3p26 is also limited by its ability to differentiate disomic tumors from tumors harboring iso disomy 3 (duplication of one remaining chromosome is equivalent to functional monosomy). Isodisomy 3 can be inferred in two patients (ID 27 and ID 32), wherein FISH-CEP3 and FISH-3p26 indicated normal chromosomal status, whereas SNP-A revealed monosomy 3. Both patients died, one with confirmed metastases (ID 32). Some of the variability in the results is influenced by differences in methodology for detection and interpretation of changes in parts of chromosome 3 (gains and losses).49,52 Data from several studies have indicated that partial loss of chromosome may not be as deleterious as total loss of chromosome 3 in influencing the risk of metastasis because tumors with partial chromosome 3 loss behave similar to tumors classified as disomic.48,49,52 Our study further underscores the need for standardization of techniques, if results from various centers are to be directly compared.
The promise of molecular prognostication lies in the possibility that it may identify the subpopulation of patients with uveal melanoma in whom the risk of metastatic disease is sufficiently high and the disease-free interval is sufficiently short to be considered for adjuvant therapy. Although the sensitivity for prediction of survival at 36 months did not reach a statistically significant difference, the sensitivity was highest with SNP-A (0.89 [0.67, 0.97]) and least with FISH-CEP3 (0.78 [0.55, 0.91]). FISH-3p26 gave intermediate sensitivity (0.83 [0.61, 0.94]). It is also important to assess technical failures (i.e., an inability to determine risk status despite a patient's desire and false-negative rates of these techniques) because a patient with such a result would be denied potentially life-saving adjuvant therapy.16 In our series, a technical failure rate of 2–4% (SNP-A analysis [2 cases]) and FISH [1 case]) was comparable to that of a commercially available gene-expression profiling technique.15 However, in both cases of SNP-A failure, tumors were positive for FISH-3p26 deletion and in a single case FISH failure, monosomy 3 was found using SNP-A. Our observations support using a combination of techniques rather than relying on a single method in an unlikely event of a technical failure.
Negative predictive values for survival at 36 months were comparable for FISH-CEP3 (0.80 ([0.58, 0.92]), FISH-3p26 (0.81 [0.57, 0.93]), and SNP-A (0.86 [0.60, 0.96]), suggesting any of these tests can be used to identify patients for adjuvant treatment trial. Our data are based on a small number of patients that had large tumors and so these observations may not be directly applicable to smaller tumors that undergo eye conservative treatments in conjunction with prognostic FNAB.48 Metastatic events may occur late53 and therefore with longer follow-up, additional metastatic events may occur in the disomy group increasing the false-negative rates. Moreover, it is always challenging to determine the exact cause of death despite all efforts.54
The shortfall in detecting tumors with poor prognosis provides an opportunity for improvements in the power of the techniques used. As we move forward in our endeavor to improve prognostication in uveal melanoma, it will be of paramount importance to use a standard methodology so that results from various study centers can be meaningfully compared.
Acknowledgments
The authors thank James Bena, MS, for assistance with statistical analysis.
Footnotes
Supported in part by National Cancer Institute/National Institutes of Health Grant R01CA136776, a Falk Medical Research Trust grant, an unrestricted grant from Research to Prevent Blindness, Department of Ophthalmology, Cleveland Clinic Learner College of Medicine, the Sondra J. and R. Hardis Endowed Chair in Cancer Genomic Medicine (Cleveland Clinic), and an F. M. Kirby Foundation grant (CE).
Disclosure: A.D. Singh, None; M.E. Aronow, None; Y. Sun, None; G. Bebek, None; Y. Saunthararajah, None; L.R. Schoenfield, None; C.V. Biscotti, None; R.R. Tubbs, None; P.L. Triozzi, None; C. Eng, None
References
- 1. Sisley K, Rennie IG, Cottam DW, Potter AM, Potter CW, Rees RC. Cytogenetic findings in six posterior uveal melanomas: involvement of chromosomes 3, 6, and 8. Genes Chromosomes Cancer. 1990;2:205–209. [DOI] [PubMed] [Google Scholar]
- 2. Sisley K, Cottam DW, Rennie IG, et al. Non-random abnormalities of chromosomes 3, 6, and 8 associated with posterior uveal melanoma. Genes Chromosomes Cancer. 1992;5:197–200. [DOI] [PubMed] [Google Scholar]
- 3. Horsthemke B, Prescher G, Bornfeld N, Becher R. Loss of chromosome 3 alleles and multiplication of chromosome 8 alleles in uveal melanoma. Genes Chromosomes Cancer. 1992;4:217–221. [DOI] [PubMed] [Google Scholar]
- 4. Prescher G, Bornfeld N, Horsthemke B, Becher R. Chromosomal aberrations defining uveal melanoma of poor prognosis. Lancet. 1992;339:691–692. [PubMed] [Google Scholar]
- 5. Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jockel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. [DOI] [PubMed] [Google Scholar]
- 6. Hughes S, Damato BE, Giddings I, Hiscott PS, Humphreys J, Houlston RS. Microarray comparative genomic hybridisation analysis of intraocular uveal melanomas identifies distinctive imbalances associated with loss of chromosome 3. Br J Cancer. 2005;93:1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naus NC, Verhoeven AC, van Drunen E, et al. Detection of genetic prognostic markers in uveal melanoma biopsies using fluorescence in situ hybridization. Clin Cancer Res. 2002;8:534–539. [PubMed] [Google Scholar]
- 8. Sandinha MT, Farquharson MA, Roberts F. Identification of monosomy 3 in choroidal melanoma by chromosome in situ hybridisation. Br J Ophthalmol. 2004;88:1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sandinha MT, Farquharson MA, McKay IC, Roberts F. Monosomy 3 predicts death but not time until death in choroidal melanoma. Invest Ophthalmol Vis Sci. 2005;46:3497–3501. [DOI] [PubMed] [Google Scholar]
- 10. Sandinha T, Farquharson M, McKay I, Roberts F. Correlation of heterogeneity for chromosome 3 copy number with cell type in choroidal melanoma of mixed-cell type. Invest Ophthalmol Vis Sci. 2006;47:5177–5180. [DOI] [PubMed] [Google Scholar]
- 11. Sisley K, Rennie IG, Parsons MA, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19:22–28. [DOI] [PubMed] [Google Scholar]
- 12. Sisley K, Nichols C, Parsons MA, Farr R, Rees RC, Rennie IG. Clinical applications of chromosome analysis, from fine needle aspiration biopsies, of posterior uveal melanomas. Eye. 1998;12:203–207. [DOI] [PubMed] [Google Scholar]
- 13. Sisley K, Parsons MA, Garnham J, et al. Association of specific chromosome alterations with tumour phenotype in posterior uveal melanoma. Br J Cancer. 2000;82:330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. White VA, McNeil BK, Horsman DE. Acquired homozygosity (isodisomy) of chromosome 3 in uveal melanoma. Cancer Genet Cytogenet. 1998;102:40–45. [DOI] [PubMed] [Google Scholar]
- 15. Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harbour JW. Molecular prognostic testing and individualized patient care in uveal melanoma. Am J Ophthalmol. 2009;148:823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cook JR. Fluorescence in situ hybridization In Tubbs RR, Stoler MH. eds Cell and Tissue Based Molecular Pathology Philadelphia: Churchill Livingstone; 2009. 104–113. [Google Scholar]
- 18. Onken MD, Worley LA, Person E, Char DH, Bowcock AM, Harbour JW. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clin Cancer Res. 2007;13:2923–2927. [DOI] [PubMed] [Google Scholar]
- 19. Bauer J, Kilic E, Vaarwater J, Bastian BC, Garbe C, de Klein A. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br J Cancer. 2009;101:813–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonaldi L, Midena E, Filippi B, et al. FISH analysis of chromosomes 3 and 6 on fine needle aspiration biopsy samples identifies distinct subgroups of uveal melanomas. J Cancer Res Clin Oncol. 2008;134:1123–1127. [DOI] [PubMed] [Google Scholar]
- 21. Cools D, Debiec-Rychter M. Parys-Van Ginderdeuren R, Hagemeijer-Hausman A, Van den Oord J, Spileers W. Preliminary results of the FRO project: chromosomal abnormalities in primary uveal melanoma. Bull Soc Belge Ophtalmol. 2000;278:67–70. [PubMed] [Google Scholar]
- 22. Coupland SE, Campbell I, Damato B. Routes of extraocular extension of uveal melanoma: risk factors and influence on survival probability. Ophthalmology. 2008;115:1778–1785. [DOI] [PubMed] [Google Scholar]
- 23. Cross NA, Ganesh A, Parpia M, Murray AK, Rennie IG, Sisley K. Multiple locations on chromosome 3 are the targets of specific deletions in uveal melanoma. Eye. 2006;20:476–481. [DOI] [PubMed] [Google Scholar]
- 24. Damato B, Duke C, Coupland SE, et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007;114:1925–1931. [DOI] [PubMed] [Google Scholar]
- 25. Damato B, Coupland SE. A reappraisal of the significance of largest basal diameter of posterior uveal melanoma. Eye. 2009;23:2152–2160 ; quiz: 2161–2162 [DOI] [PubMed] [Google Scholar]
- 26. Kilic E, Naus NC, van Gils W, et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Invest Ophthalmol Vis Sci. 2005;46:2253–2257. [DOI] [PubMed] [Google Scholar]
- 27. Lake SL, Coupland SE, Taktak AF, Damato BE. Whole-genome microarray detects deletions and loss of heterozygosity of chromosome 3 occurring exclusively in metastasizing uveal melanoma. Invest Ophthalmol Visual Sci. 2010;51:4884–4891. [DOI] [PubMed] [Google Scholar]
- 28. Maat W, Ly LV, Jordanova ES, de Wolff-Rouendaal D, Schalij-Delfos NE, Jager MJ. Monosomy of chromosome 3 and an inflammatory phenotype occur together in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:505–510. [DOI] [PubMed] [Google Scholar]
- 29. Marathe OS, Wu J, Lee SP, et al. Ocular response of choroidal melanoma with monosomy 3 versus disomy 3 after iodine-125 brachytherapy. Int J Radiat Oncol Biol Phys. 2011;81:1046–1048. [DOI] [PubMed] [Google Scholar]
- 30. McNamara M, Felix C, Davison EV, Fenton M, Kennedy SM. Assessment of chromosome 3 copy number in ocular melanoma using fluorescence in situ hybridization. Cancer Genet Cytogenet. 1997;98:4–8. [DOI] [PubMed] [Google Scholar]
- 31. Midena E, Bonaldi L, Parrozzani R, Radin PP, Boccassini B, Vujosevic S. In vivo monosomy 3 detection of posterior uveal melanoma: 3-year follow-up. Graefes Arch Clin Exp Ophthalmol. 2008;246:609–614. [DOI] [PubMed] [Google Scholar]
- 32. Mensink HW, Vaarwater J, Kilic E, et al. Chromosome 3 intratumor heterogeneity in uveal melanoma. Invest Ophthalmol Vis Sci. 2009;50:500–504. [DOI] [PubMed] [Google Scholar]
- 33. Mudhar HS, Saunders E, Rundle P, Rennie IG, Sisley K. The in vivo modulatory effects of an anterior-chamber microenvironment on uveal melanoma. Br J Ophthalmol. 2009;93:535–540. [DOI] [PubMed] [Google Scholar]
- 34. Patel KA, Edmondson ND, Talbot F, Parsons MA, Rennie IG, Sisley K. Prediction of prognosis in patients with uveal melanoma using fluorescence in situ hybridisation. Br J Ophthalmol. 2001;85:1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Radhakrishnan A, Badhrinarayanan N, Biswas J, Krishnakumar S. Analysis of chromosomal aberration (1, 3, and 8) and association of microRNAs in uveal melanoma. Mol Vis. 2009;15:2146–2154. [PMC free article] [PubMed] [Google Scholar]
- 36. White JS, Becker RL, McLean IW, Director-Myska AE, Nath J. Molecular cytogenetic evaluation of 10 uveal melanoma cell lines. Cancer Genet Cytogenet. 2006;168:11–21. [DOI] [PubMed] [Google Scholar]
- 37. Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007;13:1466–1471. [DOI] [PubMed] [Google Scholar]
- 38. Young TA, Burgess BL, Rao NP, Gorin MB, Straatsma BR. High-density genome array is superior to fluorescence in-situ hybridization analysis of monosomy 3 in choroidal melanoma fine needle aspiration biopsy. Mol Vis. 2007;13:2328–2333. [PubMed] [Google Scholar]
- 39. Young TA, Burgess BL, Rao NP, Glasgow BJ, Straatsma BR. Transscleral fine-needle aspiration biopsy of macular choroidal melanoma. Am J Ophthalmol. 2008;145:297–302. [DOI] [PubMed] [Google Scholar]
- 40. Schoenfield L, Pettay J, Tubbs RR, Singh AD. Variation of monosomy 3 status within uveal melanoma. Arch Pathol Lab Med. 2009;133:1219–1222. [DOI] [PubMed] [Google Scholar]
- 41. van Gils W, Lodder EM, Mensink HW, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008;49:4254–4262. [DOI] [PubMed] [Google Scholar]
- 42. Parrella P, Fazio VM, Gallo AP, Sidransky D, Merbs SL. Fine mapping of chromosome 3 in uveal melanoma: identification of a minimal region of deletion on chromosomal arm 3p25.1-p25.2. Cancer Res. 2003;63:8507–8510. [PubMed] [Google Scholar]
- 43. http://clinicaltrials.gov/ct2/results?term=NCT01100528++. Accessed March 30 , 2012. [Google Scholar]
- 44. Online Mendelian Inheritance in Man Web site. http://omim.org/search?index=geneMap&search=3p26. Accessed March 30, 2012. [Google Scholar]
- 45. Frater JL, Tsiftsakis EK, Hsi ED, Pettay J, Tubbs RR. Use of novel t(11;14) and t(14;18) dual-fusion fluorescence in situ hybridization probes in the differential diagnosis of lymphomas of small lymphocytes. Am J Surg Pathol. 2001;10:214–222. [DOI] [PubMed] [Google Scholar]
- 46. Collaborative Ocular Melanoma Study (COMS) Group: COMS Manual of Procedures, PB95-179693 Springfield, VA: National Technical Information Service;January1995. [Google Scholar]
- 47. Bronkhorst IH, Maat W, Jordanova ES, et al. Effect of heterogeneous distribution of monosomy 3 on prognosis in uveal melanoma. Arch Pathol Lab Med. 2011;135:1042–1047. [DOI] [PubMed] [Google Scholar]
- 48. Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, Shields JA. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2011;118:396–401. [DOI] [PubMed] [Google Scholar]
- 49. Thomas S, Putter C, Weber S, Bornfeld N, Lohmann DR, Zeschnigk M. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study. Br J Cancer. 2012;106:1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–6092. [DOI] [PubMed] [Google Scholar]
- 51. Vaarwater J, van den Bosch T, Mensink HW, et al. Multiplex ligation-dependent probe amplification equals fluorescence in-situ hybridization for the identification of patients at risk for metastatic disease in uveal melanoma. Melanoma Res. 2012;22:30–37. [DOI] [PubMed] [Google Scholar]
- 52. Abdel-Rahman MH, Christopher BN, Faramawi MF, et al. Frequency, molecular pathology and potential clinical significance of partial chromosome 3 aberrations in uveal melanoma. Mod Pathol. 2011;24:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kujala E, Makitie T, Kivela T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44:4651–4659. [DOI] [PubMed] [Google Scholar]
- 54. Moy CS, Albert DM, Diener-West M, McCaffrey LD, Scully RE, Willson JK. Cause-specific mortality coding. Methods in the Collaborative Ocular Melanoma Study COMS Report No. 14. Control Clin Trials. 2001;22:248–262. [DOI] [PubMed] [Google Scholar]