Introduction
Neurodegenerative diseases, in particular those associated with aging, represent a significant public health concern in the developed world. The prevalence of dementia doubles every five years after the age of 60, with an estimated 30 to 45 percent of those age 85 and older suffering from some form of dementia, most commonly Alzheimer’s disease [1]. As mean and maximum life expectancy have increased significantly in the last century, the prevalence of such age-associated dementias has followed course. The development of effective treatments is, however, hindered by the complex, multigenic nature of these diseases and by their relatively poorly understood molecular pathophysiology. One aspect of the biology of neurodegenerative diseases that may have important implications for better understanding the underlying molecular mechanisms causing neurodegeneration is that, in spite of the diverse clinical and cellular manifestations, mitochondrial dysfunction is a ubiquitous feature of neurodegenerative diseases, detected both in neurons and in non-neuronal cells.
There is still some discussion in the field as to whether mitochondrial dysfunction is the major cause of neurodegeneration; but the pervasive observation of dysfunctional mitochondria in affected brain regions and cells clearly indicate that mitochondria are essential for neuronal survival, and suggest that pathways involved in maintaining mitochondrial integrity function to prevent or delay the onset of neurodegeneration. This review will examine the evidence that links mitochondrial genome instability to neuronal loss and neurodegeneration. We will explore DNA damage accumulation in mitochondrial DNA (mtDNA) in several pathological conditions, and whether changes in DNA repair activities may play a role in these events (see also Yang et al, this volume, for additional discussion of mitochondrial damage and neurological pathology) . And while it is not the scope of this review, it is noteworthy that specific defects in the repair of DNA single-strand breaks in the brain have been directly associated with the pathophysiology of some types of ataxias, namely spinocerebellar ataxia with axonal neuropathy-1 (SCAN1) and ataxia oculomotor apraxia-1 (AOA1) [2] (see Lavin et al in this volume).
Mitochondrial dysfunction in neurodegeneration
The central nervous system has a very high energetic demand, used mostly to maintain ionic distribution across the membranes and for synaptic function. Glucose is virtually the sole fuel for the human brain, except during prolonged starvation. Moreover, the brain lacks fuel stores and hence requires a continuous supply of glucose. These characteristics render brain cells particularly dependent on mitochondrial function for the generation of ATP. Thus, it is not surprising that loss of mitochondrial integrity has been widely associated with neurodegeneration.
Mitochondrial dysfunction in the brain can stem from inherited or spontaneous mutations in mtDNA or in nuclear genes encoding mitochondrial proteins, or from direct damage to mtDNA caused by endogenous or exogenous toxic agents. The “Mitochondrial Theory of Aging” proposes that “normal aging” is caused by the steady accumulation of mitochondrial damage caused by endogenous agents, most likely reactive oxygen species (ROS), over an organism’s life span [3, 4]. Interestingly, diseases stemming from mitochondrial defects are characterized by a wide range of neuro-muscular phenotypes, and a large number of them are presented with at least one of several neurological symptoms, ranging from hearing loss and mild ophtalmoplegia to severe stroke, epilepsy, ataxia and neuropathy [5–7]. Moreover, the disruption of the mitochondrial respiratory chain by toxins leads to neuronal cell death and phenotypes that often resemble neurodegenerative diseases, such as Parkinson’s (PD) and Huntington’s (HD) diseases.
PD is a progressive movement disorder that is believed to be caused by the selective loss of dopaminergic neurons, particularly in the substantia nigra region of the brain. Several lines of evidence support the hypothesis that a deficiency in the mitochondrial respiratory Complex I (NADH Ubiquinone Oxireductase) is an underlying feature of PD (for review see [8–10]). This is strongly corroborated by the observation that inhibitors of Complex I induce a motor disorder that clinically and pathologically mimic PD. The effect of the compound 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP) is a classical example. MPTP-induced Parkinsonism was first identified in the late 1970’s, in drug addicts who consumed a synthetic heroine derivative [11], where MPTP was a small contaminant generated unexpectedly during the synthetic process. The toxicity of MPTP is conferred by its metabolite 1-methyl-4-phenylpyridinium ion (MPP+) [12], which accumulates selectively in mitochondria from dopaminergic neurons. MPP+ is a strong inhibitor of Complex I, resulting in decreased oxidation of NAD-linked substrates, disruption of the electron transport chain, reduced oxidative phosporylation and ATP synthesis [13–15]. Increased oxidative stress as a result of MPP+ toxicity has also been postulated to play a causative role in the loss of dopaminergic neurons, as supported by the observation that MPTP toxicity is significantly exacerbated in mice deficient for anti-oxidant enzymes including glutathione peroxidase, Cu- and Mg-Superoxide Dismutase. Interestingly, the chemical inhibition of poly-ADP-ribose polymerase (PARP) significantly attenuates MPTP toxicity [16, 17]; since PARP is activated by DNA strand breaks and oxidative damage, these findings suggest that mitochondrial-induced oxidative DNA damage also play a role in the etiology of PD (see Woodhouse et al in this volume for detailed discussion of PARP and neurological disease) . Another well known inhibitor of Complex I, rotenone, has been shown to cause PD-like pathology as well. This antibiotic is a common component of pesticides, and several epidemiological studies have implicated accidental exposure to rotenone as a risk factor in the development of PD [8].
Inhibitors of the mitochondrial respiratory Complex II (Succinate-Ubiquinol Oxireductase, or Succinate Dehydrogenase-SDH) also induce neurodegeneration in animal models (for review, see [10, 18]). Most notably, inhibition of Complex II by 3-nitropropionic acid (3-NPA) and malonate greatly resemble those seen in HD, another movement disorder characterized by cell death in the striatum and disruption of the cortical-striatal circuitry. While 3-NPA and malonate inhibit SDH by distinct molecular mechanisms, the similarities between their pathological outcomes strongly suggests that SDH inhibition is a central molecular event in HD etiology. This is consistent with Complex II and III deficiencies observed in post-mortem striatum samples [19]. Moreover, oxidative stress indicators, particularly in mitochondria, are also elevated in animal models of 3-NPA-induced toxicity as well as in HD samples [20].
The findings discussed above indicate a strong correlation between disruption of the mitochondrial electron transport chain and neurodegeneration. They suggest a scenario in which mitochondrial dysfunction, induced by toxins as discussed above, or by yet unknown mechanisms in the case of the idiopathic diseases, is an initial event in a cascade that leads to neuronal loss. This initial mitochondrial inhibition would lead to energetic crisis and elevated oxidative stress, accumulation of oxidative DNA damage and mutations, and ultimately to neuronal cell death. In this scenario, it is critical to understand how these events are linked to cellular dysfunction, since they could provide targets for clinical interventions that could block or attenuate the progression of these diseases.
Mitochondrial oxidative DNA damage accumulation
The mtDNA is situated at the inner side of the mitochondrial inner membrane. It is believed to be covalently associated with the membrane, if not always, then at least during some metabolic transactions, such as DNA replication [21]. In eukaryotic cells, the inner mitochondrial membrane is the major cellular site for generation of ROS, such as superoxide anion, hydrogen peroxide and the hydroxyl radical [22]. ROS are very reactive towards biomolecules, and can randomly attack lipids, proteins and nucleic acids. The hydroxyl radical will react with any molecule in a radius of 15 Å of its site of formation, with a rate constant only limited by diffusion [23]. The oxidative attack of biomolecules results in a variety of potentially harmful consequences, such as 1) lipid peroxidation, which leads to the generation of highly reactive aldehyde by-products; 2) oxidation of SH groups, which causes protein aggregation; 3) oxidation of amino acid side chains forming carbonyl groups, which target proteins for degradation; 4) oxidation of the sugar in nucleic acids, leading to breakage of the phosphodiester backbone; and 5) oxidation of the bases in nucleic acids, with the concomitant formation of a plethora of base modifications that can be either mutagenic or cytotoxic (reviewed in [24]).
The physical proximity of the mtDNA to the sites of ROS production in the inner mitochondrial membrane renders the mtDNA more vulnerable to being damaged by such species in comparison to nuclear DNA. Several groups, including ours, have demonstrated that the levels of oxidized bases in mtDNA are 2–3 times greater than in nuclear DNA [25, 26], likely reflecting the issue of accessibility.
From the large number of different oxidized DNA modifications detected in DNA exposed to oxidizing agents [27] , only a few have been, so far, extensively studied in regards to their biological significance. Eight-hydroxy-guanine (8-oxoG) was one of the first oxidized bases to be recognized as biologically relevant [28, 29] and has since been considered a biomarker of oxidative stress. 8-oxoG is a mutagenic lesion when present in double-stranded DNA, because in syn configuration it can mispair with adenine, leading to a G:C→T:A transversion mutation [30]. For instance, DNA polymerase gamma (pol γ), the only DNA polymerase identified in mammalian mitochondria so far (see more below), will insert a wrong base opposite 8-oxoG in the template about 27% of the time [31], resulting in a mutagenic event. Recent data, however, suggest that 8-oxoG may not be as deleterious in vivo as previously believed. Maga and colleagues showed that 8-oxoG is faithfully bypassed by the nuclear DNA polymerases lambda and eta when the auxiliary factors PCNA and RPA are present [32]. It has also been shown that the transcription machinery can bypass 8-oxoG in vitro [33]. We observed normal mitochondrial bioenergetic function in mitochondria isolated from livers and hearts of mice lacking the DNA repair enzyme Oxoguanine DNA Glycosylase (OGG1) [34]. These mice show no removal of 8-oxoG from their mtDNA and accumulate approximately 20 fold higher levels of 8-oxoG than wild type controls [35], indicating that such high levels of 8-oxoG did not compromise the integrity of the respiratory chain.
Other oxidized bases have also been detected in DNA, even in the absence of any exogenous insult. For example, thymine glycols (TG) can be detected both in genomic as well as in mtDNA [36]. TG is a polymerase-blocking lesion, causing aborted DNA and RNA synthesis [37], which could ultimately lead to cell death. In addition, we have recently shown that ring-opened formamidopyrimidine lesions (4,6-diamino-5-formamidopyrimidine, FapyA; and 2,6-diamino-4-hydroxy-5-formamidopyrimidine, FapyG) are detected in mouse liver genomic DNA at levels that are similar, or even slightly higher, than those of 8-oxoG [38]. Both FapyA and FapyG are strong mutagenic bases in vitro, directing the incorporation of adenine opposite the modified purine [39, 40].
Nonetheless, 8-oxoG quantification has been extensively used as a marker of oxidative DNA damage accumulation and several groups have demonstrated that the levels of this lesion are elevated in mtDNA both during normal aging as well as in neurodegenerative disorders. Ames and colleagues [25, 41] were the first to show that mtDNA accumulated 8-oxoG at higher levels than the nuclear DNA, and although there has since been much discussion as to the absolute levels of oxidative lesions in mtDNA from various sources [42, 43], it is well accepted that oxidative damage increases with age in the mtDNA [26, 44].
Base excision repair in mitochondria
Because mammalian cells harbor genetic material in both their mitochondrial and nuclear compartments and each genome is vulnerable to various types of spontaneous and exposure-dependent DNA lesions, organisms have evolved protective systems for both partitions. However, the spectrum of the DNA damage responses varies between the mitochondria and nucleus (Table 1), and in many cases, the precise molecular pathway and specific repair proteins involved are distinct. For the purpose of this review, we focus exclusively on the Base Excision Repair (BER) pathway in mitochondria, but refer to the following reviews for discussion on nuclear DNA repair [45] and mtDNA repair in general [46]. BER is the predominant pathway for coping with most spontaneous decay, alkylation and oxidative DNA lesions.
Table 1.
Comparison of the DNA repair pathways identified so far in the nucleus and mitochondria of mammalian cells.
| Mammalian DNA Repair Pathways | |||
|---|---|---|---|
| PATHWAY | SUBPATHWAYS | Nuclear | Mito |
| BER/SSBR | Long patch | Y | N |
| Short patch | Y | Y | |
| NER | General genome | Y | N |
| Gene-specific | Y | N | |
| Transcription associated | Y | N | |
| MMR | Y | Y? | |
| Recombination | HR | Y | ? |
| NHEJ | Y | Y | |
| Direct damage reversal | Y | Y | |
Nuclear or mitochondrial BER, like all DNA repair responses, involves a cascade of recognition and enzymatic processing steps that aim to remove the target damage and restore genome integrity. In BER is initiated by the excision of a modified or inappropriate base from the sugar-phosphate DNA backbone by a DNA glycosylase. The resulting apurinic/apyrimidinic (AP) site is subsequently cleaved by an AP endonuclease, creating a strand break with a 3’-hydroxyl end and a 5’-deoxyribose phosphate (dRP) residue. The single nucleotide gap is then filled by a specific DNA repair polymerase, and the 5’-dRP fragment is excised to create a normal 5’-phosphate terminus. To complete short patch (or single nucleotide) BER, the final nick is sealed by a DNA ligase (Figure 1). It is worth emphasizing that the mitochondrial genome does not encode DNA repair proteins, and thus, as will be discussed next, most of the mitochondrial BER proteins are splice-variants, alternative translation-initiation products, or post-translationally modified versions of the nuclear-encoded protein forms.
Figure 1.
Schematic representation of the Base Excision Repair Pathway in Mammalian Mitochondria. Each enzymatic step and the proteins involved are further described in the text.
DNA Glycosylases
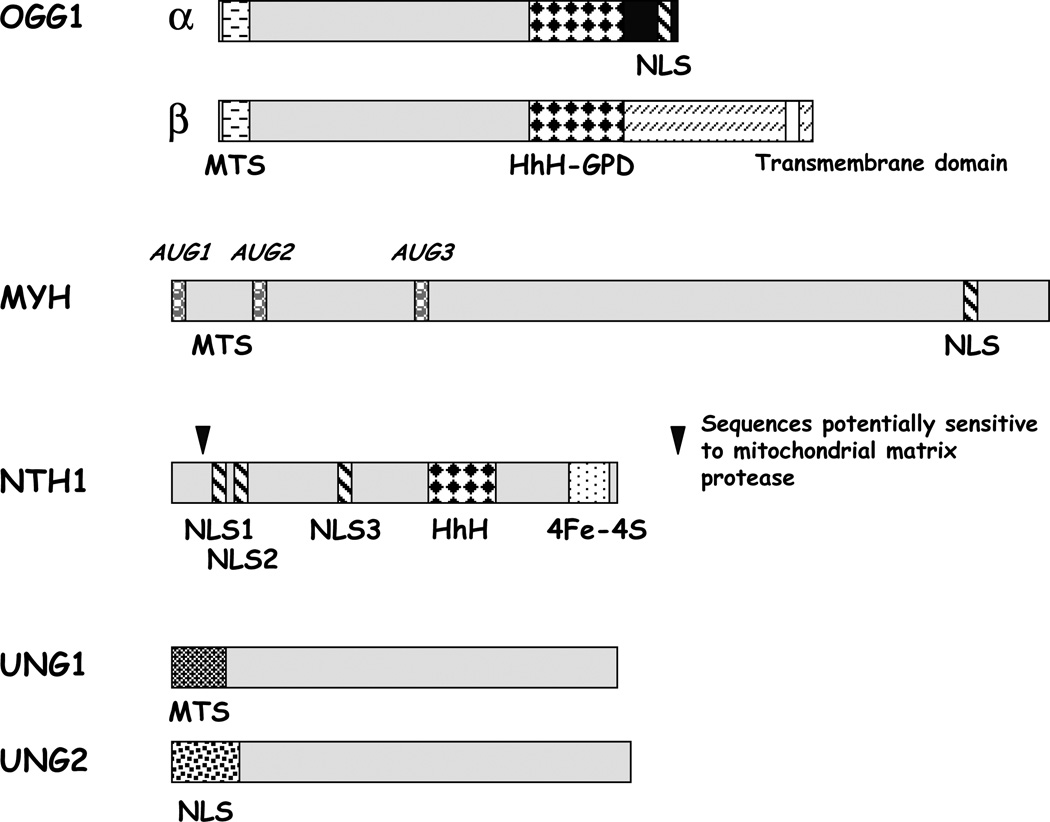
Several DNA glycosylases have been identified in mammalian mitochondria (Figure 2). The gene encoding hOGG1, which excises, among others, 8-oxoG from DNA, creates at least four mRNA isoforms by alternative splicing of distinct 3’ exons [47, 48]. Most of these isoforms, such as type 2a/OGG1β (Figure 2), which is one of the major expression products, encode proteins that localize to the mitochondria, presumably via a shared N-terminal mitochondrial targeting sequence (MTS). However, the type 1a protein (also known as OGG1α; Figure 2), which contains a unique C-terminal domain that harbors a strong nuclear localization signal (NLS), sorts mainly to the nucleus, yet to a lesser extent to the mitochondria. Recent evidence suggests that the β form is a non-active splice variant with unknown function and that OGG1α accounts for both the nuclear and mitochondrial 8-oxoG repair activities [49]. In addition to 8-oxoG [50], mammalian OGG1 has been shown to exhibit glycosylase activity for the oxidized base lesions 8-oxoA and FapyG, as well as a weak AP lyase function [51–53].
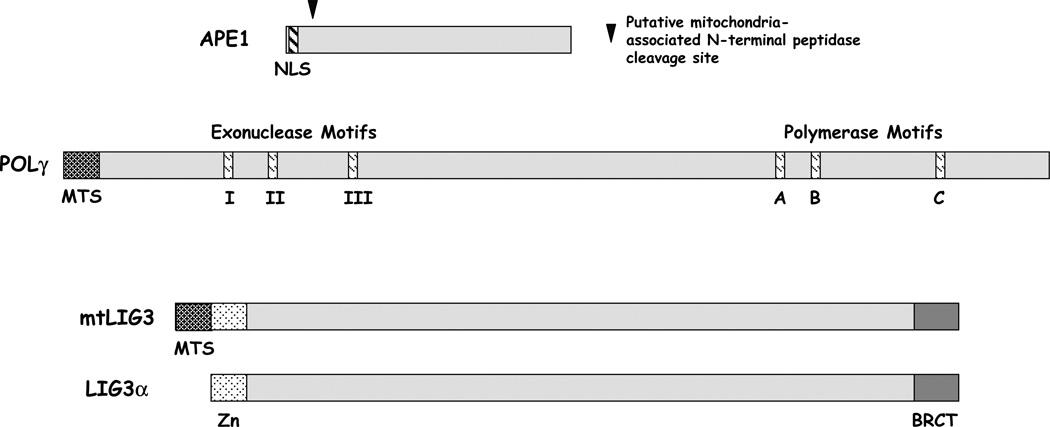
Figure 2.
Splice variants of DNA repair enzymes found in mammalian mitochondria.
Human MYH, a homolog of the Escherichia coli DNA glycosylase mutY, excises adenine opposite 8-oxoG as part of the defense system against the mutagenic consequences of this oxidative base damage (i.e. the so-called GO system [54]). The human MYH gene produces three major transcripts with unique 5’ mRNA sequences, i.e. α, β and γ, each of which is alternatively spliced to create as many as 10 different splice variants [55, 56] (see example in Figure 2). Despite initial studies indicating that hMYH resides mainly in the mitochondria [47], western blot analysis uncovered the presence of hMYH protein forms in both nuclei (p52/53) and mitochondria (p57) [56]. The p52 nuclear species has been suggested to be the product of the hMYHβ1, β3 or γ2 transcript(s). The origin of the mitochondrial MYH is likewise unclear, but studies suggest that it may be the larger protein created by the hMYHα3 mRNA (p54) or the 60 kDa species predicted to be generated by the hMYHoα1 transcript. Determination of the primary amino acid sequences of the purified MYH glycosylases from both nuclear and mitochondrial extracts will shed light on the nature of the different compartmental forms.
Mammalian NTH1, an ortholog of Escherichia coli endonuclease III (Nth), is a DNA glycosylase/AP lyase that excises a wide range of pyrimidine lesions, including TG, 5-hydroxy cytosine and fapy intermediates. Current evidence suggests that hNTH1 protein is localized almost exclusively to the nuclues, likely due to the presence of a weak MTS [47, 57, 58]. Notably, no alternative splicing of the hNTH1 gene has been reported so far, and the disparate protein isoforms of hNTH1 generated by multiple translation initiation sites do not exhibit differences in localization (Figure 2). In contrast, mouse NTH1 appears to localize mostly in mitochondria [58], which is in agreement with the abrogation of TG and 5-OHU incision activity in liver mitochondria from NTH1 knockout mice [59].
A new set of DNA glycosylases with specificity for oxidative DNA lesions has been recently identified, NEIL1, 2 and 3 [60–63]. These are mammalian homologues of the Escherichia coli DNA glycosylase Fpg/Nei, which excise primarily ring-opened oxidized purines such as FapyG and FapyA [64] . We have recently shown that NEIL1 localizes to mouse liver mitochondria, where it is likely to provide the residual incision activity observed with a FapyG containing oligomer in extracts from mice lacking both OGG1 and NTH1. The importance of NEIL1 for the maintenance of mtDNA has also been implied by the observation that mtDNA deletions accumulate in the livers of NEIL1 knockout mice, and that these mice have symptoms often associated with mitochondrial disease [63].
The uracil-DNA glycosylases (UDG) UNG1 (mitochondria) and UNG2 (nuclear) are encoded by the UNG-gene (Figure 2). These enzymes remove uracils generated by spontaneous deamination of cytosine in DNA. The mitochondrial and nuclear isoforms of UDG share a common catalytic domain, but have different N-terminal sequences for subcellular sorting as a result of the use of alternative promoters and mRNA splicing [65]. In addition to excising uracil from U:G > U:A base pairs, the UNG proteins have the ability to remove oxidized cytosines from DNA as well, including isodialuric acid, alloxan and 5-hydroxyuracil [66].
In the nucleus alkylation base damage is repaired mainly by methylpurine DNA glycosylase (MPG). While the MPG gene encodes alternative transcripts that produce proteins with putative nuclear and mitochondrial targeting sequences, MPG has not been shown explicitly to localize to the mitochondria [67]. However, efficient repair of methylated and ethylated bases in mtDNA has been documented [68–71].
AP Endonuclease
Following base excision by a DNA glycosylase, the resulting abasic site is typically incised by an AP endonuclease (Figure 1). In humans, the major, if not only significant, AP site incision activity is intrinsic to APE1. Initial studies revealed that translocation of the only known human APE1 protein species (a 318 amino acid product), which was engineered to harbor a FLAG-tag, scarcely occurred into the mitochondria of COS-7 cells [47]. APE1 harbors a NLS in its N-terminal sequence and no major alternatively-expressed forms have been reported [72]. However, endonucleolytic AP site cleavage activity had been observed in mitochondrial extracts [73], and immunohistochemistry with APE1-specific antibodies suggested the presence of an APE1-like protein in this cellular compartment [74, 75]. Mitra and colleagues recently reported that the mitochondrial AP endonuclease is derived from the main APE1 polypeptide by protease removal of the 33 N-terminal amino acid residues that contain the NLS [76] (Figure 2). It is important to emphasize that the truncated APE1 protein described above was only seen at a significant level in bovine and mouse liver and the NIH 3T3 mouse cell line, whereas only full-length APE1 was detected in a variety of human cell lines. These results may indicate that the full-length form is the major protein species in cultured cells and/or has a more pronounced role in mtDNA repair in humans. A second protein with homology to APE1, as well as its bacterial ancestor exonuclease III, APE2, has been shown to localize partially to the mitochondria, but its contribution as an AP endonuclease is in question [77, 78].
DNA Polymerase
Mitochondria possess a single DNA polymerase, POLγ (Figure 2), which assumes sole responsibility for DNA synthesis in all replication, recombination, and repair transactions involving mtDNA [79]. POLγ is homologous to the family A of DNA polymerases, which include the Klenow fragments of Escherichia coli and Bacillus stearothermophilus polymerase I and the polymerases of Thermus aquaticus and bacteriophage T7. In humans, POLγ holoenzyme is a 195 kDa heterotrimer, consisting of a catalytic subunit (p140, coded by POLG on chromosome 15q25) and two identical accessory subunits (p55, coded by POLG2 on chromosome 17q). The accessory subunit (p55) is a DNA binding factor that activates both the polymerase and exonuclease activities of the catalytic subunit, and confers high processivity by increasing the affinity of the heterotrimer for template DNA [80, 81]. The catalytic subunit (p140) possesses DNA polymerase, 3’→,5’ exonuclease activity and a 5’dRP lyase function [82], all of which are important to the accurate and productive execution of gap-filling and termini processing in mitochondrial BER (Figure 1). The accessory subunit has been shown to improve the efficiency of dRP removal (the rate-limiting step in mitochondrial BER) by increasing the lyase reaction and the ability of the enzyme to locate the damage site on DNA, presumably by enhancement of DNA binding [83].
DNA Ligase
The final step of BER is sealing of the remaining single-strand nick (Figure 1). In mitochondria, this molecular process is performed by a variant of the nuclear DNA ligase III (LIG3). In particular, the human DNA LIG3 gene encodes both nuclear and mitochondrial enzymes (Figure 2), which arise from the use of alternative-translation initiation start sites in a single mRNA transcript [84]. Interestingly, the mitochondrial DNA LIG3 protein apparently functions without XRCC1 [85], a key single-strand break repair factor required for the stability of nuclear DNA LIG3oα via its direct physical association [86].
Mitochondrial DNA instability in neurodegenerative disorders
Increased oxidative DNA damage, mutations and deletions have been observed in the aging brain and neurodegenerative diseases. Alzheimer’s disease (AD) is a progressive age-dependent neurodegenerative disease that leads to cognitive and behavioral impairment. Pathologically, AD is characterized by protein aggregates (amyloid plaques and neurofibrillary tangles) and neuronal cell death that is thought to initiate in the hippocampus and spread throughout the cortex. On the other hand, the cerebellum often does not display any of those hallmarks, at least until much later in the progression of the disease. Lovell and colleagues found that the levels of several oxidized bases in AD brains were significantly higher in frontal, parietal, and temporal lobes compared to control subjects and that mtDNA had approximately 10-fold higher levels of oxidized bases than nuclear DNA. Moreover, DNA from temporal lobe showed the most oxidative damage, whereas cerebellum was only slightly affected in the AD brains, correlating with these regions being respectively the most and least affected in human AD brain [87]. This same group also measured oxidative damage in samples from patients suffering from Mild Cognitive Impairment (MCI), the earliest clinical manifestation of AD. Eight-hydroxy-guanine was significantly higher in MCI nuclear DNA from the frontal and temporal lobe and in mtDNA from the temporal lobe compared with age-matched control subjects, and statistically significant elevations of FapyA were also observed in mtDNA of MCI temporal, frontal and parietal lobes [88]. Because MCI is considered to be a transient stage between normal aging and dementia, these results indicate that accumulation of oxidative damage in the mtDNA is an early event in AD manifestation and may have a causative role.
The results discussed above raise the question of whether this accumulation of oxidative DNA damage in AD patients is the result of enhanced damage formation or decreased repair. We addressed this question by looking at BER activities in short post-mortem brain samples from AD, MCI and age-matched control subjects. Using in vitro assays we analyzed each step of BER independently, as well as the process as a whole, and found significant changes in both AD and MCI samples [89]. We observed lower activity and protein levels of UDG, OGG1 and DNA polymerase β, as well as total BER, not only in an affected brain region (cortex) but, also in the least affected region (cerebellum). Interestingly, in MCI subjects there was an inverse correlation between total BER activity and the Braak stage, a quantification of the number of neurofibrillary tangles. Since neurofibrillary tangle pathology in AD is associated with cognitive decline [90], these findings suggest a link between BER capacity and the degree of neurological impairment, as measured by the Braak stage.
A role for oxidative DNA damage in PD pathology is much less well documented. Nonetheless, Zhang and colleagues observed increased cytoplasmic 8-oxoG immunoreactivity, indicating increased levels of this oxidized base in the mtDNA and/or in cytosolic RNA [91].
Accumulation of mutations and deletions of the mtDNA have also been associated with degenerative diseases, particularly AD and PD. However, the significance of these associations is still unclear [92]. Lin and colleagues [93] were the first to report on the association of one particular mtDNA mutation, in this case at codon 331 of NADH dehydrogenase 2, with AD, even though this finding was shortly thereafter contested [94, 95]. Since then, several reports have associated mtDNA mutations [96–100] and deletions [101–103] with AD. On the other hand, other studies found none or inconclusive associations between mtDNA instability and neurodegenerative diseases [104–108]. This conflicting range of results has led to the speculation that this variability may be due to the misleading amplification of ancient mtDNA sequences present in the nuclear DNA [109] or to great mosaicism of mtDNA among single cells in the brain [110].
While several mutations in genes with yet unclear function (alpha-synuclein, parkin, DJ-1, PINK-1 and LRRK2) have been associated with PD, the vast majority of cases are sporadic, without any relevant family history [111]. Mutations in the mtDNA have also been found in brains from PD patients [112–116], however recent results suggest that mtDNA deletions play a more significant role in the pathology of the disease [117]. Two studies published in parallel [118, 119] report very high levels of deleted mtDNA genomes in the substantia nigra region of brains from PD patients. These deletions are clonally expanded and are particularly enriched in cytochrome oxidase-negative cells, indicating that they may be directly responsible for impaired cellular respiration.
While the findings discussed above are correlative, and do not address the issue of causation directly, some results obtained with transgenic mouse models lend further support for a role of mtDNA damage accumulation in neuronal loss. For instance, alpha-synuclein (the primary component of Lewy bodies, which are seen in the brains of patients suffering from several neurodegenerations [120]) mutations have been linked to familial PD and transgenic mice expressing human mutated genes develop a severe motor disorder (for review, see [121]). Interestingly, in one of these mouse models (the A53T mutant), the mitochondria in a subset of neurons from the neocortex, brainstem and spinal cord ventral horn were positive for deoxynucleotidyl transferase-mediated UTP nick end labeling, indicating accumulation of mtDNA damage [122]. Using another approach, mice with a conditional knockout of the mitochondrial transcription factor A (TFAM) in dopaminergic (DA) neurons show reduced mtDNA expression and respiratory chain deficiency in midbrain DA neurons, which, in turn, lead to parkinsonism with adult onset of slow, progressive impairment of motor function accompanied by formation of intraneuronal inclusions and dopamine nerve cell death [123].
Changes in BER with age and in neurodegeneration
The “Mitochondrial Theory of Aging” has been extensively discussed lately, with conflicting observations emerging. Two independent groups [124, 125] have generated transgenic mice expressing a mutated, proof-reading deficient POLγ. These animals exhibit severely elevated levels of mtDNA mutations and deletions and a premature aging phenotype that is strikingly similar to normal aging. These observations were initially hailed as “definitive proof” that mtDNA mutations cause aging. However, further investigation found that animals heterozygous for the mutated POLγ accumulate nearly as many mutations as homozygous littermates, but show no clear aging phenotypes, thus dissociating the mutational load from the premature aging outcome [126]. One implication of this theory is that the age-associated accumulation of mtDNA damage and mutations leads to increased oxidative stress because of faulty expression of mitochondrial genes that encode for components of the mitochondrial respiratory complexes. There is a great deal of experimental evidence that age is associated with increased mitochondrial generation of ROS [127], but, again, the recent observation that the mitochondrial POLγ mutant mice do not show enhanced oxidative stress [128], despite the elevated mutations, casts doubt on the direct relationship between these events. Nonetheless, oxidative damage in the mtDNA has been shown to increase significantly with age and yeast require BER activities to attain a full chronological lifespan, implying that accumulation of damage hinders life span [129].
Since the first demonstration by Hart & Setlow that DNA repair activities are inversely correlated to life span in mammals [130], the general consensus had been that DNA repair capacity decreases with age. However, we demonstrated that OGG1 activity rather increases with age in liver and heart mitochondria and does not change significantly in the nuclear extracts of rats [131] and mice [132]. Interestingly, the rodent mtUDG activity did not change with age in either compartment, indicating that the increased capacity to repair 8-oxodG does not reflect a general up-regulation of DNA repair in mitochondria with age in these tissues. Thus, a steady-state accumulation of 8-oxoG in mtDNA in mouse liver and heart is therefore likely to be caused by an increased rate of damage formation, which exceeds the mtDNA repair capacity. The induction of the mitochondrial OGG1 activity may represent a cellular response in an attempt to counteract the increased DNA damage formation. Interestingly, Szczesny and Mitra demonstrated that mouse hepatocyte AP endonuclease activity is increased in nuclei as well as in mitochondria with age, whereas the total APE1 activity/level was unchanged [133].
A high level of oxidative damage can be particularly deleterious in post-mitotic tissue, such as the heart and the brain, because they cannot self-renew through cell proliferation. When nuclear and mitochondrial BER activities were evaluated in five distinct mouse brain regions, a significant age-dependent decrease in mitochondrial OGG1, UDG and NTH1 activity was observed for all regions [134]. Another study of rat cerebral cortices showed that mitochondrial BER activities gradually declined with age, and this decline in activity parallels decreased expression of repair enzymes, such as OGG1 and POLγ [135].
Together these results point to a very important aspect in aging research, which is that there is a great degree of variability among different tissues in an organism. While the implication that different cell types will show specific responses to DNA damage must be appreciated, these results also suggest that changes in mitochondrial BER with age may contribute to the accumulation of DNA damage, and the increase in oxidative damage to mtDNA might contribute to age-related decline, particularly in the brain.
Only a few studies have been performed to address age-associated changes in BER in humans. However, using a biotin-containing aldehyde-reactive probe for measuring AP sites in living cells, Ames and co-workers have shown that leukocytes from old human donors possess reduced AP site repair and a reduced activity of DNA glycosylases that remove methylated bases [136]. Moreover, cultured human fibroblasts showed decreased BER activity both in the nucleus and mitochondria with higher passage number [137], suggesting that senescence is also associated with decreased BER. However, in vitro cellular senescence may not fully emulate the molecular changes that take place in vivo, and this issue still needs to be further addressed using human samples.
Genetically inherited premature aging syndromes are very valuable tools when aiming to understand the normal aging process in humans. Cell lines from patients suffering from such syndromes have been established and isogenic cell lines with the functional gene transfected back into the deficient line are employed as controls. In the human disorder Cockayne syndrome, complementation group B (CSB), which is characterized by segmental premature aging and neurological dysfunction, there appears to be a deficiency in 8-oxoG repair. In particular, whole cell extracts from CSB-deficient cells display reduced 8-oxoG incision [138] and CSB mutant cells exhibit impaired in vivo repair of oxidative damage in nuclear genes [139]. More recently, we reported that the mitochondrial repair of 8-oxoG is also deficient in CSB-deficient cells [140]. While this deficiency in repairing oxidative damage, especially from mtDNA, may be a major underlying cause of the disease, the precise molecular role of CSB in the BER pathway is, however, still unclear. Based on identified interaction partners of the CSB protein, it has been proposed that CSB is an auxiliary factor in BER [141].
Work from several groups has proposed that changes in BER play a causative role in neurodegeneration. Iida and colleagues found changes in expression of BER enzymes, including the mitochondrial β-OGG1 in neuronal cytoplasm and reduced Polβ in midtemporal cortex samples of affected AD tissue [142]. Lower OGG1 activity was also reported for nuclear lysates from affected human AD brain regions using a sodium borohydride trapping assay, which, however, only measures the robustness of the AP lyase activity of OGG1 [143]. Moreover, the same group recently reported that mutations in the OGG1 gene that affect its activity are found preferentially in AD patients [144].
The results discussed here provide strong support to the hypothesis that mtDNA instability contributes to neurodegeneration. Our results showing lower BER activity in AD brains further suggest that lower BER may be a predisposing factor in the development of AD. Together, these shed some light in the sequence of events and factors that may be involved in the complex cascade that leads to neurodegeneration, and hopefully contribute to the development of risk assessment tools as well as preventive drug therapy.
Acknowledgements
We thank Drs. Maria Aamann and Dharmendra Singh for critical reading of this manuscript. Part of the work reviewed here was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Cummings JL, Jeste DV. Alzheimer’s disease and its management in the year 2010. Psychiatr. Serv. 1999;50:1173–1177. doi: 10.1176/ps.50.9.1173. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DM, III, Mattson MP. Neurodegeneration: nicked to death. Curr. Biol. 2007;17:R55–R58. doi: 10.1016/j.cub.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 4.Vina J, Borras C, Miquel J. Theories of ageing. IUBMB. Life. 2007;59:249–254. doi: 10.1080/15216540601178067. [DOI] [PubMed] [Google Scholar]
- 5.McFarland R, Taylor RW, Turnbull DM. Mitochondrial disease--its impact, etiology, and pathology. Curr. Top. Dev. Biol. 2007;77:113–155. doi: 10.1016/S0070-2153(06)77005-3. [DOI] [PubMed] [Google Scholar]
- 6.Wong LJ. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion. 2007;7:45–52. doi: 10.1016/j.mito.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Morava E, van den HL, Hol F, de Vries MC, Hogeveen M, Rodenburg RJ, Smeitink JA. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 8.Greenamyre JT, Sherer TB, Betarbet R, Panov AV. Complex I and Parkinson’s disease. IUBMB. Life. 2001;52:135–141. doi: 10.1080/15216540152845939. [DOI] [PubMed] [Google Scholar]
- 9.Schapira AH. Mitochondrial dysfunction in Parkinson’s disease. Cell Death. Differ. 2007;14:1261–1266. doi: 10.1038/sj.cdd.4402160. [DOI] [PubMed] [Google Scholar]
- 10.Browne SE, Beal MF. Toxin-induced mitochondrial dysfunction. Int. Rev. Neurobiol. 2002;53:243–279. doi: 10.1016/s0074-7742(02)53010-5. [DOI] [PubMed] [Google Scholar]
- 11.Davis GC, Williams AC, Markey SP, Ebert MH, Caine ED, Reichert CM, Kopin IJ. Chronic Parkinsonism secondary to intravenous injection of meperidine analogues. Psychiatry Res. 1979;1:249–254. doi: 10.1016/0165-1781(79)90006-4. [DOI] [PubMed] [Google Scholar]
- 12.Chiba K, Trevor A, Castagnoli N., Jr Metabolism of the neurotoxic tertiary amine, MPTP, by brain monoamine oxidase. Biochem. Biophys. Res. Commun. 1984;120:574–578. doi: 10.1016/0006-291x(84)91293-2. [DOI] [PubMed] [Google Scholar]
- 13.Nicklas WJ, Vyas I, Heikkila RE. Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenyl-pyridine, a metabolite of the neurotoxin, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci. 1985;36:2503–2508. doi: 10.1016/0024-3205(85)90146-8. [DOI] [PubMed] [Google Scholar]
- 14.Heikkila RE, Nicklas WJ, Vyas I, Duvoisin RC. Dopaminergic toxicity of rotenone and the 1-methyl-4-phenylpyridinium ion after their stereotaxic administration to rats: implication for the mechanism of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine toxicity. Neurosci. Lett. 1985;62:389–394. doi: 10.1016/0304-3940(85)90580-4. [DOI] [PubMed] [Google Scholar]
- 15.Ramsay RR, Salach JI, Dadgar J, Singer TP. Inhibition of mitochondrial NADH dehydrogenase by pyridine derivatives and its possible relation to experimental and idiopathic parkinsonism. Biochem. Biophys. Res. Commun. 1986;135:269–275. doi: 10.1016/0006-291x(86)90972-1. [DOI] [PubMed] [Google Scholar]
- 16.Cosi C, Colpaert F, Koek W, Degryse A, Marien M. Poly(ADP-ribose) polymerase inhibitors protect against MPTP-induced depletions of striatal dopamine and cortical noradrenaline in C57B1/6 mice. Brain Res. 1996;729:264–269. [PubMed] [Google Scholar]
- 17.Cosi C, Marien M. Implication of poly (ADP-ribose) polymerase (PARP) in neurodegeneration brain energy metabolism Decreases in mouse brain NAD+ and ATP caused by MPTP are prevented by the PARP inhibitor benzamide. Ann. N. Y. Acad. Sci. 1999;890:227–239. doi: 10.1111/j.1749-6632.1999.tb07998.x. [DOI] [PubMed] [Google Scholar]
- 18.Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem. Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 19.Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann. Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 20.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. U. S. A. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 23.Ward JF. Biochemistry of DNA lesions. Radiat. Res. Suppl. 1985;8:S103–S111. [PubMed] [Google Scholar]
- 24.Cadenas E. Biochemistry of oxygen toxicity. Annu. Rev. Biochem. 1989;58:79–110. doi: 10.1146/annurev.bi.58.070189.000455. [DOI] [PubMed] [Google Scholar]
- 25.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic. Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 27.Mori T, Hori Y, Dizdaroglu M. DNA base damage generated in vivo in hepatic chromatin of mice upon whole body gamma-irradiation. Int. J. Radiat. Biol. 1993;64:645–650. doi: 10.1080/09553009314551881. [DOI] [PubMed] [Google Scholar]
- 28.Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 29.Kohda K, Tada M, Kasai H, Nishimura S, Kawazoe Y. Formation of 8-hydroxyguanine residues in cellular DNA exposed to the carcinogen 4-nitroquinoline 1-oxide. Biochem Biophys Res Commun. 1986;139:626–632. doi: 10.1016/s0006-291x(86)80036-5. [DOI] [PubMed] [Google Scholar]
- 30.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 31.Pinz KG, Shibutani S, Bogenhagen DF. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J Biol Chem. 1995;270:9202–9206. doi: 10.1074/jbc.270.16.9202. [DOI] [PubMed] [Google Scholar]
- 32.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 33.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuart JA, Bourque BM, Souza-Pinto NC, Bohr VA. No evidence of mitochondrial respiratory dysfunction in OGG1-null mice deficient in removal of 8-oxodeoxyguanine from mitochondrial DNA. Free Radic. Biol. Med. 2005;38:737–745. doi: 10.1016/j.freeradbiomed.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 36.Wirtz M, Schumann CA, Schellentrager M, Gab S, Vom BJ, Podeschwa MA, Altenbach HJ, Oscier D, Schmitz OJ. Capillary electrophoresis-laser induced fluorescence analysis of endogenous damage in mitochondrial and genomic DNA. Electrophoresis. 2005;26:2599–2607. doi: 10.1002/elps.200410397. [DOI] [PubMed] [Google Scholar]
- 37.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic. Acids. Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J, Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1 and NEIL1 enzymes. J. Biol. Chem. 2005 doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 39.Wiederholt CJ, Greenberg MM. Fapy.dG instructs Klenow exo(−) to misincorporate deoxyadenosine. J. Am. Chem. Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 40.Delaney MO, Wiederholt CJ, Greenberg MM. Fapy.dA induces nucleotide misincorporation translesionally by a DNA polymerase. Angew. Chem. Int. Ed Engl. 2002;41:771–773. doi: 10.1002/1521-3773(20020301)41:5<771::aid-anie771>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 41.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higuchi YLS. Purification of all forms of HeLa cell mitochondrial DNA and assessment of damage to it caused by hydrogen peroxide treatment of mitochondria or cells. J. Biol. Chem. 1995;270:7950–7956. doi: 10.1074/jbc.270.14.7950. [DOI] [PubMed] [Google Scholar]
- 43.Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29:2117–2126. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- 45.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 46.Hashiguchi K, Bohr VA, Souza-Pinto NC. Oxidative stress and mitochondrial DNA repair: implications for NRTIs induced DNA damage. Mitochondrion. 2004;4:215–222. doi: 10.1016/j.mito.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashiguchi K, Stuart JA, Souza-Pinto NC, Bohr VA. The C-terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–5608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 53.Jensen A, Calvayrac G, Karahalil B, Bohr VA, Stevnsner T. Mammalian 8-oxoguanine DNA glycosylase 1 incises 8-oxoadenine opposite cytosine in nuclei and mitochondria, while a different glycosylase incises 8-oxoadenine opposite guanine in nuclei. J. Biol. Chem. 2003;278:19541–19548. doi: 10.1074/jbc.M301504200. [DOI] [PubMed] [Google Scholar]
- 54.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luna L, Bjoras M, Hoff E, Rognes T, Seeberg E. Cell-cycle regulation, intracellular sorting and induced overexpression of the human NTH1 DNA glycosylase involved in removal of formamidopyrimidine residues from DNA. Mutat. Res. 2000;460:95–104. doi: 10.1016/s0921-8777(00)00015-x. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda S, Kohmoto T, Tabata R, Seki Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA Repair (Amst) 2002;1:847–854. doi: 10.1016/s1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- 59.Karahalil B, Souza-Pinto NC, Parsons JL, Elder RH, Bohr VA. Compromised incision of oxidized pyrimidines in liver mitochondria of mice deficient in NTH1 and OGG1 glycosylases. J. Biol. Chem. 2003;278:33701–33707. doi: 10.1074/jbc.M301617200. [DOI] [PubMed] [Google Scholar]
- 60.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 62.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 63.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 65.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;21:2579–2584. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dizdaroglu M, Karakaya A, Jaruga P, Slupphaug G, Krokan HE. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Izumi T, Tatsuka M, Tano K, Asano M, Mitra S. Molecular cloning and characterization of the promoter of the human N-methylpurine-DNA glycosylase (MPG) gene. Carcinogenesis. 1997;18:1837–1839. doi: 10.1093/carcin/18.9.1837. [DOI] [PubMed] [Google Scholar]
- 68.Satoh MS, Huh N, Rajewsky MF, Kuroki T. Enzymatic removal of O6-ethylguanine from mitochondrial DNA in rat tissues exposed to N-ethyl-N-nitrosourea in vivo. J Biol Chem. 1988;263:6854–6856. [PubMed] [Google Scholar]
- 69.Myers KA, Saffhill R, O Connor PJ. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988;9:285–292. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- 70.Pettepher CC, LeDoux SP, Bohr VA, Wilson GL. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J. Biol. Chem. 1991;266:3113–3117. [PubMed] [Google Scholar]
- 71.LeDoux SP, Patton NJ, Avery LJ, Wilson GL. Repair of N-methylpurines in the mitochondrial DNA of xeroderma pigmentosum complementation group D cells. Carcinogenesis. 1993;14:913–917. doi: 10.1093/carcin/14.5.913. [DOI] [PubMed] [Google Scholar]
- 72.Jackson EB, Theriot CA, Chattopadhyay R, Mitra S, Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1) Nucleic Acids Res. 2005;33:3303–3312. doi: 10.1093/nar/gki641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomkinson AE, Bonk RT, Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- 74.Tell G, Crivellato E, Pines A, Paron I, Pucillo C, Manzini G, Bandiera A, Kelley MR, Di Loreto C, Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat. Res. 2001;485:143–152. doi: 10.1016/s0921-8777(00)00068-9. [DOI] [PubMed] [Google Scholar]
- 75.Frossi B, Tell G, Spessotto P, Colombatti A, Vitale G, Pucillo C. H(2)O(2) induces translocation of APE/Ref-1 to mitochondria in the Raji B-cell line. J. Cell Physiol. 2002;193:180–186. doi: 10.1002/jcp.10159. [DOI] [PubMed] [Google Scholar]
- 76.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amst) 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 79.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 80.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 81.Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39:1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- 82.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5’-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro [In Process Citation] Proc Natl Acad Sci U S A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pinz KG, Bogenhagen DF. The influence of the DNA polymerase gamma accessory subunit on base excision repair by the catalytic subunit. DNA Repair (Amst) 2006;5:121–128. doi: 10.1016/j.dnarep.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 84.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutat. Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 87.Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J. Neurochem. 2005;93:953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 88.Wang J, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. J. Neurochem. 2006;96:825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- 89.Weissman L, Jo DG, Sorensen MM, Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bancher C, Braak H, Fischer P, Jellinger KA. Neuropathological staging of Alzheimer lesions and intellectual status in Alzheimer’s and Parkinson’s disease patients. Neurosci. Lett. 1993;162:179–182. doi: 10.1016/0304-3940(93)90590-h. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am. J. Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Howell N, Elson JL, Chinnery PF, Turnbull DM. mtDNA mutations and common neurodegenerative disorders. Trends Genet. 2005;21:583–586. doi: 10.1016/j.tig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 93.Lin FH, Lin R, Wisniewski HM, Hwang YW, Grundke-Iqbal I, Healy-Louie G, Iqbal K. Detection of point mutations in codon 331 of mitochondrial NADH dehydrogenase subunit 2 in Alzheimer’s brains. Biochem. Biophys. Res. Commun. 1992;182:238–246. doi: 10.1016/s0006-291x(05)80136-6. [DOI] [PubMed] [Google Scholar]
- 94.Petruzzella V, Chen X, Schon EA. Is a point mutation in the mitochondrial ND2 gene associated with Alzheimer’s disease. Biochem. Biophys. Res. Commun. 1992;186:491–497. doi: 10.1016/s0006-291x(05)80834-4. [DOI] [PubMed] [Google Scholar]
- 95.Kosel S, Egensperger R, Mehraein P, Graeber MB. No association of mutations at nucleotide 5460 of mitochondrial NADH dehydrogenase with Alzheimer’s disease. Biochem. Biophys. Res. Commun. 1994;203:745–749. doi: 10.1006/bbrc.1994.2245. [DOI] [PubMed] [Google Scholar]
- 96.Chang SW, Zhang D, Chung HD, Zassenhaus HP. The frequency of point mutations in mitochondrial DNA is elevated in the Alzheimer’s brain. Biochem. Biophys. Res. Commun. 2000;273:203–208. doi: 10.1006/bbrc.2000.2885. [DOI] [PubMed] [Google Scholar]
- 97.Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MF, Mirra SS, Beal MF, Yang CC, Gearing M, Salvo R. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 98.Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davis RE, Miller S, Herrnstadt C, Ghosh SS, Fahy E, Shinobu LA, Galasko D, Thal LJ, Beal MF, Howell N, Parker WD., Jr Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blanchard BJ, Park T, Fripp WJ, Lerman LS, Ingram VM. A mitochondrial DNA deletion in normally aging and in Alzheimer brain tissue. Neuroreport. 1993;4:799–802. doi: 10.1097/00001756-199306000-00051. [DOI] [PubMed] [Google Scholar]
- 102.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet Nature Genetics J1 - Nat Gen J2 - Nat. Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 103.Hamblet NS, Castora FJ. Elevated levels of the Kearns-Sayre syndrome mitochondrial DNA deletion in temporal cortex of Alzheimer’s patients. Mutat. Res. 1997;379:253–262. doi: 10.1016/s0027-5107(97)00158-9. [DOI] [PubMed] [Google Scholar]
- 104.Lezza AM, Mecocci P, Cormio A, Beal MF, Cherubini A, Cantatore P, Senin U, Gadaleta MN. Mitochondrial DNA 4977 bp deletion and OH8dG levels correlate in the brain of aged subjects but not Alzheimer’s disease patients. FASEB J. 1999;13:1083–1088. doi: 10.1096/fasebj.13.9.1083. [DOI] [PubMed] [Google Scholar]
- 105.Payami H, Hoffbuhr K. Lack of evidence for maternal effect in familial Alzheimer’s disease. Genet. Epidemiol. 1993;10:461–464. doi: 10.1002/gepi.1370100622. [DOI] [PubMed] [Google Scholar]
- 106.Janetzky B, Schmid C, Bischof F, Frolich L, Gsell W, Kalaria RN, Riederer P, Reichmann H. Investigations on the point mutations at nt 5460 of the mtDNA in different neurodegenerative and neuromuscular diseases. Eur. Neurol. 1996;36:149–153. doi: 10.1159/000117233. [DOI] [PubMed] [Google Scholar]
- 107.Zsurka G, Kalman J, Csaszar A, Rasko I, Janka Z, Venetianer P. No mitochondrial haplotype was found to increase risk for Alzheimer’s disease. Biol. Psychiatry. 1998;44:371–373. doi: 10.1016/s0006-3223(97)00461-7. [DOI] [PubMed] [Google Scholar]
- 108.Garcia-Lozano JR, Mir P, Alberca R, Aguilera I, Gil NE, Fernandez-Lopez O, Cayuela A, Nunez-Roldan A. Mitochondrial DNA A4336G mutation in Alzheimer’s and Parkinson’s diseases. Eur. Neurol. 2002;48:34–36. doi: 10.1159/000064955. [DOI] [PubMed] [Google Scholar]
- 109.Wallace DC, Stugard C, Murdock D, Schurr T, Brown MD. Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14900–14905. doi: 10.1073/pnas.94.26.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cantuti-Castelvetri I, Lin MT, Zheng K, Keller-McGandy CE, Betensky RA, Johns DR, Beal MF, Standaert DG, Simon DK. Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol. Aging. 2005;26:1343–1355. doi: 10.1016/j.neurobiolaging.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 111.Thomas B, Beal MF. Parkinson’s disease. Hum. Mol. Genet. 16 Spec No. 2007;2:R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 112.Ikebe S, Tanaka M, Ozawa T. Point mutations of mitochondrial genome in Parkinson’s disease. Brain Res. Mol. Brain Res. 1995;28:281–295. doi: 10.1016/0169-328x(94)00209-w. [DOI] [PubMed] [Google Scholar]
- 113.Ozawa T, Tanaka M, Ino H, Ohno K, Sano T, Wada Y, Yoneda M, Tanno Y, Miyatake T, Tanaka T. Distinct clustering of point mutations in mitochondrial DNA among patients with mitochondrial encephalomyopathies and with Parkinson’s disease. Biochem. Biophys. Res. Commun. 1991;176:938–946. doi: 10.1016/s0006-291x(05)80276-1. [DOI] [PubMed] [Google Scholar]
- 114.Mayr-Wohlfart U, Rodel G, Henneberg A. Mitochondrial tRNA(Gln) and tRNA(Thr) gene variants in Parkinson’s disease. Eur. J. Med. Res. 1997;2:111–113. [PubMed] [Google Scholar]
- 115.Autere J, Moilanen JS, Finnila S, Soininen H, Mannermaa A, Hartikainen P, Hallikainen M, Majamaa K. Mitochondrial DNA polymorphisms as risk factors for Parkinson’s disease and Parkinson’s disease dementia. Hum. Genet. 2004;115:29–35. doi: 10.1007/s00439-004-1123-9. [DOI] [PubMed] [Google Scholar]
- 116.Smigrodzki R, Parks J, Parker WD. High frequency of mitochondrial complex I mutations in Parkinson’s disease and aging. Neurobiol. Aging. 2004;25:1273–1281. doi: 10.1016/j.neurobiolaging.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 117.Manfredi G. mtDNA clock runs out for dopaminergic neurons. Nat. Genet. 2006;38:507–508. doi: 10.1038/ng0506-507. [DOI] [PubMed] [Google Scholar]
- 118.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 119.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 120.Ferman TJ, Boeve BF. Dementia with Lewy bodies. Neurol. Clin. 2007;25:741–760. vii. doi: 10.1016/j.ncl.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martin LJ. Transgenic mice with human mutant genes causing Parkinson’s disease and amyotrophic lateral sclerosis provide common insight into mechanisms of motor neuron selective vulnerability to degeneration. Rev. Neurosci. 2007;18:115–136. doi: 10.1515/revneuro.2007.18.2.115. [DOI] [PubMed] [Google Scholar]
- 122.Martin LJ, Pan Y, Price AC, Sterling W, Copeland NG, Jenkins NA, Price DL, Lee MK. Parkinson’s disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J. Neurosci. 2006;26:41–50. doi: 10.1523/JNEUROSCI.4308-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson FS, Trifunovic A, Hoffer B, Cullheim S, Mohammed AH, Olson L, Larsson NG. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 125.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 126.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 127.Orrenius S, Gogvadze V, Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annu. Rev. Pharmacol. Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 128.Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maclean MJ, Aamodt R, Harris N, Alseth I, Seeberg E, Bjoras M, Piper PW. Base excision repair activities required for yeast to attain a full chronological life span. Aging Cell. 2003;2:93–104. doi: 10.1046/j.1474-9728.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 130.Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision repair and life span in a number of mammalian species. Proc Natl Acad Sci USA Proceedings of the National Academy of Sciences of the United States of America J1 - Proceedings of the National Academy of Sciences USA J2 - PNAS. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Souza-Pinto NC, Croteau DL, Hudson EK, Hansford RG, Bohr VA. Age-associated increase in 8-oxo-deoxyguanosine glycosylase/AP lyase activity in rat mitochondria. Nucleic Acids. Res. 1999;27:1935–1942. doi: 10.1093/nar/27.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Souza-Pinto NC, Hogue BA, Bohr VA. DNA repair and aging in mouse liver: 8-oxodG glycosylase activity increase in mitochondrial but not in nuclear extracts. Free Radic. Biol. Med. 2001;30:916–923. doi: 10.1016/s0891-5849(01)00483-x. [DOI] [PubMed] [Google Scholar]
- 133.Szczesny B, Mitra S. Effect of aging on intracellular distribution of abasic (AP) endonuclease 1 in the mouse liver. Mech. Ageing Dev. 2005;126:1071–1078. doi: 10.1016/j.mad.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 134.Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner, Neurobiol. Aging. 2005 doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 135.Chen D, Cao G, Hastings T, Feng Y, Pei W, O’Horo C, Chen J. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J. Neurochem. 2002;81:1273–1284. doi: 10.1046/j.1471-4159.2002.00916.x. [DOI] [PubMed] [Google Scholar]
- 136.Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shen GP, Galick H, Inoue M, Wallace SS. Decline of nuclear and mitochondrial oxidative base excision repair activity in late passage human diploid fibroblasts. DNA Repair (Amst) 2003;2:673–693. doi: 10.1016/s1568-7864(03)00006-5. [DOI] [PubMed] [Google Scholar]
- 138.Dianov G, Bischoff C, Sunesen M, Bohr VA. Repair of 8-oxoguanine in DNA is deficient inCockayne syndrome group B cells. Nucleic Acids. Res. 1999;27:1365–1368. doi: 10.1093/nar/27.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sunesen M, Stevnsner T, Brosh RM, Jr, Dianov GL, Bohr VA. Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene. 2002;21:3571–3578. doi: 10.1038/sj.onc.1205443. [DOI] [PubMed] [Google Scholar]
- 140.Stevnsner T, Nyaga S, Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–8682. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- 141.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., III Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Iida T, Furuta A, Nishioka K, Nakabeppu Y, Iwaki T. Expression of 8-oxoguanine DNA glycosylase is reduced and associated with neurofibrillary tangles in Alzheimer’s disease brain. Acta Neuropathol. (Berl) 2002;103:20–25. doi: 10.1007/s004010100418. [DOI] [PubMed] [Google Scholar]
- 143.Lovell MA, Xie C, Markesbery WR. Decreased base excision repair and increased helicase activity in Alzheimer’s disease brain. Brain Res. 2000;855:116–123. doi: 10.1016/s0006-8993(99)02335-5. [DOI] [PubMed] [Google Scholar]
- 144.Mao G, Pan X, Zhu BB, Zhang Y, Yuan F, Huang J, Lovell MA, Lee MP, Markesbery WR, Li GM, Gu L. Identification and characterization of OGG1 mutations in patients with Alzheimer’s disease. Nucleic Acids Res. 2007;35:2759–2766. doi: 10.1093/nar/gkm189. [DOI] [PMC free article] [PubMed] [Google Scholar]