Abstract
O-GlcNAcylation is a dynamic protein modification which has been studied mainly in metazoans. We reported previously that an Arabidopsis thaliana O-GlcNAc transferase modifies at least two threonine residues of the Plum pox virus (PPV) capsid protein (CP). Now, six additional residues were shown to be involved in O-GlcNAc modification of PPV CP. CP O-GlcNAcylation was abolished in the PPV CP7-T/A mutant, in which seven threonines were mutated. PPV CP7-T/A infected Nicotiana clevelandii, Nicotiana benthamiana, and Prunus persica without noticeable defects. However, defects in infection of A. thaliana were readily apparent. In mixed infections of wild-type arabidopsis, the CP7-T/A mutant was out-competed by wild-type virus. These results indicate that CP O-GlcNAcylation has a major role in the infection process. O-GlcNAc modification may have a role in virion assembly and/or stability as the CP of PPV CP7-T/A was more sensitive to protease digestion than that of the wild-type virus.
Keywords: O-GlcNAcylation, Plum pox virus, Potyvirus, Capsid protein, Mass spectrometry
Introduction
In the sophisticated network of mechanisms that regulates gene expression and gene product function, a major role is played by a large number of post-translational modifications. Among them, glycosylation is one of the most varied, complex and widespread (Spiro, 2002). At least 41 different sugar–amino acid linkages are known. These glycopeptide bonds have been arranged in five distinct groups (Spiro, 2002). Whereas the most usual protein glycosylations add complex oligosaccharides to asparagine residues in the endoplasmic reticulum and the Golgi apparatus, O-GlcNAcylation is much simpler, involving modification of serine or threonine residues of nuclear and cytoplasmic proteins with a D-N-acetylglucosamine monosaccharide (Hart et al., 2011).
Although O-GlcNAcylation went unnoticed until the early 1980s, it has been revealed as an abundant post-translational modification that is involved in a broad array of cellular processes in of most organisms (Butkinaree et al., 2010; Hanover et al., 2010; Hart et al., 2011). O-GlcNAcylation is a dynamic process that is often linked to phosphorylation. O-GlcNAcylation is involved in sensing environmental signals and nutrient status to regulate signaling cascades. Defects in O-GlcNAcylation underlie several important diseases, especially diabetes and neurodegenerative disorders (Butkinaree et al., 2010; Hanover et al., 2010; Hart et al., 2011).
As of 2012, O-GlcNAcylation had been experimentally demonstrated for ~800 proteins of more than 80 organisms (http://cbsb.lombardi.georgetown.edu/hulab/OGAP.html, Wang et al., 2011). Information on ~400 O-GlcNAcylation sites identified in 172 proteins has been used to develop an O-GlcNAcylation site prediction system, but a well-defined sequence motif specific for O-GlcNAc-modified sites has not been found (Wang et al., 2011). O-GlcNAcylation is carried out in metazoans by a single and generally essential O-GlcNAc transferase (OGT) (Shafi et al., 2000). In Caenorhabditis elegans, while deletion of the unique OGT causes severe defect in metabolism, it is not lethal (Hanover et al., 2005). Two OGTs, SECRET AGENT (SEC) and SPINDLY (SPY) have been identified in Arabidopsis thaliana (Thornton et al., 1999; Hartweck et al., 2002). SEC and SPY are related to OGTs of animals and fungi, and bacteria (Olszewski et al., 2010). SEC and SPY have both unique and overlapping functions in A. thaliana (Olszewski et al., 2010). The fact that double mutants affected in both SEC and SPY are lethal, indicates that O-GlcNAc modification is essential in plants (Hartweck et al., 2002).
Very little is known about proteins targeted by plant OGTs. None of the 798 O-GlcNAcylated proteins experimentally validated deposited in the dbOGAP database (http://cbsb.lombardi.georgetown.edu/hulab/OGAP.html) is a plant protein. However, protein overlay assays and coimmunoprecipation experiments allowed Taoka et al. (2007) to establish that Nicotiana tabacum NON-CELL-AUTONOMOUS PATHWAY PROTEIN 1 and several non-cell autonomous proteins (NCAPs) of Cucurbita maxima were phosphorylated and O-GlcNAcylated. More detailed analysis of one NCAP, Cm-PP16-1 identified a serine residue that could be both phosphorylated and O-GlcNAc-modified (Taoka et al., 2007). Moreover, O-glycosylation with terminal GlcNAc modification has been described for nuclear pore complex proteins of tobacco, but, in contrast with typical O-GlcNAcylations, the modifications were oligosacharides rather than single O-GlcNAc residues (Heese-Peck et al., 1995; Heese-Peck and Raikhel, 1998). It is not known whether SEC- or SPY-type OGTs are involved in the glycosylation of these plant proteins.
The best characterized target of a plant OGT is the capsid protein (CP) of the potyvirus Plum pox virus (PPV), which is also phosphorylated (Fernández-Fernández et al., 2002). Genetic studies suggest that SEC but not SPY modifies PPV CP (Chen et al., 2005). Thr19 and Thr24 have been identified as two of the PPV CP residues that are O-GlcNAc-modified in virions purified from infected plants (Pérez et al., 2006) and O-GlcNAcylation of these two residues as well as three other residues, Thr41, Thr53, Ser65, by SEC was demonstrated using a heterologous Escherichia coli-based co-expression system (Scott et al., 2006; Kim et al., 2011). Several proteins from different animal and human viruses have been shown to be O-GlcNAcylated (Benko et al., 1988; Caillet-Boudin et al., 1989; Mullis et al., 1990; Gonzalez and Burrone, 1991; Whitford and Faulkner, 1992; Greis et al., 1994; Medina et al., 1998), but there is little information about the relevance of O-GlcNAc modification for viral infections. While O-GlcNAcylation by SEC was non-essential for PPV infection in A. thaliana, PPV titer and movement are reduced in SEC-deficient sec mutants (Chen et al., 2005). In this paper, we identify one serine and seven threonine residues that are modified or influence the modification of other residues, and demonstrate that mutating the seven threonine residues to alanine blocks PPV CP modification. Using the non-modifiable mutant, we demonstrate that O-GlcNAc modification of the PPV CP increases its infectivity. We also demonstrate that the non-modifiable mutant and wild-type PPV are similarly infectious to sec plants indicating that modification of host proteins has little or no role in the infection process. Finally, we present evidence that unmodified CP is more sensitive to proteases suggesting O-GlcNAc modification might affect virion structure.
Results
Mapping of the PPV CP O-GlcNAcylation sites occurring in Nicotiana clevelandii
Previously MALDI-TOF analysis of purified virions from the potyvirus PPV identified a tryptic peptide (aa 1–39) from the N-terminal region of the viral CP that was O-GlcNAcylated (Fernández-Fernández et al., 2002) (Fig. 1) and a mutagenesis analysis identified threonines T19 and T24 as the modified residues (Pérez et al., 2006). Mutational mapping using a heterologous E. coli system indicated that in addition to modifying T19 and T24 SEC also modified T41 and S43 (Scott et al., 2006). A new MALDI-TOF analysis of virions isolated from plants to identify additional O-GlcNAc modification sites detected a second O-GlcNAcylated tryptic peptide (aa 40–93) and suggested that at least four additional residues were modified by O-GlcNAcylation within the first 93 aa (Fig. 1). Since the modifications made by SEC in E. coli occurred in the first 43 aa, a mutational mapping approach was employed to determine if these sites are also modified in plants.
Fig. 1.
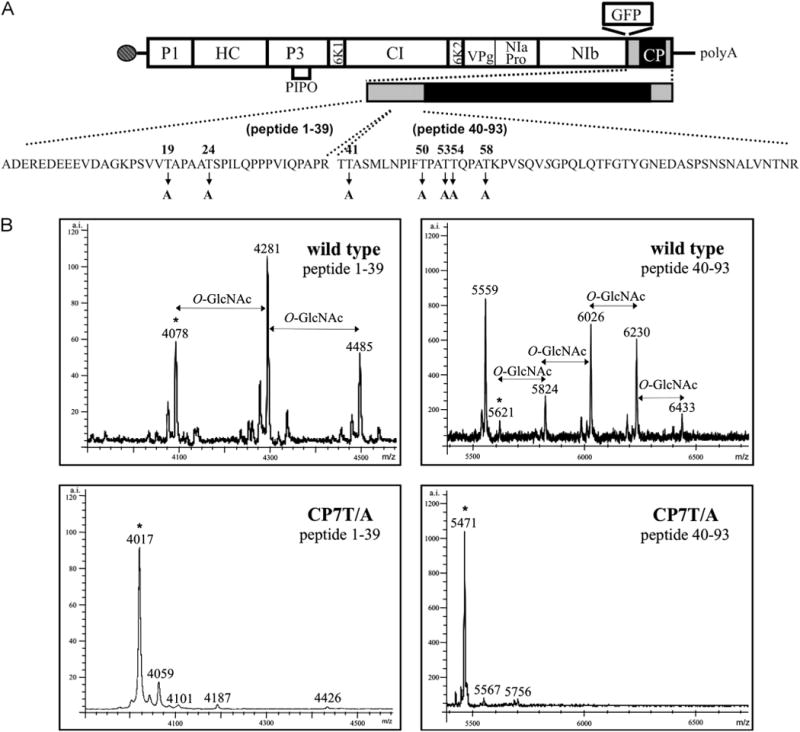
MALDI-TOF analysis of trypsin-digested virions of wild type PPV and the CP7T/A PPV mutant. (A) Localization of the tryptic peptides analyzed and the residues mutated in PPV-CP7T/A in the CP gene and in the genome of PPV. The different PPV proteins are shown in the box representing the PPV open reading frame. Dark gray and black boxes represent the N-and C-terminal regions and the core region, respectively, of PPV CP. The sequences of the peptides 1–39 and 40–93, with the mutations introduced in PPV-CP7T/A are shown below. Serine 65, which was shown to be O-GlcNAc-modified in an E. coli expression system (Kim et al., 2011), and in this study (Table 1) is in italics (B) MALDI-TOF spectra in the regions corresponding to the peptides 1–39 and 40–93. The mass/charge ratio (m/z, in Daltons) assigned to relevant peaks is indicated above them. The asterisks signal the peaks corresponding to the non-glycosylated 1–39 and 40–93 tryptic peptides. O-GlcNAc, O-GlcNAc modification; a.i., arbitrary intensity.
A series of mutations were engineered into the PPV infectious cDNA clones pICPPV-NK-lGFP (to infect N. clevelandii) and pICPPV-5′BDGFP (to infect Prunus persica). The first mutant (MG) had all Ser and Thr residues in the first 43 aa of PPV CP replaced with Ala (Supplementary Fig. S1) and the remaining mutants restored Thr 40 (MG-T40), Thr 41 (MG-T41) or Ser 43 (MG-S43) individually to the MG mutant. MALDI-TOF analysis of virions purified from plants infected with the PPV mutants MG, MG-T40 and MG-S43 indicated that three sites of the 40–93 peptide were modified, whereas four sites were modified in the MG-T41 mutant (Supplementary Fig. S1). These results indicate that additional modification sites in the 40–93 peptide remain to be identified and strongly suggest that T41, but not T40 or S43, is O-GlcNAcylated.
While this work was in progress, Kim et al. (2011) showed by ETD MS/MS that T19, T24, T41, T53 and S65 were O-GlcNAcylated by SEC in E. coli. To ensure maximal coverage both the mutational approach and ETD MS/MS (Zhao et al., 2011) were used to map the CP O-GlcNAc modifications sites produced in plants. Threonine residues occurring in the first 93 aa not previously analyzed were individually replaced by Ala in the pICPPV-NK-lGFP full-length cDNA clone and the effects of these mutations on modification were determined. The T71A, T74A and T91A mutations did not reduce the number of O-GlcNAc modifications (Supplementary Fig. S2). In contrast, the T50A, T53A, T54A and T58A mutations reduced the number of modifications of the 40–93 peptide from four to two or three. The T50A and T53A mutations had the most drastic effect on the modification profile: in the MALDI-TOF spectrum the ion intensity peak corresponding to the di-O-GlcNAcylated form, the most abundant peak from wild type CP, was lower than that of the mono O-GlcNAcylated peaks in the spectra of CP-T50 and CP-T53 and the abundance of the triply-O-GlcNAcylated peptide peak was greatly reduced in CP-T50 and undetectable in CP-T53 (Supplementary Fig. S2). In contrast, the T54A and T58A mutations appear to affect only one O-GlcNAcylation event, since the peak of triply-O-GlcNAcylated form of the 40–93 peptide was clearly detectable (Supplementary Fig. S2). All together the mutational mapping experiments indicate that Thr 19, 24, 41, 50, 53, 54 and 58 are modified or influence modification at other sites.
When peptides obtained by digestion of wild type PPV virions with trypsin and LysC proteinases were analyzed by LC-MS/MS, peptides from the 1–39 and 40–93 regions with up to two and five O-GlcNAc moieties, respectively, were detected. Analysis of selected peptides by ETD MS/MS demonstrated that T19, T24, T41, T53, S65 and T54 and/or T58 are O-GlcNAcylated (Fig. 3 and Table 1).
Fig. 3.
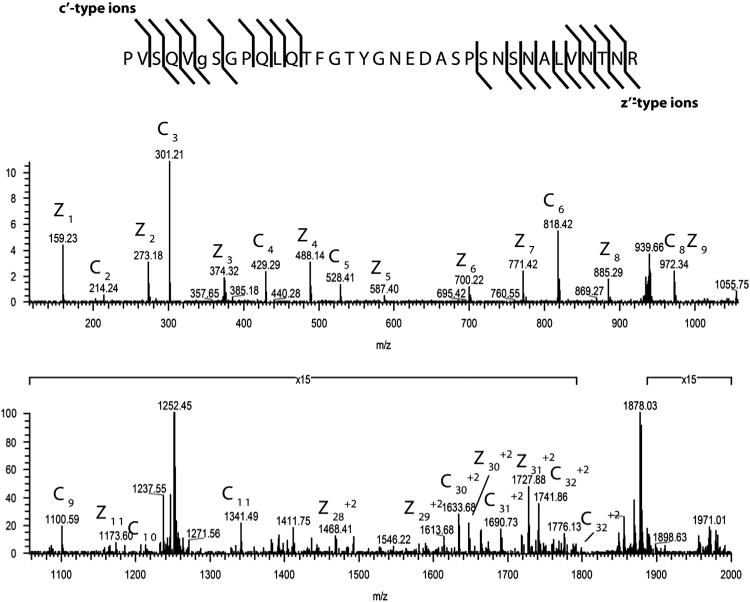
ETD MS/MS spectrum recorded on [M+4H]+4 ions (m/z 939.46) corresponding to O-GlcNAc-modified PPV-CP peptide PVSQVSGPQLQTFGTYGNEDASPSNSNALVNTNR. To acquire this spectrum, an ETD-enabled LTQ mass spectrometer was operated in the data dependent mode. Singly and doubly charged fragment ions that were observed and labeled in the spectrum indicate that the O-GlcNAc moieties are located on S65 of PPV.
Table 1.
Locations of O-GlcNAc modifications on PPV CP identified using ETD MS.
| Modification | Protein site | Modified peptide |
|---|---|---|
| GlcNAc | T19 | ADEREDEEEVDAGKPSVVT#APAATSPILQPPPVIQPAPR |
| GlcNAc | T24 | ADEREDEEEVDAGKPSVVTAPAAT#SPILQPPPVIQPAPR |
| 2GlcNAc | T19 & T24 | ADEREDEEEVDAGKPSVVT#APAAT#SPILQPPPVIQPAPR |
| GlcNAc | (T40/T41)^ | (TT)#^ASMLNPIFTPATTQPATK |
| 2GlcNAc | (T40/T41)^ & T53 | (TT)#^ASMLNPIFTPAT#TQPATK |
| GlcNAc | S65 | PVSQVS#GPQLQTFGTYGNEDASPSNSNALVNTNR |
| GlcNAc | T41 | TT#ASMLNPIFTPATTQPATKPVSQVSGPQLQTFGTYGNEDASPSNSNALVNTNR |
| 3GlcNAc | T41 & T53 & S65 | TT#ASMLNPIFTPAT#TQPATKPVSQVS#GPQLQTFGTYGNEDASPSNSNALVNTNR |
| 4GlcNAc | T41 & T53 & (T54/T58)^ & S65 | TT#ASMLNPIFTPAT#(TQPAT)#^KPVSQVS#GPQLQTFGTYGNEDASPSNSNALVNTNR |
=O-GlcNAcylated residue.
=data do not allow to discriminate which of two threonines is O-GlcNAcylated.
O-GlcNAcylation of CP differentially affects PPV infection of Nicotiana benthamiana, N. clevelandii, P. persica and A. thaliana
When the infectivity of all of the mutants used in the mapping studies was assessed, none of the mutations affected infectivity, pathogenicity or virus accumulation in N. clevelandii, and RT-PCR analysis showed that all mutations were stably maintained in the progeny virus (data not shown). Since none of these mutations completely block O-GlcNAc modification, it is possible that modification must be reduced further to cause a detectable defect in infection.
To further reduce O-GlcNAcylation of PPV CP, all threonines whose individual mutations had been shown to affect O-GlcNAcylation of PPV-CP (T19, T24, T41, T50, T53, T54 and T58) were replaced by alanines (CP7T/A mutation) in the pICPPV-NK-lGFP and pICPPV-5′BDGFP full-length cDNA clones. MALDI-TOF analysis of virions purified from N. clevelandii plants infected with PPV-CP7T/A showed that the seven T to A changes in the CP7T/A mutant abolished O-GlcNAcylation of both the 1–39 and 40–93 peptides (Fig. 1). Since S65 is still present but the CP is not modified, modification of this site must be dependent on primary sequence determinants altered in the CP7T/A mutant or requires O-GlcNAcylation of other CP residues.
The pICPPV-NK-lGFP-derived mutant (PPV-CP7T/A) infected N. clevelandii and N. benthamiana very efficiently, with no differences in symptoms or virus accumulation detectable between the plants infected with the mutant or wild type virus (Fig. 2A and B). The CP7T/A mutation did not to have a drastic effect on virus infection in P. persica: The infectivity and pathogenicity of PPV5′BD-CP7T/A in this host was similar to that of the wild type virus, and although the level of accumulation of the mutant virus was somewhat lower than that of wild type PPV5′BD, the differences were not statistically significant (P value 0.289) (Fig. 2C). RT-PCR analysis of N. clevelandii and P. persica infected plants showed that the CP7T/A was stably maintained in the progeny virus.
Fig. 2.
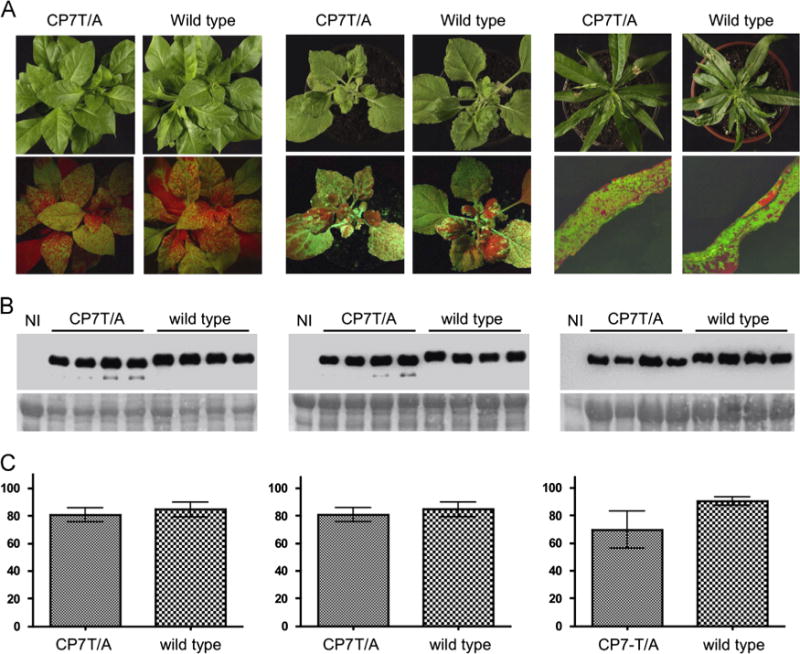
Infection of PPV-CP7T/A in N. clevelandii and N. benthamiana and of PPV-5′BD-CPT/A in P. persica. (A) Pictures of the plants infected with the indicated viruses, taken at 21 days post inoculation (dpi) under visible light (upper panels) or UV illumination (lower panels) with a hand lamp (N. clevelandii or N. benthamiana) and with an epifluorescence microscope (P. persica). (B) Western blot analysis of extracts of plants infected with the virus indicated above each lane, or of non-inoculated plants (NI), probed with antiserum to PPV CP. Samples were collected at 21 dpi. The blots stained with Ponceau red showing the Rubisco are included as loading controls. (C) Densitometric analysis of the Western blots shown in (B). Values are percentages of the density of the most intense band in the blot. Each bar shows the average value and the standard deviation of the four infected plants analyzed. Data of N. clevelandii, N. benthaniana and P. persica plants are shown in the left, central and right panels, respectively.
Since PPV infection in A. thaliana is compromised by mutations affecting SEC, A. thaliana Col-0 plants were biolistically inoculated with pICPPV-NK-lGFP wild type or the CP7T/A mutant. The infectivity of the two viruses was similar (100% and 90% for PPV wild type and CP7T/A, respectively, in two independent experiments), however, much lower virus accumulation was observed in the plants infected with the O-GlcNAcylation-deficient mutant, especially at early times of the infection (Fig. 4).
Fig. 4.
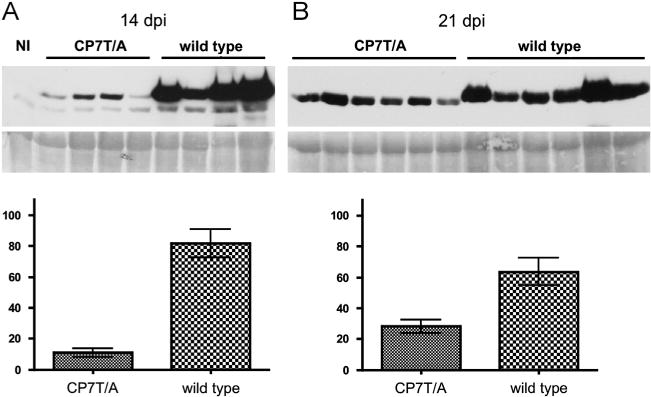
Effect of O-GlcNAcylation of CP on PPV accumulation in A. thaliana. Upper panels show a Western blot analysis of extracts of plants of A. thaliana Col-0 infected with the virus indicated above each lane, or of non-inoculated plants (NI), probed with antiserum to PPV CP. Samples were collected at 14 (A) or 21 (B) days post inoculation. The blots stained with Ponceau red showing the Rubisco are included as loading controls. Lower panels show the densitometric analysis of the Western blots. Values are percentages of the density of the most intense band in the blot. Each bar shows the average value and the standard deviation of the four or six infected plants analyzed in (A) or (B), respectively.
To further investigate the defects in infection by the PPV-CP7T/A mutant, competition experiments were conducted by coinoculating A. thaliana and N. clevelandii plants with mixtures of DNA of wild type pICPPV-NK-lGFP and a slightly higher amount of pICPPV-NK-lGFP CP7T/A. At 22 days post inoculation persistence of the competing viruses was assessed by immune capture and RT-PCR with primers external to the mutated region, followed by digestion at polymorphic restriction sites. In agreement with the similar infection efficiency of PPV wild type and CP7T/A in N. clevelandii, both viruses still coexisted in the four plants analyzed (Fig. 5A). In contrast, only wild type virus was detected in all coinfected A. thaliana Col-0 plants (Fig. 5B), confirming the lower fitness of the CP7T/A mutant in this host.
Fig. 5.
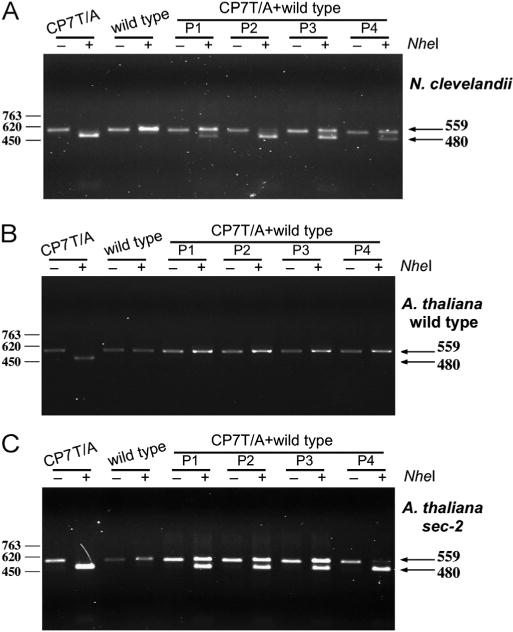
Competition of PPV wild type and CP7T/A in different host plants. N. clevelandii (A), A. thaliana Col-0 wild type (B), and A. thaliana Col-0 sec2 mutant (C) were biolistically inoculated with a 1.25:1 mixture of DNA of pICPPV-NK-lGFP and pICPPV-NK-lGFP-CP7T/A. Specific cDNA fragments were amplified by IC-RT-PCR from infected tissue of four plants (P1–P4) collected at 22 days post inoculation. The amplified fragments undigested, or digested with NheI, which leaves intact the wild type fragment (559 nt) but cleaves the CP7T/A fragment in two pieces (480 nt and 79 nt). The position of the fragments of 559 nt and 480 nt are signaled by arrows. Samples of plants singly infected with either PPV wild type or PPV-CP7T/A were also analyzed as controls. The size of fragments of DNA of the phage ø29 digested with HindIII used of size markers are shown at the left of the panels.
In order to verify that the infection defect of the CP7T/A mutant in A. thaliana is a consequence of its O-GlcNAcylation deficiency and not due to another effect of the seven T to A mutations, a competition experiment was conducted in the sec-2 mutant. In sec-2 plants, in which neither the wild type nor CP7T/A CP were O-GlcNAcylated, both viruses coexisted (Fig. 5C), suggesting that the seven T to A substitutions do not affect infection of A. thaliana per se.
O-GlcNAcylation deficiency increases the protease sensitivity of PPV CP
The impairment of infection due to mutations blocking CP O-GlcNAcylation could be due to defects in virion assembly or stability. Therefore, we tested the sensitivity of CP to plant proteases. Cell-free extracts prepared from plants infected with wild type PPV or CP7T/A were prepared and incubated at 24 °C for varying lengths of time and then CP intactness and abundance was assessed by Western blot analysis. Both wild type and CP7T/A CP were very stable in N. clevelandii and P. persica extracts, however, some degradation products accumulated to a greater extent in the CP7T/A sample (Supplementary Fig. S3). The effect of the lack of O-GlcNAc modification was much more noticeable with A. thaliana extracts. Wild type CP was very stable, but less than 30% of the CP7T/A CP remained intact after 60 min of incubation (Fig. 6A).
Fig. 6.
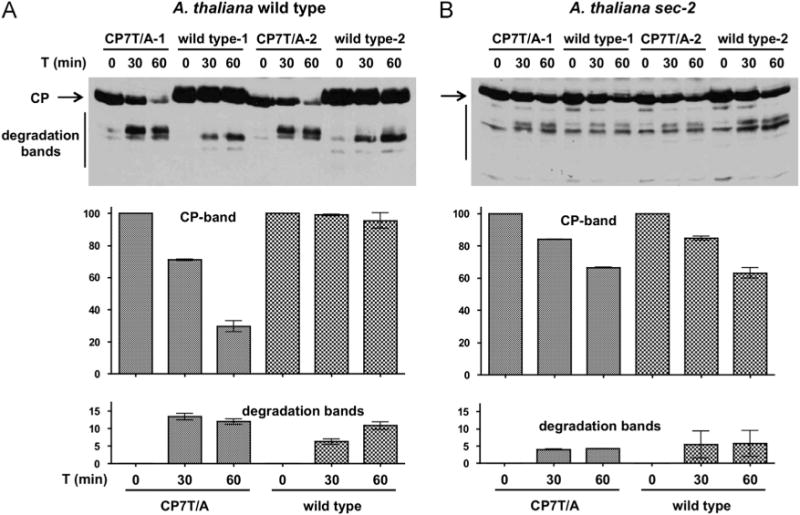
Effect of O-GlcNAcylation on CP stability in extracts of PPV-infected A. thaliana. Two pools of extracts of 3 plants of A. thaliana Col-0 wild type (A) or sec-2 (B) biolistically inoculated with PPV wild type or CP7T/A and collected at 21 days post inoculation were incubated at room temperature for the indicated times, and subjected to Western blot analysis with antiserum to PPV CP (top panels). Lower panels show the densitometric analysis of the CP band and the CP degradation bands of the Western blots. Values are percentages of the density of the CP band of each pool at time 0. Each bar shows the average and the standard deviation of the pools analyzed for each virus.
In order to differentiate between effects due to O-GlcNAcylation deficiency and other defects caused by the CP7T/A mutations, we studied the CP stability in extracts of the A. thaliana sec-2 mutant. In this mutant, the rates of degradation of wild type and CP7T/A CP were similar (Fig. 6B), indicating that in a plant unable to O-GlcNAcylate PPV CP, the T to A substitutions of CP7T/A do not have a noticeable effect on protein stability. Surprisingly, the decrease of CP stability caused by the sec mutation in the infected plant was less marked than that caused by the CP7T/A mutation in wild type plants (Fig. 6B), suggesting that sec mutation could produce additional stabilizing effects partially compensating for the destabilizing effects of the O-GlcNAcylation deficiency.
Discussion
Whereas the identification of O-GlcNAcylated proteins and O-GlcNAc modification sites in mammals has been rapidly growing during the last years, equivalent information from plants is still very limited (Olszewski et al., 2010; Hart et al., 2011; Wang et al., 2011). In this paper, we extend the information available about O-GlcNAcylation target sites in the CP of the plant potyvirus PPV. Moreover we demonstrate that impairment of PPV CP O-GlcNAcylation causes defects in virus infection and CP stability.
Although our analyses do not allow an accurate quantification of the level of O-GlcNAcylation of PPV CP, the rather high intensity of the signals corresponding to the peptides 1–39 and 40–93 modified with several O-GlcNAc residues in the MS spectra suggest that PPV CP is extensively, but not homogeneously, O-GlcNAcylated (Fig. 1 and Supplementary Fig. S1). In the PPV MG mutant all serines and threonines in the first 43 aa of CP have been replaced by alanines. The detection of three O-GlcNAcylated forms of the tryptic peptide 40–93 in the MALDI-TOF analysis of CP of virions purified from plants infected with this mutant, rather than the four O-GlcNAc-modified peptides detected for wild type CP (Supplementary Fig. S1), demonstrates that three O-GlcNAcylation target sites are located between the threonines 50 and 91, and strongly suggest that either T40, T41 or S43 is O-GlcNAc-modified. The fact that the restoration of T41 in the MG mutant background, but not those of T40 or S43, recovers the fourth glycosylation peak (Supplementary Fig. S1) supports the suggestion that T41 is the O-GlcNAcylation site. However, these results do not allow us formally rule out the possibility that the T41A mutation alters the protein structure and disturbs the O-GlcNAcylation of another position.
The T to A mutations at T71, T74 and T91 did not change the O-GlcNAcylation pattern of the 40–93 peptide (Supplementary Fig. S2), which demonstrates that these threonines are not modified. In contrast, mutating the threonines between positions 50 and 58 to alanine affected O-GlcNAcylation of PPV CP (Supplementary Fig. S2). These data do not allow us to discriminate whether these threonines are indeed O-GlcNAc modified or if they are important for modification of neighboring residues. However, ETD MS/MS analysis confirmed that, in addition to the previously identified O-GlcNAcylation sites at T19 and T24 (Pérez et al., 2006), the CP of virions is O-GlcNAc modified at T41, T53 and T54 and/or T58, which is in good agreement with the mutagenesis analysis. In addition, S65, which was not mutated, was also shown to be O-GlcNAc-modified. Since the ETD analysis detected five O-GlcNAc modifications on the 40–93 peptide, two residues, in addition to T41, T53 and S65 can be O-GlcNAcylated. Taking into account the results of the mutagenesis analysis the most likely possibilities are that both T54 and T58 or one of these residues and T50 are glycosylated. Since the T50A and T53A mutations disturbed two O-GlcNAc (Supplementary Fig. S2), some T to A mutations affect the O-GlcNAcylation of adjacent residues.
Our results are in good agreement with previous ETD MS/MS data showing O-GlcNAc-modification of PPV CP by the A. thaliana OGT SEC at T19, T24, T41, T53 and S65 in an E. coli expression system (Fig. 3 and Table 1). The coincidence of five O-GlcNAcylation sites in PPV CP expressed in infected plants and in E. coli, further supports the previously suggested conclusion that the specificity of SEC recognition does not depend on the cellular environment in which it takes place (Kim et al., 2011). The fact that the O-GlcNAc modifications at T50 and T54 or T58 were not identified in the ETD-MS/MS analysis of PPV CP O-GlcNAcylated in E. coli is probably due to the lower O-GlcNAcylation efficiency of this heterologous system (Kim et al., 2011). The mammalian OGT is known to be post-translationally modified and to form part of multicomponent complexes (Hart et al., 2011); if SEC behaves in a similar manner in plant cells, the weak O-GlcNAcylation of PPV CP detected in E. coli may be due to the absence of some accessory factors that increase the enzymatic activity. Although there is not a strict consensus sequence for O-GlcNAcylation sites, they appear to share some common sequence features, mainly the proximity of a proline residue and a serine/threonine-rich environment. A site prediction system, OGlcNAcScan, has been developed on the basis of a training data set of 373 experimental O-GlcNAcylation sites, mainly from mammalian proteins (Wang et al., 2011). OGlcNAcScan was able to identify, although with low scores, T19, T24 and T53, as putative O-GlcNAc modification sites, with a possible false positive prediction for S62. Taking into account the limitation of this prediction algorithm, the result suggests that modification site selection is similar for mammalian OGTs and SEC, as it has been previously suggested (Pérez et al., 2006; Kim et al., 2011).
Seven T to A mutations affecting O-GlcNAcylation were integrated in a multiple mutant, CP7T/A. We were unable to detect any O-GlcNAcylation of CP7T/A virion CP (Fig. 1) in spite of the fact that S65, which was found to be modified in wild type virions (Table 1), was not mutated. This further supports the remote effect of at least one of the threonine mutations (see above), however, we cannot discern between direct disturbance of the SEC recognition sequence and some requirement of O-GlcNAcylation at other position(s).
It has been reported that mutation of T19 and T24 to alanine does not impair PPV infection in N. clevelandii and P. persica (Pérez et al., 2006). The same result is observed now for the CP7T/A virus, which has seven T to A mutations and abolishes O-GlcNAc modification of PPV CP (Fig. 2). The small effect of O-GlcNAcylation of CP in PPV infection efficiency in N. clevelandii is further supported by the persistence of both wild type and CP7T/A PPV in a competition experiment where plants are co-infected with both viruses (Fig. 5A). In contrast, the efficiency of infection of the CP7T/A mutant and its competitiveness against wild type PPV is low in A. thaliana (Figs. 4 and 5B). This result suggests that the impairment in PPV infection of A. thaliana sec mutants that has been previously reported (Chen et al., 2005) is consequence of the lack of O-GlcNAcylation of the viral CP. This is further supported by the persistence of both wild type and CP7T/A PPV when competition experiments are performed using the sec mutant (Fig. 5C), which also suggests that the T to A mutations of CP7T/A do not directly confer a fitness cost. The specific selection of the wild type virus in the mixed infection of wild type A. thaliana also demonstrates that, although heteroencapsidation has been observed in potyviral infections (Varrelmann and Maiss, 2000), the CP7T/A mutant cannot be rescued by this process. We have previously reported the failure of heteroencapsidation to rescue a PPV CP mutation during a competition experiment between wild type PPV and the NAT mutant (Salvador et al., 2008). The lack of complementation of CP defects could be due to restrictions for several RNA molecules to initiate replication in the same cell, as it can be inferred from the observation that most primary infection foci in potyviral mixed infections derive from single genotypes (Zwart et al., 2011), or to some preference of the CP to interact with the RNA from which it was translated.
The absence of O-GlcNAcylation appears to cause some instability to PPV CP in the three plant species assayed, but the effect was greatest in A. thaliana, which could explain the reduced infection efficiency of PPV-CP7T/A in this host (Fig. 6A and supplementary Fig. S3). Although the in vitro behavior does not necessarily reproduce what happens in the infected cells, our results suggest an important role of O-GlcNAcylation on virions assembly or stability, or CP stability. The fact that the stability of wild type and CP7T/A CP are similar in extracts of SEC-deficient A. thaliana (Fig. 6B) suggests that it is the lack of O-GlcNAc modification, rather than the T to A mutations per se that directly or indirectly promotes CP instability. Interestingly, the stability of the CPs of PPV wild type and CP7T/A expressed in A. thaliana sec is higher than that of PPV CP7T/A expressed in A. thaliana wild type (Fig. 6B and C). This suggests that O-GlcNAcylation of unidentified viral or host factors may promote CP degradation. Further research is needed to unravel how O-GlcNAc modification affects virion structure/stability and/or CP stability and their relevance for the different steps of the reproductive cycle of PPV. Another unresolved question raised by this research is why the relative importance of O-GlcNAcylation on infection is different in different host species. While the assay of competition used here failed to detect fitness defects of PPV-CP7T/A in N. clevelandii (Fig. 5), CP stability was affected in all host examined (Figs. 6 and S3) and PPV-CP7T/A accumulation, appeared to be slightly reduced with respect to that of wild type PPV in N. clevelandii, N. benthamiana and P. persica (Fig. 2), suggesting that O-GlcNAcylation deficiency could cause defects in infection not only in A. thaliana, but in all PPV hosts. Since O-GlcNAcylation is influenced by metabolic status and protects against some stresses, it will be interesting to learn if the importance of O-GlcNAc modification in the infection process varies as the growth conditions of the host plant are varied.
Materials and methods
Virus infection and purification
Young plants of N. clevelandii, N. benthamiana and P. persica (4–6 leaf stage) and A. thaliana ecotype Columbia (Col-0) (wild-type or sec-2 mutant (Hartweck et al., 2002)), (8 leaf stage) were inoculated by bombardment with microgold particles coated with cDNA of pICPPV-NK-lGFP- or pIC-PPV-5′BDGFP-derived plasmids using a Helios gene gun device (Bio-Rad) (López-Moya and García, 2000). Microcarrier cartridges were prepared from two different clones per construct, with 1.0-μm gold particles coated at a DNA loading ratio of 2 μg/mg gold and a microcarrier loading amount of 0.5 mg/shooting. Helium pressures of 7.5 bar and 10 bar were used for inoculations of herbaceous plants and P. persica seedlings, respectively. For Nicotiana or P. persica plants, one or two cartridges were each shot twice onto two leaves of each plant. For A. thaliana one cartridge was shot twice onto two groups of three leaves.
N. clevelandii, N. benthamiana and P. persica plants were maintained in a glasshouse with 16 h of light by supplementary illumination and kept between 19 and 24 °C. A. thaliana plants were maintained in a climate-controlled chamber with 14 h of light and 22 °C.
PPV-NK-lGFP-derived viruses were purified from infected N. clevelandii plants according to Laín et al. (1988).
PPV mutagenesis
Mutations affecting putative O-GlcNAcylation target sites at the N-terminal region of PPV CP were engineered in PPV full-length cDNA clones suited for the infection of herbaceous plants (pICPPV-NK-lGFP) or P. persica seedlings (pIC-PPV5′BDGFP). pICPPV-NK-lGFP, also named pICPPV-NK-GFPn, derives from the cDNA clone of PPV-Rankovic genome pICPPV-NK (Fernández-Fernández et al., 2001) with the GFP gene from pTXS.GFP (Baulcombe et al., 1995) inserted between their NaeI and KpnI sites (P. Sáenz, M.R. Fernández-Fernández and J.A.G., unpublished results). pICPPV-5′BDGFP is a chimeric clone derived from pICPPV-NK-GFP (Fernández-Fernández et al., 2001) and cDNA from a D-type PPV isolate that infects very efficiently Prunus plants (Salvador et al., 2008).
As a first step to introduce the mutations MG, MG-T40, MG-T41 and MG-S43 (Scott et al., 2006) in the full-length cDNA clones (Supplementary Fig. S1), CP mutated cDNA was amplified from pp4-2MG, pp4-2MG-T40, pp4-2MG-T41 and pp4-2MG-S43 (Scott et al., 2006) by PCR using as primers ETGFP-1 forward (5′-GTGCACCAAGCTGACGAAAGAGAAGAC-3′) and ETGFP-2 reverse (5′-CACCTGTGAAACTGGTTTTGTTGC-3′). The products of the first PCRs together with an EcoRV-BpiI fragment of pICPPV-NK-1GFP were used as template for a second round of PCRs with Oligo 272 (5′-TTTAACGATGATGGTG-3′) and ETGFP-2 reverse as primers. The products of the second PCRs together with a BpiI-Bsh13651 fragment of pICPPV-NK-1GFP were used as template for new PCRs with Oligo 272 and Oligo 55 (5′-CTATGCACCAAACC-3′). The products of the last PCR were digested with KpnI and SacI and used to replace the corresponding fragments from pICPPV-NK-1GFP and pICPPV-5′BDGFP.
Point mutations T50A, T53A, T54A, T58A, T71A, T74A and T91A (Fig. 1) were created by a two-steps PCR-based mutagenesis method (Herlitze and Koenen, 1990), using as mutator oligodeox-inucleotides Oligo T50A (5′-GCTGGCGCGAAAATGGG-3′), T53A (5′-GAGTTGCTGCTGGCGTG-3′), T54A (5′-TTGAGCTGTTGCTGGCGT-3′), T58A (5′-GTTTTGCTGCTGGTTGAG-3′), T71A (5′-CAAAAGCTTGCAGTTGAGG-3′), T74A (5′-CCATATGCTCCAAAAGTTTG-3′) and T91A (5′-TGTTTGCGTTGACTAGCG-3′), and as flanking primers Oligo 80 (5′-TTGGGTTCTTGAACAAGC-3′) and Oligo 55. First PCRs used pICPPV-NK as template, and their DNA products together with a BsaBI-EcoRV fragment of pICPPV-NK were the template for the second PCRs. The products of these second PCRs were digested with KpnI and SacI and used to replace the corresponding fragment from pICPPV-NK-1GFP.
The mutagenesis to create the mutant CP7T/A, in which Thr-19, Thr-24, Thr-41, Thr-50, Thr-53, Thr-54 and Thr-58 from PPV CP (Fig. 1) were replaced by alanines, was performed by gene synthesis (Genscript Corporation Piscatway, NJ, USA) of the PPV cDNA fragment corresponding to the 9482-9953 region of the PPV genome, which was supplied with flanking KpnI and SacI restriction sites and cloned into pUC57. The KpnI and SacI fragment of the resulting plasmid was used to replace the corresponding fragments from pICPPV-NK-1GFP and pICPPV-5′BDGFP.
MALDI-TOF analysis of PPV CP
Purified PPV virions (5 μg) were digested with 50 ng of Modified Porcine Trypsin (Promega) at room temperature for 10 min and purified as previously described (Chen et al., 2005).
About 0.7 μl of matrix solution were deposited onto a 600 μm AnchorChipTM MALDI target (Bruker Daltonics) and allowed to dry at room temperature. Then, 0.7 μl of the tryptic peptide mixture were applied to the plate, and peptide mass fingerprinting spectra were acquired as previously described (Chen et al., 2005). Processing of the spectra and data analysis were performed with the Bruker Daltonics FlexAnalysis 2.4 and Biotools 2.1 Software.
LC-MS/MS analysis of PPV CP
Purified PPV virions were reduced and alkylated as previously described (Udeshi et al., 2008) and digested using an enzyme to substrate ratio of 1:20 with endoproteinase LysC (Roche) and/or Trypsin (Promega). Peptides were reconstituted in 0.1% acetic acid and loaded onto a self-packed 360 μm o.d. × 75 μm i.d. C18 (5–20 μm diameter, 120 Å) microcapillary precolumn. The sample was desalted by washing the precolumn with 0.1% acetic acid. After washing, the precolumn was butt-connected to a 360 μm o. d. × 50 μm i.d. self-packed C18 (5 μm diameter, 120 Å) analytical column equipped with an integrated electrospray emitter tip (Martin et al., 2000). Peptides were gradient-eluted into the mass spectrometer at a flow rate of 60 nL/min using the HPLC gradient, 0–60% solvent B in 60 min (A = 0.1M acetic acid, B = 70% acetonitrile, 0.1M acetic acid). An LTQ-XL mass spectrometer was utilized to record ETD MS/MS spectra using the following parameters: reaction time=150 milliseconds, full AGC target=1E4 ion counts, MSn AGC target=1E4 ion counts, isolation window=3 m/z, reagent AGC target=3E5 ion counts, ETD reagent=fluoranthene. All PPV-CP peptide sequences were confirmed by manual interpretation of the corresponding ETD MS/MS spectra.
Assessment of virus infection
PPV-expressed GFP was observed either under a long-wavelength UV lamp (Black Ray model B 100 AP) and photographed with a Nikon D1X digital camera, or under a Leica MZ FLIII fluorescence microscope with excitation and barrier filters of 480/40 nm and 510 nm, respectively, and photographed with an Olympus DP70 digital camera.
PPV accumulation was determined by Western blot analysis. Systemically infected leaves (N. clevelandii, N. benthamiana and P. persica) or whole plants (A. thaliana) were grinded in disruption buffer (125 mM Tris–HCl pH 7.5, 2% SDS, 0.1% bromophenol blue, 6 M urea, and 5% ß-mercaptoethanol) [2 ml (g tissue)−1]. Samples were boiled for 10 min and cell debris removed by centrifugation. Supernatants were resolved on 12.5% SDS-PAGE, electroblotted to nitrocellulose membrane and subjected to immunodetection using as primary antibody a polyclonal serum obtained from a rabbit immunized with purified wild type PPV virions, which reacts with similar efficiency with wild type and O-GlcNAc-deficient PPV CP (dilutions 1:100000 for N. clevelandii, N. benthamiana and P. persica, and 1:50000 for A. thaliana), and as secondary antibody peroxidase-conjugated goat anti-rabbit IgG purchased from Jackson ImmunoResearch Laboratories (1:10000). The immunostained proteins were visualized by enhanced chemiluminescense detection with LiteAblot (Euroclone). The intensity of the bands was quantified by densitometry using Quantity One-4.2.0 (Bio-Rad) and the data were analyzed with Prism 4.0 (GraphPad Software, Inc.). Ponceau red staining was used to check the global proteins content of the samples.
The prevalence of different viral species in competition experiments was analyzed by comparing the restriction enzyme digestion patterns of RT-PCR amplification products produced from RNA isolated from immunocaptured virions. Systemically infected leaves (N. clevelandii) or whole infected plants (A. thaliana) were homogenized in 5 mM sodium phosphate buffer, pH 7.2 [2 ml (g tissue)−1] and incubated in tubes previously coated with anti-PPV IgGs overnight. The immunocaptured viral RNA was amplified by RT-PCR using the Titan one Tube system (Roche) with primers ETGFP-1 forward and Oligo 55, which flank the coding region of the N-terminal region of PPV CP. The amplification products were digested with the restriction enzyme NheI, which has a target site in the sequence of the CP7T/A mutant that is absent in the wild type PPV sequence, resolved on 1% agarose gels and stained with ethidium bromide.
Assessment of CP stability
Systemically infected leaves (N. clevelandii and P. persica) or whole plants (A. thaliana) were ground in 5 mM phosphate buffer, pH 7.2 [2 ml (g tissue)−1] and incubated at room temperature (24 °C). Aliquots were removed after different incubation times and subjected to Western blot analysis as explained above. The quantification of the intact CP and their degradation products was performed by densitometry using Quantity One-4.2.0 Bio-Rad, and the data were analyzed with Prism 4.0 (GraphPad Software, Inc.).
Acknowledgments
We thank Elvira Domínguez and Beatriz García García for technical assistance. This work was supported by Grants BIO2010-18541 from Spanish MCINN, SAL/0185/2006 from Comunidad de Madrid, and KBBE-204429 (SharCo) from the European Union to JAG, 2005 × 747_3 from the Spanish National Proteomics Institute (ProteoRed-ISC III) to SC and SJ, MCB-0112826 from National Science Foundation and DE-FG02-02ER15353 from US Department of Energy to NEO, and GM037537 from U.S. Public Health Service to DFH.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2013.03.029.
References
- Baulcombe D, Chapman S, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- Benko DM, Haltiwanger RS, Hart GW, Gibson W. Virion basic phosphoprotein from human cytomegalovirus contains O- linked N-acetylglucosamine. Proc Natl Acad Sci USA. 1988;85:2573–2577. doi: 10.1073/pnas.85.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Boudin ML, Strecker G, Michalski JC. O-linked GlcNAc in serotype-2 adenovirus fibre. Eur J Biochem. 1989;184:205–211. doi: 10.1111/j.1432-1033.1989.tb15008.x. [DOI] [PubMed] [Google Scholar]
- Chen D, Juárez S, Hartweck L, Alamillo JM, Simón-Mateo C, Pérez JJ, Fernández-Fernández MR, Olszewski NE, García JA. Identification of secret agent as the O-GlcNAc transferase that participates in Plum Pox virus infection. J Virol. 2005;79:9381–9387. doi: 10.1128/JVI.79.15.9381-9387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Fernández MR, Camafeita E, Bonay P, Méndez E, Albar JP, García JA. The capsid protein of a plant single-stranded RNA virus is modified by O-linked N-acetylglucosamine. J Biol Chem. 2002;277:135–140. doi: 10.1074/jbc.M106883200. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández MR, Mouriño M, Rivera J, Rodríguez F, Plana-Durán J, García JA. Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus-based vector. Virology. 2001;280:283–291. doi: 10.1006/viro.2000.0762. [DOI] [PubMed] [Google Scholar]
- Gonzalez SA, Burrone OR. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology. 1991;182:8–16. doi: 10.1016/0042-6822(91)90642-o. [DOI] [PubMed] [Google Scholar]
- Greis KD, Gibson W, Hart GW. Site-specific glycosylation of the human cytomegalovirus tegument basic phosphoprotein (UL32) at serine 921 and serine 952. J Virol. 1994;68:8339–8849. doi: 10.1128/jvi.68.12.8339-8349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. Two O-Linked N-Acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV. Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell. 1995;7:1459–1471. doi: 10.1105/tpc.7.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck A, Raikhel NV. A glycoprotein modified with terminal N-acetylglucosamine and localized at the nuclear rim shows sequence similarity to aldose-1-epimerases. Plant Cell. 1998;10:599–612. doi: 10.1105/tpc.10.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlitze S, Koenen M. A general and rapid mutagenesis method using polymerase chain reaction. Gene. 1990;91:143–147. doi: 10.1016/0378-1119(90)90177-s. [DOI] [PubMed] [Google Scholar]
- Kim YC, Udeshi ND, Balsbaugh JL, Shabanowitz J, Hunt DF, Olszewski NE. O-GlcNAcylation of the Plum pox virus capsid protein catalyzed by SECRET AGENT: characterization of O-GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids. 2011;40:869–876. doi: 10.1007/s00726-010-0706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laín S, Riechmann JL, Méndez E, García JA. Nucleotide sequence of the 3′ terminal region of plum pox potyvirus RNA. Virus Res. 1988;10:325–342. doi: 10.1016/0168-1702(89)90013-0. [DOI] [PubMed] [Google Scholar]
- López-Moya JJ, García JA. Construction of a stable and highly infectious intron-containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 2000;68:99–107. doi: 10.1016/s0168-1702(00)00161-1. [DOI] [PubMed] [Google Scholar]
- Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- Medina L, Grove K, Haltiwanger RS. SV40 large T antigen is modified with O-linked N-acetylglucosamine but not with other forms of glycosylation. Glycobiology. 1998;8:383–391. doi: 10.1093/glycob/8.4.383. [DOI] [PubMed] [Google Scholar]
- Mullis KG, Haltiwanger RS, Hart GW, Marchase RB, Engler JA. Relative accessibility of N-acetylglucosamine in trimers of the adenovirus types 2 and 5 fiber proteins. J Virol. 1990;64:5317–5323. doi: 10.1128/jvi.64.11.5317-5323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM. O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta-Gen Subj. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez JJ, Juárez S, Chen D, Scott CL, Hartweck LM, Olszewski NE, García JA. Mapping of two O-GlcNAc modification sites in the capsid protein of the potyvirus Plum pox virus. FEBS Lett. 2006;580:5822–5828. doi: 10.1016/j.febslet.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Salvador B, Delgadillo MO, Saénz P, García JA, Simón-Mateo C. Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol Plant Microbe Interact. 2008;21:20–29. doi: 10.1094/MPMI-21-1-0020. [DOI] [PubMed] [Google Scholar]
- Scott CL, Hartweck LM, Pérez JdJ, Chen D, García JA, Olszewski NE. SECRET AGENT, an Arabidopsis thaliana O-GlcNAc transferase, modifies the Plum pox virus capsid protein. FEBS Lett. 2006;580:5829–5835. doi: 10.1016/j.febslet.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ham BK, Xoconostle-Cazares B, Rojas MR, Lucas WJ. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TM, Kreppel L, Hart GW, Olszewski NE. Genetic and biochemical analysis of Arabidopsis SPY. In: Altman A, Ziv M, Izhar S, editors. Plant Biotechnology and In Vitro Biology in the 21st Century. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1999. pp. 445–448. [Google Scholar]
- Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protocols. 2008;3:1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrelmann M, Maiss E. Mutations in the coat protein gene of plum pox virus suppress particle assembly, heterologous encapsidation and complementation in transgenic plants of Nicotiana benthamiana. J Gen Virol. 2000;81:567–576. doi: 10.1099/0022-1317-81-3-567. [DOI] [PubMed] [Google Scholar]
- Wang J, Torii M, Liu H, Hart GW, Hu ZZ. dbOGAP—an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinf. 2011;12:91. doi: 10.1186/1471-2105-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford M, Faulkner P. A structural polypeptide of the baculovirus Autographa californica nuclear polyhedrosis virus contains O-linked N-acetylglucosamine. J Virol. 1992;66:3324–3329. doi: 10.1128/jvi.66.6.3324-3329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Viner R, Teo CF, Boons GJ, Horn D, Wells L. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J Proteome Res. 2011;10:4088–4104. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Daros JA, Elena SF. One is enough: in vivo effective population size is dose-dependent for a plant RNA virus. PLoS Pathog. 2011;7:e1002122. doi: 10.1371/journal.ppat.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]


