Abstract
Background
Transcript dosage imbalance may influence the transcriptome. To gain insight into the role of altered gene expression in hereditary colorectal polyposis predisposition, in the present study we analyzed absolute and allele-specific expression (ASE) of adenomatous polyposis coli (APC) and mutY Homolog (MUTYH) genes.
Methods
We analyzed DNA and RNA extracted from peripheral blood mononuclear cells (PBMC) of 49 familial polyposis patients and 42 healthy blood donors selected according similar gender and age. Patients were studied for germline alterations in both genes using dHPLC, MLPA and automated sequencing. APC and MUTYH mRNA expression levels were investigated by quantitative Real-Time PCR (qRT-PCR) analysis using TaqMan assay and by ASE assays using dHPLC-based primer extension.
Results
Twenty out of 49 patients showed germline mutations: 14 in APC gene and six in MUTYH gene. Twenty-nine patients did not show mutations in both genes. Results from qRT-PCR indicated that gene expression of both APC and MUTYH was reduced in patients analyzed. In particular, a significant reduction in APC expression was observed in patients without APC germline mutation vs control group (P < 0.05) while APC expression in the mutation carrier patients, although lower compared to control individuals, did not show statistical significance. On the other hand a significant reduced MUTYH expression was detected in patients with MUTYH mutations vs control group (P < 0.05). Altered ASE of APC was detected in four out of eight APC mutation carriers. In particular one case showed a complete loss of one allele. Among APC mutation negative cases, 4 out of 13 showed a moderate ASE. ASE of MUTYH did not show any altered expression in the cases analyzed. Spearman’s Rho Test analysis showed a positive and significant correlation between APC and MUTYH genes both in cases and in controls (P = 0.020 and P < 0.001).
Conclusions
APC and MUTYH showed a reduced germline expression, not always corresponding to gene mutation. Expression of APC is decreased in mutation negative cases and this appears to be a promising indicator of FAP predisposition, while for MUTYH gene, mutation is associated to reduced mRNA expression. This study could improve the predictive genetic diagnosis of at-risk individuals belonging to families with reduced mRNA expression regardless of presence of mutation.
Electronic supplementary material
The online version of this article (doi:10.1186/s13046-015-0244-4) contains supplementary material, which is available to authorized users.
Keywords: APC, MUTYH, Mutation, Adenomatous polyposis, Colon cancer, Phenotype, Gene expression, qRT-PCR, ASE, Predisposition
Introduction
Colorectal cancer (CRC) is one of the most frequent causes of cancer death worldwide [1, 2]. Inherited forms of CRC account for as much as 20–30 % of all CRC cases whereas hereditary colorectal polyposis syndromes account for about 1 % of all cases of CRC [3]. The identification of mechanisms predisposing to hereditary colorectal polyposis is necessary for direct genetic diagnosis of carrier status in cases without detectable mutations in known genes. Mendelian predisposition syndromes, like familial adenomatous polyposis (FAP), associated with mutations in known genes account for 5–10 % of the overall incidence of the disease [4, 5]. The genes mostly implicated in the inheritance of adenomatous polyposis, a condition that leads to colorectal cancer, are adenomatous polyposis coli (APC) involved in FAP (OMIM #175100) and mutY Homolog (MUTYH) involved in MUTYH-associated polyposis (MAP) (OMIM#608456). The tumor suppressor APC gene, a component of Wnt pathway, encodes a multifunctional protein that regulates many cellular processes, as differentiation, proliferation, life and death decision; it is involved in ephithelial turn-over, then APC loss is an early event in gut epithelium tumorigenesis [6]. APC germline mutations are detected in the majority of FAP patients, even if in a relevant subset of cases the mutations cannot be identified. A fraction (7–23 %) of APC mutation negative cases with phenotypes overlapping with attenuated FAP (AFAP) or classical FAP, is associated with biallelic germline variants of the MUTYH gene. MUTYH together with OGG1 and MTH1 is a component of DNA Base Excision Repair (BER) pathway that removes damaged bases generated by reactive oxygen species (ROS), capable to induce mutations commonly observed in cancerogenesis [7–9]. A relevant phenotypic pre-cancerous variability has been observed in kindred carrying the same germline mutation and literature data suggest that gene expression dosage can play a role as a new genetic mechanism for colorectal cancer susceptibility [10, 11]. The variation in gene expression contributes to phenotypic variability, plays an important role in the etiology of diseases and may affect absolute and allele specific expression [12–14]. In up to 50 % of polyposis families no germline mutation is identified and about 10–15 % of FAP patients could have a reduced APC expression with similar phenotype to patients with truncating APC mutation [15, 16]. Germline altered allele specific expression (ASE) is an indicator of genetic imbalances and a useful marker of predisposition to polyposis and/or colorectal cancer [10, 11, 17–19]. The dosage of the APC transcripts may modulate the disease resulting in classic or attenuated phenotype in cases with and without pathogenic mutations [17, 18, 20–22]. To gain insight in the correlation between APC and MUTYH mutations and altered expression, in the present study we investigated the role of dosage imbalance influencing the transcriptome of these two colon cancer-predisposing genes, performing an analysis of absolute and allele-specific expression in patients with different degrees of penetrance of hereditary colorectal disease. We analyzed peripheral blood mononuclear cells (PBMCs) from patients with and without mutations and compared the gene expression with control individuals. Finally, to understand the interaction between APC and BER pathway, we investigated the possible mutual modulation. This exploratory study on correlation among mutational spectrum, gene dosage and phenotype, could improve the genetic diagnosis performing predictive testing of at-risk individuals belonging to families with reduced mRNA expression regardless of presence of mutation. The identification of carriers of mutation or/and reduced mRNA expression may be helpful for the appropriate monitoring and clinical management of patients.
Patients and methods
Patients and nucleic acid preparation
We analyzed 49 polyposis patients recruited in different collaborating Italian Institutions including the Units of Clinical Physiopathology and Medical Genetics of the University of Florence, Medical Genetics of the University “G. d’Annunzio” of Chieti-Pescara, Gastroenterology and Digestive Endoscopy of the “Regina Elena” National Cancer Institute of Rome. This series consisted of patients affected by classical FAP, AFAP, multiple colorectal polyposis (with more than five polyps) [23] and familial colon cancer. Sixteen cases represented a research-based cohort from Aceto et al. [24]. Patients were selected based on availability of RNA and on the presence of at least 2–5 polyps at diagnosis in the probands and/or their relatives. One hundred and fifty healthy blood donors who reported no personal or family history of adenomas or colorectal cancer have been employed to identify and filter transcripts that exhibit altered transcript expression in the unaffected population. All study participants gave written informed consent after verbal counseling and the study was approved by the Ethics Committee of the University “G.d’Annunzio” of Chieti. Nucleic acids extraction from PBMCs and synthesis of complementary DNA (cDNA) from 3.5 μg of total RNA were performed as previously described [24]. Samples with poor quality or insufficient quantity of target in the cDNA template were not included in the mRNA expression analyses.
Screening for sequence variants
Mutation screening of APC and MUTYH genes for patients analyzed in previous study was conducted with different PCR-based techniques as previously described [24]. Additional screening was conducted in this study using denaturing high performance liquid chromatography (dHPLC) and automated sequencing. For APC also multiplex ligation-dependent probe amplification (MLPA) analysis was performed using the SALSA P043 APC MLPA kit (MCR-Holland, Amsterdam, The Netherlands) to detect large gene-rearrangements. Mutations are listed in the International Society for Gastrointestinal Hereditary Tumours (InSiGHT, http://www.insight-group.org/variants/database/) database.
Real-time quantitative PCR analysis (qRT-PCR)
The level of APC and MUTYH messenger RNA (mRNA) expression in PBMCs was investigated by TaqMan quantitative real-time PCR (qRT-PCR) analysis using StepOne™ 2.0 (Applied Biosystems). Data were analyzed using the comparative Ct method and were graphically indicated as 2−∆Ct ± SE. In accordance with the method, the mRNA amounts of the target genes (APC and MUTYH #Hs01568269_m1, #Hs01014856_m1 respectively, Applied Biosystems) were normalized to the endogenous housekeeping gene GUSB (#Hs99999908_m1, Applied Biosystems) [25]. Target and reference genes were amplified separately in triplicate for both cases and controls in a volume of 10 μl containing 1 μl template cDNA diluted 1:10, 0.5 μl of primers and probes mixture (20X FAM-labeled Assay-on-Demand Gene Expression Assay Mix) and 5 μl of 2X TaqMan Universal Master Mix (Applied Biosystems). The cycling conditions were performed as follows: 2 min at 50 °C, 10 min at 95 °C and 40 cycles of 15 s at 95 °C followed by 1 min at 60 °C.
Allele-specific expression (ASE) analysis
ASE analyses were performed by dHPLC-based single nucleotide primer extension (SNuPE) [26]. This technique allows to measure the relative allele expression comparing the heights of the peaks corresponding to the two alleles obtained by PCR-amplified gDNA and cDNA templates. Overall mean ratio obtained by analyzing gDNA templates for the corresponding variant was used to normalize cDNAs. These normalized cDNA/gDNA ratios were designed as ASE values. ASE measures were performed multiple times and coefficient of variation (CV) was calculated to test for assay reproducibility. Single-nucleotide polymorphism (SNP) genotyping analysis was performed to detect allelic markers of heterozygosity useful to ASE analysis. The frequent and common SNPs rs2229992 (c.1458C > T) (minor allele frequency, MAF = 0.49) in exon 12 of the APC gene and rs3219489 (c.1014G > C) (MAF = 0.31) in exon 12 of MUTYH gene were selected. An additional specific assay on exon 7 frequent pathogenic mutation of MUTYH gene (rs34612342, c.536A > G) was also designed (Additional file 1: Table S1).
Statistical analysis
The one-way analysis of variance (ANOVA) in the absolute expression of mRNA levels was performed to test the difference in means between different groups. Bonferroni post-hoc test allowed us to identify which group resulted significantly different based on the comparison of multiple values simultaneously.
ASE values were summarized as median, mean and standard deviation (SD) separately for the two groups (cases and controls). Shapiro-Wilk test was used to evaluate the normality in the distribution within each group. To assess differences of ASE values between cases and controls the Kruskall-Wallis test was performed followed by other tests carried out to determine if different proportions of individuals in cases and controls group were at a standard distance from the overall mean (1.0 SD, about 68 % of the distribution) by Chi-square test or Fisher’s exact test. To assess a possible correlation between the two genes Spearman’s rho correlation coefficient was evaluated. All analyses were performed with STATA software (version 10).
Results
Cases were analyzed for APC and MUTYH germline mutations, absolute expression and ASE. All analyses were performed and compared to a control population.
Screening for APC and MUTYH germline mutations
Mutation analysis detected pathogenic mutations in 20 patients out of 49 analyzed (Table 1). For APC gene 14 mutations introducing frameshift were detected, of which 12 previously published [2] and 2 detected in the present study, including a deletion of entire exon 15 and the p. Leu878_CysfsX916. Among these mutations only one (the p.His393_PhefsX396) fell in the gene portion associated to an attenuated phenotype [8, 21, 27]. This mutation was found in case GD37, and was located in the alternative splicing region of exon 10. Mutation analysis of MUTYH gene was performed for cases negative at APC mutation screening and detected sequence variants in six patients: two carriers of heterozygous variants (GD72 and GD155), three compound heterozygotes (GD68, GD82#1, GD82#2), and one carrier of exon 12 homozygous deletion (GD91) (Table 1). Four mutations were detected in a previous study [24] and 2 in the present study. Twenty-nine out of 49 patients did not show mutations in both genes (Table 2). Basic clinical features are reported in Tables 1 and 2; as noted polyposis patients without APC mutations manifested similar phenotype to patients with truncating APC mutations.
Table 1.
Clinical and molecular characteristics of patients with mutations
| Patients with APC mutations | |||||||
|---|---|---|---|---|---|---|---|
| Casea | Age at diagnosis | Phenotype | Family History (transmission) | Additional informations | APC Mutation | Exon | Effect |
| GD22 | 45 | FAP | yes (vertical) | osteomas, desmoids, duodenal cancer | c.646-1 G > Ab | 7 | Criptic splice site fsX292 |
| GD23 | n.a. | FAP | yes | c.4666dupb | 16 | p.Thr1556_AsnfsX1558 | |
| GD31 | 72 | FAP | yes (vertical) | >1000 polyps, colon cancer | c.3183_3187delb | 16 | p.Lys1061_LysfsX1062 |
| GD33 | n.a | FAP | yes (vertical) | c.904 C > Tb | 9 | p.Arg302X | |
| GD37 | 41 | AFAP | yes (vertical) | ileal polyps, colon cancer, | c.1176_1177insTb | 10 | p.His393_PhefsX396 |
| GD38 | 35 | FAP | yes (vertical) | CHRPE, osteomas | c.4717 G > Tb | 16 | p.Glu1573X |
| GD41 | 24 | FAP | yes (vertical) | gastric and ileal polyps | c.2684 C > Ab | 16 | p.Ser895X |
| GD48 | 33 | FAP | yes (vertical) | duodenal polyps | c.2758_2759delb | 16 | p.Asp920_CysfsX922 |
| GD57 | 22 | FAP | no | c.2299 C > Tb | 16 | p.Gln767X | |
| GD58 | 48 | FAP | no | c.4393_4394delb | 16 | p.Ser1465_TrpfsX1467 | |
| GD59 | 31 | FAP | yes (vertical) | c.4192_4193delb | 16 | p.Ser1398_SerfsX1407 | |
| GD74 | 27 | FAP | yes (vertical) | >100 polyps, desmoids, colon cancer | ex15del | 15 | ∆ 15 |
| GD103 | 55 | FAP | no | c.694 C > Tb | 7 | p.Arg232X | |
| GD119 | 26 | FAP | yes (vertical) | duodenal polyps, rectal cancer | c.2633delT | 16 | p.Leu878_CysfsX916 |
| ᅟ | ᅟ | ᅟ | ᅟ | ᅟ | ᅟ | ᅟ | ᅟ |
| Patients with MUTYH mutations | |||||||
| Casea | Age at diagnosis | Phenotype | Family History (transmission) | Additional informations | MUTYH Mutation | Exon | Effect |
| GD68 | 45 | AFAP | no | 40 polyps | [c.536A > G]+ | 7, 12 | [p.Tyr179Cys]+ |
| [c.1163 T > C]b | [p.Leu388Pro] | ||||||
| GD72 | 29 | FAP | yes (vertical) | [c.536A > G] + [c.=]b | 7 | [p.Tyr179Cys] + [p.=] | |
| GD82#1 | 49 | Multiple polyposis | yes (horizontal) | caecum cancer | [c.536A > G]+ | 7, 10 | [p.Tyr179Cys]+ |
| [c.820C > T]b | [p.Arg274Trp] | ||||||
| GD82#2 | 54 | Colon cancer | yes (horizontal) | [c.536A > G]+ | 7, 10 | [p.Tyr179Cys]+ | |
| [c.820C > T] | [p.Arg274Trp] | ||||||
| GD91 | 30 | Multiple polyposis | no | 100 polyps | [c.1145del]+ | 12 | [p.Ala382AlafsX407]+ |
| [c.1145del]b | |||||||
| [p.Ala382AlafsX407] | |||||||
| GD155 | 70 | AFAP | no | [c.536A > G] + [c.=] | 7 | [p.Tyr179Cys] + [p.=] | |
aIdentification number is followed by number of individual if several members were investigated per family
bData from [24]
n.a. = not available
Table 2.
Clinical and molecular characteristics of patients without mutations
| Patients without APC and MUTYH mutations | ||||
|---|---|---|---|---|
| Casea | Age at diagnosis | Phenotype | Family History (transmission) | Additional informations |
| GD70 | 43 | AFAP | yes (vertical) | |
| GD78 | 35 | Multiple polyposis | yes (vertical) | >10 polyps |
| GD80 | 45 | Multiple polyposis | yes (horizontal) | |
| GD81 | 36 | AFAP | yes (vertical) | |
| GD83 | 36 | FAP | yes (horizontal) | |
| GD84 | 32 | FAP | no | desmoids |
| GD86 | 17 | FAP | yes (vertical) | 143 microadenomas and 2 polyps |
| GD87 | 22 | FAP | no | rectal polyps, desmoids |
| GD92 | 58 | AFAP | n.a. | |
| GD94 | 65 | AFAP | yes (vertical) | 5 polyps, colon cancer |
| GD102 | n.a. | Multiple polyposis | n.a. | cystadenocarcinoma (unknown site) |
| GD106 | 50 | Multiple polyposis | n.a. | |
| GD107 | 42 | Multiple polyposis | yes (horizontal) | |
| GD109 | 52 | Multiple polyposis | no | 36 polyps, colon cancer |
| GD112#1 | 37 | FAP | yes (vertical) | colon, gastric, duodenal and rectal polyps |
| GD112#2 | 14 | FAP | yes (vertical) | colon and gastric polyps |
| GD117 | 63 | Multiple polyposis | yes (horizontal) | 10 polyps, gastric cancer |
| GD118 | 71 | AFAP | yes (vertical) | 10 polyps, colon cancer |
| GD121 | 41 | AFAP | n.a. | |
| GD122 | 59 | Colon cancer | yes (vertical) | |
| GD123 | 60 | AFAP | no | colon cancer |
| GD140 | 37 | AFAP | no | gastric polyps |
| GD146 | 44 | AFAP | no | colon and breast cancer |
| GD153 | 47 | AFAP | yes (horizontal) | hyperplastic polyps, colon cancer |
| GD154 | 44 | FAP | no | 836 polyps |
| GD157 | 53 | FAP | no | colon cancer |
| GD158 | 52 | FAP | yes (vertical) | |
| GD159 | 26 | FAP | yes (vertical) | rectal cancer |
| GD160 | 52 | AFAP | no | 10 polyps |
Analyses of APC and MUTYH mRNA expression
Patients analyzed by qRT-PCR belonged to three different subgroups: carriers of APC mutations (APC+, n. 13), carriers of MUTYH mutations (MUTYH+, n. 5) and patients negative at mutation screening for both genes (APC-/MUTYH-, n. 21). 2−∆Ct mean values of cases were compared to those of 42 healthy blood donors selected according similar gender and age in order to have a comparable sample size. RNA amounts from ten patients were used up for qRT-PCR analysis.
Analysis of APC gene expression by qRT-PCR showed a statistically significant reduced expression in the 21 APC-/MUTYH- patients (mean value of 0.18 ± 0.07 2−∆Ct) as compared to the control group (mean value of 0.32 ± 0.12 2−∆Ct) (P < 0.05) (Fig. 1a). On the other hand, APC expression in the 13 APC+ and 5 MUTYH+ patients, although lower compared to control individuals, did not show statistical significance (Fig. 1a).
Fig. 1.
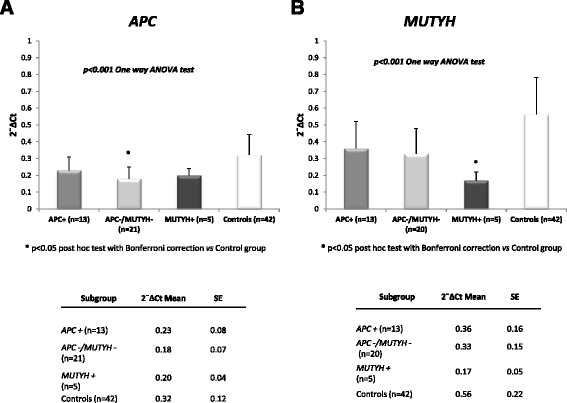
Germline expression by qRT-PCR of the APC and MUTYH genes. The figure shows the results from qRT-PCR analysis conducted in 39 patients carrying either APC mutation, MUTYH mutation or none mutations in both genes, compared to 42 controls. mRNA isolated from PBMCs was quantified by qRT-PCR using GUSB gene as endogenous control. Data were analyzed using the comparative Ct method and are expressed as mean and standard error (SE) of relative expression, which corresponds to the 2−ΔCt value; *P < 0.05 versus control group with post hoc test. a Relative expression levels of APC gene and associated table showing 2−∆Ct mean values ± SE of each subgroup. b Relative expression levels of MUTYH gene and associated table showing 2−∆Ct mean values ± SE of each subgroup
Regarding MUTYH gene expression, qRT-PCR analysis showed a statistically significant reduced expression in the 5 MUTYH+ patients (mean value of 0.17 ± 0.05 2−∆Ct) as compared to the control group (mean value of 0.56 ± 0.22 2−∆Ct) (P < 0.05), while the 13 APC+ and the 20 APC-/MUTYH- patients did not show significant MUTYH reduction (Fig. 1b). One case, GD117, resulted not informative by MUTYH qRT-PCR (Additional file 1: Table S1).
Altogether, these data suggest that reduced APC expression may be detected even in the absence of APC germline mutation and that MUTYH mutations may be associated with reduced MUTYH expression.
ASE analysis of APC and MUTYH genes
Germline ASE of APC was performed in 20 (out of 49) cases heterozygous for the SNP rs2229992 (c.1458C > T), of which 7 carrying APC mutation (Additional file 1: Table S1). To investigate if variations in ASE values can contribute to forms of colorectal disease with different degree of penetrance, the values were compared to data obtained from 53 consecutive CRC patients previously analyzed by the same assay [11]. We included one CRC case (case 19) turned out to be a de novo FAP in previous study, among APC mutation positive cases. A total of 21 polyposis and 52 consecutive CRC patients, were compared to 68 previously analyzed control individuals [11]. Results of ASE analysis showed in one case, GD41, bearing the p.Ser895X mutation, a germline allelic loss. The mean and median values are reported in Table 3, excluding the case GD41 without a quantifiable ASE value. The distribution of ASE values was tested for normality (Shapiro-Wilk normality test) and both APC mutation-positive and -negative FAP cases were distributed normally (respectively P = 0.499 and 0.682), whereas CRC cases and controls were not (Table 3). Only APC mutation-negative FAP cases had median and mean ASE values slightly, but significantly lower than controls by Kruskal-Wallis test (Table 3). Four out of 8 APC mutation-positive cases showed an imbalanced ASE, with values more than 1 SD from the overall mean (<0.99 and >1.53) compared to controls (Table 4; P = 0.022). Furthermore the group of the CRC cases deviating more than 1 SD from the overall mean was significantly larger than controls (Table 4; P = 0.001), as noted before [11]. Altogether, as regards APC mRNA expression, four patients with imbalanced ASE (GD22, GD41, GD72, GD82#1) showed also low levels of APC expression by qRT-PCR (Additional file 1: Table S1).
Table 3.
Mean and Median ASE values of cases and controls for APC c.1458C > T assay
| Status | N | Mean (±SD) | Median | Min | Max | Shapiro-Wilk | Multiple comparison of Kruskall-Wallis vs control |
|---|---|---|---|---|---|---|---|
| Controlsb | 68 | 1.25 (0.21) | 1.20 | 0.89 | 1.92 | 0.014 | |
| APC + polyposis cases | 7a | 1.20 (0.41) | 1.38 | 0.43 | 1.60 | 0.499 | 0.560 |
| APC - polyposis cases | 13 | 1.11 (0.18) | 1.11 | 0.83 | 1.51 | 0.682 | 0.029 |
| CRC casesb | 52 | 1.30 (0.32) | 1.28 | 0.68 | 2.46 | 0.041 | 0.350 |
| Overall | 140 | 1.26 (0.27) | 1.22 | 0.43 | 2.46 |
aCase GD41 was not considered in the table because it showed monoallelic expression
bPreviously published [11]
Bold values indicate statistical significance
Table 4.
Distribution of cases and controls at 1.0 SD from the overall mean ASE for APC c.1458C > T assay
| More than 1.0 SD from the overall mean (<0.99 and >1.53) | ||||
|---|---|---|---|---|
| Status | Within 1 SD | Out of 1 SD | Total | Chi-square or Fisher’s Exact p-value vs control |
| Controlsb | 57 | 11 | 68 | |
| APC + polyposis cases | 4 | 4a | 8 | 0.022 |
| APC - polyposis cases | 9 | 4 | 13 | 0.214 |
| CRC casesb | 30 | 22 | 52 | 0.001 |
| Overall | 100 | 41 | 141 | |
aCase GD41, with monoallelic expression, was included in the table among cases deviating more than 1.0 SD from the overall mean
bPreviously published [11]
Bold values indicate statistical significance
As regards MUTYH, all APC negative cases, heterozygous at MUTYH c.1014G > C variant, and cases bearing the c.536A > G (Y179C) pathogenic variant, were analyzed for ASE using 2 primer extension assays developed in this study. The ASE assay for MUTYH c.1014G > C variant was performed in 14 out of 49 cases (Additional file 1: Table S1) and 34 control individuals resulted heterozygous for the variant. The ASE assay for MUTYH c.536A > G (Y179C) pathogenic variant was performed in 5 cases (Additional file 1: Table S1). One case (GD68) has been analysed for both MUTYH assays. Considering that the means of ASE assays using the c.1014G > C and the c.536A > G variants were very similar (CV 4.28 % of the two means) we calculated the overall mean of all ASE values measured in cases and controls with both assays and that was 0.96 (SD ± 0.14; CV 14.51 %); the range of variation ± 1 SD was <0.82 and >1.10 (Additional file 2: Tables S2 and Additional file 3: Table S3). There were slight differences in mean and median ASE values in cases and controls. None of the individuals showed monoallelic expression or marked ASE. Results from MUTYH ASE analysis showed modest imbalances of values deviating more than 1 SD from the mean, in several individuals among cases and controls, but the differences were not statistical (Additional file 3: Table S3).
We also correlated the APC and MUTYH gene expression in 39 cases and 42 controls by Spearman’s test. As reported in Fig. 2, the two genes were positively correlated both in cases and in controls (respectively Rho = 0.381, P = 0.020 e Rho = 0.708, P < 0.001). This correlation was more strengthened among controls.
Fig. 2.
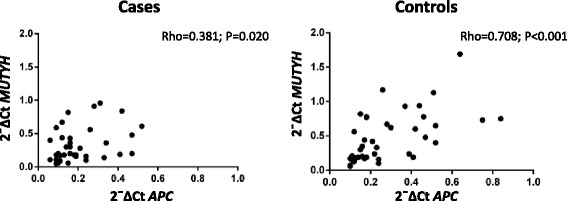
Correlation between APC and MUTYH gene expression. The figure shows APC and MUTYH absolute mRNA expression values measured in 39 cases (left) and 42 controls (right) correlated by Spearman’s test: correlation coefficient (rho) and P values (p)
Discussion
In this study we analyzed mutational status, transcript dosage and disease phenotype, to investigate the role of altered gene expression in the pathogenesis of familial colorectal tumors. The results of this study showed that at germline level gene expression of both APC and MUTYH was reduced in hereditary colorectal polyposis patients compared to healthy donors. In particular a reduced APC expression was observed even in patients without APC germline mutation meanwhile a reduced MUTYH expression has been detected in patients with MUTYH mutations. The statistically significant reduction in APC expression detected by qRT-PCR in 21 cases with APC-/MUTYH- genotype (Fig. 1a) is in agreement with previous studies [16, 28]. As far as ASE of APC, marked imbalances (>2fold) were detected only in two APC+ patients included in this study. Other patients had only modest degrees of imbalances of APC, which, as previously discussed, increase the risk of colorectal cancer [11]. Among the 14 cases that could be analyzed by both qRT-PCR and ASE, 6 showed no altered expression by both methods, 4 (GD22, GD41, GD72, GD82#1) showed reduced expression and altered ASE, 2 showed (GD68, GD84) reduced expression by qRT-PCR with a balanced allelic expression, likely related to decreased expression of both alleles, and 2 had only modest degrees of imbalanced expression which might reflect a small reduction in the expression of one allele compensated by a small increase in the expression of the other (GD33, GD123) (Additional file 1: Table S1, Additional file 3: Table S3). A noteworthy observation comes from the allelic expression analysis of case GD41, carrier of the p.Ser895X APC mutation. In this case ASE identified the complete loss of an allele that could not be seen with the simple observation by qRT-PCR even if this case, with monoallelic expression, showed an approximately 50 % reduction in the overall expression of APC (Additional file 1: Table S1). Among patients with MUTYH+ genotype two cases, GD82#1 and GD82#2, belonging to the same family, compound heterozygotes and carriers of the same mutations, showed different levels of APC expression (Table 1, Additional file 1: Table S1). GD82#1, affected by multiple polyposis and ciecum carcinoma, showed a lower level of APC expression; for GD82#2, presenting colorectal carcinoma without polyposis, the expression level for APC gene was higher. It can be assumed that the difference in APC expression in the two affected brothers might be associated with their different phenotype.
The cause of mRNA reduced expression is not known and could be related to mutations in regions of the gene not analyzed by mutational screening. Because APC is an oncosuppressor with a dominant pattern of hereditary transmission, its decreased expression is expected to contribute to FAP predisposition in these mutation negative cases. Thus decreased expression of APC in mutation negative cases appears to be a promising indicator of FAP predisposition.
Regarding MUTYH, five out of six patients with mutations, analyzed by qRT-PCR, showed a statistically reduced expression of the MUTYH gene and four out of five were carriers of MUTYH Y179C mutation. Patients belonging to this group manifested an attenuated phenotype (with absence of family history or horizontal transmission), except case GD72, who developed a classical FAP with vertical transmission (Table 1); these findings are in line with several studies [9, 27, 29].
Case GD72, carrier of a monoallelic Y179C mutation and showing by qRT-PCR a reduced expression (0.06 ± 0.01, Additional file 1: Table S1), has been analyzed by ASE of MUTYH and APC genes. In MUTYH the two alleles showed a quantitatively similar expression (1.08); as regarding APC, we found a moderately reduced relative expression (0.83) (Additional file 1: Table S1). In this patient both MUTYH alleles showed a low but equal expression with a reduction in absolute mRNA expression. This might explain FAP phenotype in a carrier of monoallelic MUTYH mutation. It is noteworthy that case GD72 manifested the disease at the age of 29 years. Individuals carrying monoallelic mutations tend to have a later presentation of disease of at least 5 years compared to biallelic ones but patients with heterozygous Y179C mutation have usually a more severe phenotype with earlier onset as compared with the other frequent G396D mutation [9, 30]. The missense Y179C mutation, situated in the catalytic domain of MUTYH, led to the formation of an inactive protein, as it can remove normal interactions with other proteins [31] and causes a quantitative reduction of mRNA, which would lead to reduced protein levels. Further studies on protein in those cases with both mutations and reduced expression could support our data on MUTYH protein functionality.
The risk of colorectal cancer in heterozygous carriers of MUTYH mutations has been considered comparable with that of first-degree relatives of patients with sporadic colorectal cancer by some authors [32] or considered increased due to a synergism with alterations in other genes involved in BER or other reparative pathways [9, 33]. Other authors assume that a slightly increased risk of cancer related to the presence of a monoallelic inactivation of MUTYH could be attributed to a situation of haploinsufficiency [34]. A complete picture of the effect of monoallelic mutations on the variability of clinical phenotype linked to MUTYH is still to be defined. Studies of absolute and relative gene expression applied to a larger group of monoallelic mutation carriers could provide a clue of possible involved pathways.
Correlation between germline altered gene expression and disease, sometimes in absence of mutations, have been reported in chronic inflammatory diseases and in forms of intellectual disability, all compared to control individuals [35–37]. In spite of several evidences that showed as different phenotypes can be classified based on degree of severity of polyposis and site of APC gene mutation [8, 21, 27, 38, 39], there is a scarcity of data concerning correlations between disease manifestations and APC germline reduced expression. The observations in relatives GD82#1 and GD82#2 might be the first evidence of this kind of correlations. It will be very interesting to extend the study to other polyposis families with a larger number of relatives. If confirmed in further studies, the correlation between germline reduced expression and disease manifestations could improve the predictive genetic diagnosis of at-risk individuals belonging to families with reduced mRNA expression regardless of presence of mutation.
Among the various cellular functions of APC gene, it was recently discovered its involvement in DNA repair mediated by BER, as it is able to modulate the activity of this pathway [40]. Therefore we searched for possible mutual modulation between APC and MUTYH expression in 39 cases and 42 controls by Spearman’s test. As reported in Fig. 2, the two genes were positively correlated both in cases and in controls although the significance seemed to be stronger among controls. A modulation between APC and MUTYH was also observed in Fig. 1: carriers of mutation compared to cases without mutations in one gene showed a higher expression of the other gene.
Conclusions
In this study the most significant result comes from the analysis of APC and MUTYH expression levels in hereditary colorectal polyposis patients carrying either APC mutation, MUTYH mutation or none mutations in both genes, compared to 42 controls. Both genes showed a reduced germline expression, not always corresponding to gene mutation. Expression of APC is decreased in mutation negative cases and this appears to be a promising indicator of FAP predisposition, while for MUTYH gene, reduced mRNA expression is associated to mutation. This is the first evidence of ASE analysis of MUTYH but, as the recessive pattern of hereditary transmission of this gene, altered ASE could be masked by contemporary reduced expression of both alleles. This study could improve the predictive genetic diagnosis of at-risk individuals belonging to families with reduced mRNA expression regardless of presence of mutation. It will be very interesting to extend the study to other genes of the Wnt and BER pathways or to other known cancer predisposing genes.
Acknowledgments
Funding
This study was supported by the Italian Ministry of Instruction, University and Research (MCC, PB, AC).
Abbreviations
- CRC
Colorectal cancer
- ASE
Allele-specific expression
- APC
Adenomatous polyposis coli
- MUTYH
Mut Y homolog
- PBMCs
Peripheral blood mononuclear cells
- qRT-PCR
Quantitative real-time PCR
- FAP
Familial adenomatous polyposis
- MAP
MUTYH-associated polyposis
- BER
Base excision repair
- ROS
Reactive oxygen species
- cDNA
Complementary DNA
- dHPLC
Denaturing high performance liquid chromatography
- MLPA
Multiplex ligation-dependent probe amplification
- mRNA
Messenger RNA
- GUSB
B-Glucuronidase
- SNP
Single nucleotide polymorphism
- SD
Standard deviation
- MAF
Minor allele frequency
- gDNA
Genomic DNA
Additional files
qRT-PCR and ASE results from 50 cases investigated. (DOCX 37 kb)
Mean and median ASE values of cases and controls after combining both MUTYH assays. (DOCX 18 kb)
Distribution of cases and controls at 1 SD from the overall mean ASE after combining both MUTYH assays. (DOCX 41 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GMA contributed to the study design and performed mutation screening. FF performed mutation screening, qRT-PCR and ASE analyses. SDI performed mutation screening and ASE analysis. GP, RV, PDG, VS and MG collected and evaluated the clinical data. MDN performed the statistical calculations. PB and AC reviewed all the genetic data. AC helped to draft the manuscript. MCC conceived and coordinated the study, reviewed all genetic and clinical data and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Gitana Maria Aceto, Email: gaceto@unich.it.
Fabiana Fantini, Email: f.fantini@unich.it.
Sabrina De Iure, Email: s.deiure@unich.it.
Marta Di Nicola, Email: m.dinicola@unich.it.
Giandomenico Palka, Email: gdpalka@unich.it.
Rosa Valanzano, Email: rosa.valanzano@unifi.it.
Patrizia Di Gregorio, Email: patrizia.digregorio@asl2abruzzo.it.
Vittoria Stigliano, Email: stigliano@ifo.it.
Maurizio Genuardi, Email: m.genuardi@rm.unicatt.it.
Pasquale Battista, Email: p.battista@unich.it.
Alessandro Cama, Email: cama@unich.it.
Maria Cristina Curia, Email: mc.curia@unich.it.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.GLOBOCAN. Estimated cancer incidence, mortality and prevalence worldwide in 2012. 2012. http://globocan.iarc.fr/Default.aspx
- 3.Aretz S. The differential diagnosis and surveillance of hereditary gastrointestinal polyposis syndromes. Dtsch Arztebl Int. 2010;107:163–73. doi: 10.3238/arztebl.2010.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 5.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–58. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson S, Näthke IS. Interactions and functions of the adenomatous polyposis coli (APC) protein at a glance. J Cell Sci. 2013;126:873–7. doi: 10.1242/jcs.100479. [DOI] [PubMed] [Google Scholar]
- 7.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, et al. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-- > T:A mutations. Hum Mol Genet. 2002;11:2961–7. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 8.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–9. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 9.Morak M, Laner A, Bacher U, Keiling C, Holinski-Feder E. MUTYH-associated polyposis - variability of the clinical phenotype in patients with biallelic and monoallelic MUTYH mutations and report on novel mutations. Clin Genet. 2010;78:353–63. doi: 10.1111/j.1399-0004.2010.01478.x. [DOI] [PubMed] [Google Scholar]
- 10.Castellví-Bel S, Castells A. Allele-specific expression as a new genetic susceptibility mechanism for colorectal cancer. Gastroenterology. 2009;136:2397–9. doi: 10.1053/j.gastro.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 11.Curia MC, De Iure S, De Lellis L, Veschi S, Mammarella S, White MJ, et al. Increased variance in germline allele-specific expression of APC associates with colorectal cancer. Gastroenterology. 2012;142:71–77. doi: 10.1053/j.gastro.2011.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–7. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 14.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laken SJ, Papadopoulos N, Petersen GM, Gruber SB, Hamilton SR, Giardiello FM, et al. Analysis of masked mutations in familial adenomatous polyposis. Proc Natl Acad Sci U S A. 1999;96:2322–6. doi: 10.1073/pnas.96.5.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanter-Smoler G, Fritzell K, Rohlin A, Engwall Y, Hallberg B, Bergman A, et al. Clinical characterization and the mutation spectrum in Swedish adenomatous polyposis families. BMC Med. 2008;6:10. doi: 10.1186/1741-7015-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renkonen ET, Nieminen P, Abdel-Rahman WM, Moisio AL, Järvelä I, Arte S, et al. Adenomatous polyposis families that screen APCmutation-negative by conventional methods are genetically heterogeneous. JClin Oncol. 2005;23:5651–9. doi: 10.1200/JCO.2005.14.712. [DOI] [PubMed] [Google Scholar]
- 18.Castellsagué E, González S, Guinó E, Stevens KN, Borràs E, Raymond VM, et al. Allele-specific expression of APC in adenomatous polyposis families. Gastroenterology. 2010;139:439–47. doi: 10.1053/j.gastro.2010.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Sun R, Liang Y, Pan X, Li Z, Bai P, et al. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J Exp Clin Cancer Res. 2013;32:104. doi: 10.1186/1756-9966-32-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell SM, Petersen GM, Krush AJ, Booker S, Jen J, Giardiello FM, et al. Molecular diagnosis of familial adenomatous polyposis. N Engl J Med. 1993;329:1982–7. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 21.Curia MC, Esposito DL, Aceto G, Palmirotta R, Crognale S, Valanzano R, et al. Transcript dosage effect in familial adenomatous polyposis: model offered by two kindreds with exon 9 APC gene mutations. Hum Mutat. 1998;11:197–201. doi: 10.1002/(SICI)1098-1004(1998)11:3<197::AID-HUMU3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 22.Yan H, Dobbie Z, Gruber SB, Markowitz S, Romans K, Giardiello FM, et al. Small changes in expression affect predisposition to tumorigenesis. Nat Genet. 2002;30:25–6. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- 23.Mongin C, Coulet F, Lefevre JH, Colas C, Svrcek M, Eyries M, et al. Unexplained polyposis: a challenge for geneticists, pathologists and gastroenterologists. Clin Genet. 2012;81:38–46. doi: 10.1111/j.1399-0004.2011.01676.x. [DOI] [PubMed] [Google Scholar]
- 24.Aceto G, Curia MC, Veschi S, De Lellis L, Mammarella S, Catalano T, et al. Mutations of APC and MYH in unrelated Italian patients with adenomatous polyposis coli. Hum Mutat. 2005;26:394. doi: 10.1002/humu.9370. [DOI] [PubMed] [Google Scholar]
- 25.Sørby LA, Andersen SN, Bukholm IR, Jacobsen MB. Evaluation of suitable reference genes for normalization of real-time reverse transcription PCR analysis in colon cancer. J Exp Clin Cancer Res. 2010;29:144. doi: 10.1186/1756-9966-29-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aceto GM, De Lellis L, Catalano T, Veschi S, Radice P, Di Iorio A, et al. Nonfluorescent denaturing HPLC-based primer-extension method for allele-specific expression: application to analysis of mismatch repair genes. Clin Chem. 2009;55:1711–8. doi: 10.1373/clinchem.2009.126300. [DOI] [PubMed] [Google Scholar]
- 27.Nieuwenhuis MH, Vasen HF. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol. 2007;5:374–8. doi: 10.1016/j.critrevonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Venesio T, Balsamo A, Rondo-Spaudo M, Varesco L, Risio M, Ranzani GN. APC haploinsufficiency, but not CTNNB1 or CDH1 gene mutations, accounts for a fraction of familial adenomatous polyposis patients without APC truncating mutations. Lab Invest. 2003;83:1859–66. doi: 10.1097/01.LAB.0000106722.37873.8D. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen M, Morreau H, Vasen HF, Hes FJ. MUTYH-associated polyposis (MAP) Crit Rev Oncol Hematol. 2011;79:1–16. doi: 10.1016/j.critrevonc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen M. Joerink-van de Beld MC, Jones N, Vogt S, Tops CM, Vasen HF, et al. Analysis of MUTYH genotypes and colorectal phenotypes in patients with MUTYH-associated polyposis. Gastroenterology. 2009;136:471–6. doi: 10.1053/j.gastro.2008.10.056. [DOI] [PubMed] [Google Scholar]
- 31.Ali M, Kim H, Cleary S, Cupples C, Gallinger S, Bristow R. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135:499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones N, Vogt S, Nielsen M, Christian D, Wark PA, Eccles D, et al. Increased colorectal cancer incidence in obligate carriers of heterozygous mutations in MUTYH. Gastroenterology. 2009;137:489–94. doi: 10.1053/j.gastro.2009.04.047. [DOI] [PubMed] [Google Scholar]
- 33.Morak M, Massdorf T, Sykora H, Kerscher M, Holinski-Feder E. First evidence for digenic inheritance in hereditary colorectal cancer by mutations in the base excision repair genes. Eur J Cancer. 2011;47:1046–55. doi: 10.1016/j.ejca.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Plotz G, Casper M, Raedle J, Hinrichsen I, Heckel V, Brieger A, et al. MUTYH gene expression and alternative splicing in controls and polyposis patients. Hum Mutat. 2012;33:1067–74. doi: 10.1002/humu.22059. [DOI] [PubMed] [Google Scholar]
- 35.Mesko B, Poliska S, Szegedi A, Szekanecz Z, Palatka K, Papp M, et al. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med Genomics. 2010;3:15. doi: 10.1186/1755-8794-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentilini D, Perino A, Viganò P, Chiodo I, Cucinella G, Vignali M, et al. Gene expression profiling of peripheral blood mononuclear cells in endometriosis identifies genes altered in non-gynaecologic chronic inflammatory diseases. Hum Reprod. 2011;26:3109–17. doi: 10.1093/humrep/der270. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–25. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 38.Ficari F, Cama A, Valanzano R, Curia MC, Palmirotta R, Aceto G, et al. APC gene mutations and colorectal adenomatosis in familial adenomatous polyposis. Br J Cancer. 2000;82:348–53. doi: 10.1054/bjoc.1999.0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedl W, Caspari R, Sengteller M, Uhlhaas S, Lamberti C, Jungck M, et al. Can APC mutation analysis contribute to therapeutic decisions in familial adenomatous polyposis? Experience from 680 FAP families. Gut. 2001;48:515–21. doi: 10.1136/gut.48.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaiswal AS, Narayan S. A novel function of adenomatous polyposis coli (APC) in regulating DNA repair. Cancer Lett. 2008;271:272–80. doi: 10.1016/j.canlet.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


