Loss of Meis3 leads to defects in enteric neural crest cell migration, number, and proliferation during colonization of the gut. This leads to colonic aganglionosis, in which the hindgut is devoid of neurons, identifying it as a novel candidate factor in the etiology of Hirschsprung’s disease during enteric nervous system development.
Abstract
During development, vagal neural crest cells fated to contribute to the enteric nervous system migrate ventrally away from the neural tube toward and along the primitive gut. The molecular mechanisms that regulate their early migration en route to and entry into the gut remain elusive. Here we show that the transcription factor meis3 is expressed along vagal neural crest pathways. Meis3 loss of function results in a reduction in migration efficiency, cell number, and the mitotic activity of neural crest cells in the vicinity of the gut but has no effect on neural crest or gut specification. Later, during enteric nervous system differentiation, Meis3-depleted embryos exhibit colonic aganglionosis, a disorder in which the hindgut is devoid of neurons. Accordingly, the expression of Shh pathway components, previously shown to have a role in the etiology of Hirschsprung’s disease, was misregulated within the gut after loss of Meis3. Taken together, these findings support a model in which Meis3 is required for neural crest proliferation, migration into, and colonization of the gut such that its loss leads to severe defects in enteric nervous system development.
INTRODUCTION
The enteric nervous system (ENS) is derived from the neural crest, a multipotent migratory stem cell population that develops into a variety of cell types, ranging from craniofacial cartilage and pigment cells to neurons and glia of sensory, sympathetic, and enteric ganglia (for review, see Rogers et al., 2012). The ENS comprises an autonomous network of thousands of interconnected ganglia, containing a diverse array of neurons and glia, embedded within the walls of the digestive tract. Enteric ganglia regulate muscle contraction, gut secretion, and water balance within the gastrointestinal tract (Furness, 2006). Throughout life, the ENS functions to regulate peristalsis during digestion, a key aspect of gastrointestinal function. Defects in neural crest migration lead to a spectrum of developmental disorders (Noisa and Raivio, 2014), including Hirschsprung’s disease, characterized by a lack of ganglia formation in the hindgut. This in turn leads to megacolon, in which decreased innervation leads to accumulation of digested food and an enlarged and nonfunctional colon requiring surgical repair (Bergeron et al., 2013).
During development, the ENS mostly derives from the “vagal” subpopulation of the neural crest, which originates adjacent to somite pairs 1–7 (Le Douarin and Teillet, 1973; Epstein et al., 1994; Burns et al., 2000; Kuo and Erickson, 2011). To colonize the gut, vagal neural crest cells emigrate from the hindbrain, enter the foregut mesenchyme, and migrate caudally such that they eventually populate the entire gut, a migration process that takes days and is the longest of any embryonic cell migration (Le Douarin and Teillet, 1973; Burns, 2005; Anderson et al., 2006). In chicken embryos, the colonization process is largely complete by day 8 of development (Le Douarin and Teillet, 1973), in mouse embryos by embryonic day 15 (Druckenbrod and Epstein, 2005), and in humans by 7 wk of gestation (Fu et al., 2003). In zebrafish embryos, the process of migration is complete by the end of 3 d postfertilization (dpf) (Olden et al., 2008). Because of their rapid development, transparency, and simple gut development (Shepherd and Eisen, 2011), zebrafish embryos offer an attractive model for studying the cellular and molecular mechanisms underlying ENS development.
In the developing zebrafish embryo, vagal neural crest cells first enter the foregut at 32 h postfertilization (hpf) as two migratory streams from the left and right postotic vagal regions (Kelsh and Eisen, 2000; Dutton et al., 2001; Shepherd et al., 2001; Olden et al., 2008). Once they reach their final destinations, enteric neural crest cells differentiate into neuronal subtypes or glia, subsequently forming interconnected ganglia within the myenteric and submucosal plexuses that run within the mucosa and smooth muscle layers in zebrafish (Olsson et al., 2008; Shepherd and Eisen, 2011).
Studies aimed at investigating the molecular underpinnings of Hirschsprung’s disease have demonstrated a role for several extrinsic signaling pathways and intrinsic effectors in ENS development (for review, see Heanue and Pachnis, 2007). Among these, the receptor tyrosine kinase (Ret), glial cell line–derived neurotrophic factor (GDNF), Sonic Hedgehog (Shh), and the endothelin signaling pathways play important roles in the migration, proliferation, and survival of enteric precursors (Taraviras et al., 1999; Natarajan et al., 2002; Shepherd et al., 2004; Landman et al., 2007). Despite many studies, however, little is known about the early stages of vagal and enteric neural crest development. Important remaining questions include the following: What confers their “enteric” identity? Why do vagal neural crest enter the gut at a particular location? How do they know where to stop migrating along the gut?
Meis cofactors are three–amino acid loop extension (TALE)–family homeodomain proteins that control accessibility at Hox-regulated promoters. They are expressed widely in various tissues and perform a spectrum of functions during embryonic development (for review, see Cerdá-Esteban and Spagnoli, 2014). The three Meis family members (Meis1–3) control accessibility at Hox-regulated promoters (Choe et al., 2009) by forming dimeric or trimeric DNA-binding complexes with Pbx and/or Hox proteins (Vlachakis et al., 2000; Waskiewicz et al., 2001) and thus regulate transcriptional output in a cell-specific manner. Meis and Pbx proteins associate with each other via conserved N-terminal domains, thereby controlling their nuclear localization and ability to participate in DNA-dependent Hox complex activities (Berthelsen 1998; Mann et al., 2009). Several studies have highlighted the role of Meis proteins in the regulation of progenitor cell maintenance, proliferation, and differentiation during organogenesis. For example, Meis1 serves an essential role in progenitor/stem cell development in various developmental and postembryonic contexts, such as neural stem cell development (Barber et al., 2013), hematopoiesis (Ariki et al., 2014), and thymic epithelial cell progenitor maintenance (Hirayama et al., 2014). Meis2 regulates limb outgrowth (Capdevila et al., 1999), olfactory neurogenesis (Agoston et al., 2014), human cardiac development (Paige et al., 2012), and the differentiation of craniofacial cartilage in zebrafish embryos (Melvin et al., 2013).
In contrast to Meis1 and 2, about which much is known, little is known about the function of Meis3 other than that it influences posterior hindbrain patterning in zebrafish (Waskiewicz et al., 2001; Vlachakis et al., 2001) and neural induction and neuronal differentiation in the frog hindbrain (Dibner et al., 2001; Aamar and Frank, 2004; Elkouby et al., 2012). Of interest, Meis3 is expressed in the zebrafish foregut environment at 36 hpf, within the lateral plate mesoderm (Manfroid et al., 2007), and has been implicated in early foregut patterning at 24 hpf (diIorio et al., 2007). Given the integral roles of Meis proteins in diverse patterning and growth events and the role of Meis3 in the foregut, we asked whether Meis3 might influence enteric neural crest migration and/or ENS development in zebrafish embryos. The results reveal novel in vivo roles for Meis3 during ENS development.
RESULTS
meis3 is expressed in the developing foregut environment during vagal and enteric neural crest cell migratory phases
At 33 hpf, meis3 is expressed in the hindbrain, as well as in the posterior branchial arches and the developing fin buds (arrowhead and arrow, respectively, in Figure 1A). meis3 is also expressed medially on the ventral side of the embryo, near the foregut (arrow in Figure 1B). This is consistent with previously observed meis3 expression in the posterior lateral plate mesoderm, which lies adjacent to and overlaps the foregut endoderm at 36 hpf (Manfroid et al., 2007). In serial transverse sections at 33 hpf, meis3 is expressed in the mesoderm that dorsally surrounds and flanks the developing gut (arrows in Figure 1, C and C′). At this stage, the gut endoderm is a simple epithelium before lumen formation (Field et al., 2003; Wallace and Pack, 2003; Ng et al. 2005). Postotic vagal neural crest cells migrate toward the developing ventral midline and along the anterior foregut between 32 and 36 hpf (Shepherd et al., 2004; Olden et al., 2008).
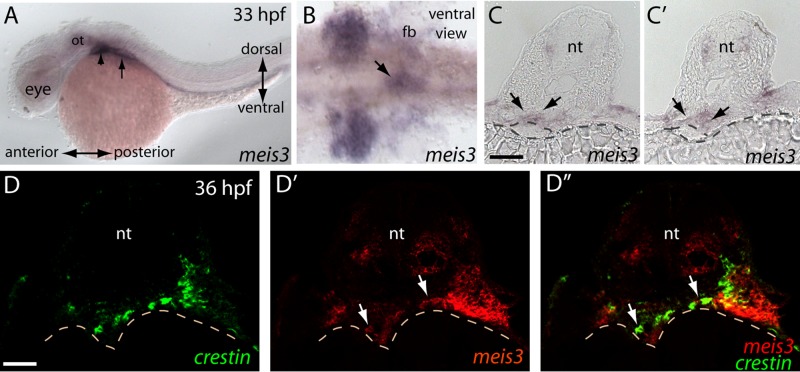
FIGURE 1:
meis3 is expressed with the vagal and enteric neural crest migratory environments during early phases of ENS development. (A) meis3 transcript within the postotic branchial arches (arrowhead) and the fin buds (arrow) at 33 hpf. (B) After yolk sac removal, a ventral view shows meis3 expression field within the posterior branchial arches and the fin buds and within the foregut (arrow). (C, D) Serial adjacent sections reveal meis3 localization within the hindbrain neural tube and mesenchymal layer of the developing gut (arrows), just above the developing gut endoderm (dashed circle above yolk). (D–D′′) Double fluorescence in situ hybridization reveals that meis3 (red) is expressed in the gut and its surrounding environment at 36 hpf and that crestin+ neural crest cells (green) migrate into and localize within the meis3+ region. fb, fin bud; nt, neural tube; ot, otic. Scale bars, 50 μm (C, C′), 40 μm (D–D′′).
In transverse section, double fluorescence in situ hybridization for meis3 and the panneural crest marker crestin at 36 hpf revealed the presence of crestin-positive cells migrating from the lateral mesoderm toward the developing gut (Figure 1, D and D′′, arrows). meis3 was detected in the mesoderm surrounding the gut and in a subset of the crestin+ neural crest cells migrating toward the gut (Figure 1, D′ and D′′, arrows). Taken together, these data indicate that meis3 is present in the developing vagal and foregut regions during neural crest cell entry.
Loss of Meis3 leads to delayed migration of vagal neural crest cells into the developing foregut
The spatial expression pattern of meis3 near the foregut suggested that it might play a functional role during enteric neural crest and/or foregut development. To test this, we performed loss-of- function experiments by using two strategies to knock down Meis3 at the one-cell stage in Tg(-4.9sox10:GFP) transgenic embryos, hereafter referred to as sox10:GFP: 1) by use of a translation-blocking morpholino (diIorio et al., 2007) or 2) by injection of mRNA encoding a dominant-negative Pbx4 construct (pbcab) that prevents nuclear entry of Meis proteins (Choe et al., 2002). At 25 hpf, Meis3 morphants and pbcab-injected embryos exhibited reduced brain and eye size compared with control-injected embryos (Supplemental Figure S1, A–C), in agreement with previous studies demonstrating a role for Meis proteins in eye and brain development (Bessa et al., 2008; Heine et al., 2008). To examine the localization of sox10:green fluorescent protein (GFP)+ neural crest cells after Meis3 knockdown, we performed whole-mount immunohistochemistry with a GFP antibody. In control embryos at 28 and 36 hpf, sox10:GFP+ neural crest cells migrated anteriorly around the eye, into the hyoid arch, and into the branchial arches (Supplemental Figure S1D and Figure 2A; n = 20/20). In Meis3 morphants (Supplemental Figure S1F and Figure 2B; n = 18/20) and pbcab-injected embryos (Supplemental Figure S1E; n = 17/20), sox10:GFP+ neural crest cells also migrated around the eye, into the hyoid arch, and into the branchial arches, indicating that neural crest specification was unaffected. Demonstrating the specificity of the morpholino, Meis3 protein levels significantly decreased after Meis3 morpholino treatment and were rescued after coinjection of meis3 mRNA, which cannot be bound by Meis3 MO, with Meis3 MO (Supplemental Figure S1G).
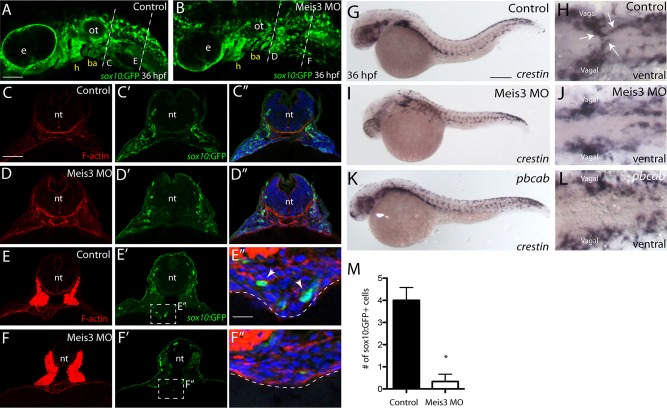
FIGURE 2:
Meis3 is required for the timely migration of vagal neural crest to the developing foregut during early phases of ENS development. (A) Control-injected and (B) Meis3 MO–injected embryos at 36 hpf display sox10:GFP+ neural crest cells around the eye, the hyoid, the otic, and the branchial arches. (C–C′′) Transverse sections depict sox10:GFP+ cells located laterally within the postotic vagal regions of control embryos and (D–D′′) within Meis3 MO–injected embryos. (E–E′′) Control sox10:GFP+ vagal neural crest cells are detected within the foregut environment at 36 hpf (E′, E′′; arrows). (F–F′′) virtually no sox10:GFP+ cells are observed within Meis3 MO–injected embryos around the foregut. F-actin in red (C–F), sox10:GFP in green (C′, D, E′, F′) and merge, with 4′,6-diamidino-2-phenylindole (DAPI) in blue (C′′, D′′, E′′, F′′). (G, H) Control-injected embryos contain crestin+ neural crest cells within the postotic vagal region and ventrally within two streams of migratory cells emanating toward the ventral midline along the foregut at 36 hpf (H; arrows). (I–L) Although Meis3 MO–injected and pbcab-injected embryos contain crestin+ neural crest ventrally within the postotic vagal regions, no streams of neural crest are detected migrating toward the ventral midline. (M) Bar graph quantifying the number of sox10:GFP+ neural crest cells near the gut in control and Meis3 MO-injected embryos at 36 hpf, *p < 0.05. ba, branchial arch; e, eye; h, hyoid; ot, otic. Scale bars, 70 μm (A, B), 50 μm (C–F), 20 μm (E′′, F′′), 100 μm (G, I, K).
To examine whether enteric neural crest localization was altered after loss of Meis3, we analyzed serial transverse sections from the postotic and foregut levels at 36 hpf. In control embryos, at the postotic level, sox10:GFP+ neural crest cells localized laterally (Figure 2, C′ and C′′; n = 10/10), whereas posterior sections at the level of the foregut showed sox10:GFP+ enteric neural crest localized around the gut (arrows in Figure 2, E′ and E′′; n = 10/10). Although postotic vagal neural crest cells were present in lateral regions after loss of Meis3 (Figure 2, D′ and D′′), they were virtually absent along the foregut (Figure 2, F′ and F′′; n = 8/10). Control embryos exhibited an average of 4 sox10:GFP+ cells along the foregut, whereas Meis3 morphants contained only an average of 0.33 sox10:GFP+ cells (Figure 2M; p = 0.0053). To assess qualitatively neural crest cell distribution along the gut in whole mount, we performed crestin in situ hybridization (Figure 2, G–L). In control embryos at 36 hpf, a ventral view revealed two chains of crestin-positive cells migrating from the left and right vagal regions to the ventral midline (Figure 2H, arrows; n = 20/20). In Meis3 MO and pbcab-injected embryos, although crestin-positive cells resided within the vagal regions, no migratory chains toward the midline were detected (Figure 2, J and L; n = 18/20 and 20/20, respectively). Taken together, these results suggest that Meis3 may be required for the timely migration of vagal neural crest cells into the developing ventral midline and foregut environment.
During the initial phases of ENS development, the endoderm is required for migration of vagal neural crest cells toward the ventral midline (Reichenbach et al., 2008). One possibility is that the delay of vagal neural crest migration to the ventral midline in Meis3 morphants actually might reflect an absence of foregut endoderm. To examine this possibility, we assayed the presence of gut endoderm by in situ hybridization at 36 hpf using the digestive endoderm marker foxa1 (Odenthal and Nüsslein-Volhard, 1998). Similar to control-injected embryos, Meis3 morphants possessed foxa1-positive gut endoderm territory (Supplemental Figure S2, A–D), as observed in both whole-mount (Supplemental Figure S2, A and B, n = 20/20) and sectioned embryos (Supplemental Figure S2, C and D, n = 6/6), suggesting that Meis3 is not required for the presence of gut endoderm. To assess whether Meis3 loss alters the specification of pancreatic endodermal progenitors, we also examined the expression of pdx1 in control and Meis3 morpholino (MO)–injected embryos. During zebrafish pancreas specification, pdx1-positive endoderm gives rise to the dorsal pancreatic bud and the ventral pancreatic anlage by 32 hpf (Biemar et al., 2001; Roy et al., 2001; Field et al., 2003). No significant difference in the expression domain of pdx1 was observed between control and Meis3 MO–injected embryos at 36 hpf (Supplemental Figure S2, E and F; arrows; n = 20/20). Thus Meis3 is likely not required for the specification or survival of gut and pancreatic endoderm, suggesting that vagal neural crest migration defects in Meis3-depleted embryos likely do not stem from a lack of foregut endoderm tissue.
Mesoderm-derived hand2 is required for the efficient migration of enteric precursors along the gut in zebrafish embryos during ENS development (Reichenbach et al., 2008). To investigate whether hand2 localization was altered after loss of Meis3, possibly contributing to defects in enteric neural crest migration, we also examined hand2 localization at 52 hpf by whole-mount in situ hybridization. Compared with control-injected embryos (Supplemental Figure S2G; arrows; n = 20/20), Meis3 morphants and pbcab-injected embryos exhibited largely normal hand2 expression domains along the gut (Supplemental Figure S2, H and I; arrows; n = 18/20 and 17/20, respectively). These results suggest that Meis3 is not required for specification of hand2-positive mesoderm.
Meis3 is required for the efficient migration of enteric neural crest along the developing gut
During early zebrafish ENS development, enteric neural crest cells migrate as two chains from the postotic vagal regions and subsequently along the left and right sides of the developing gut tube (Shepherd et al., 2004; Olden et al., 2008). Making their way caudally along it, they colonize more than one-fourth of the length of the gut by 54 hpf, and by 66 hpf, they reach the caudal end of the hindgut (Olden et al., 2008). These migratory enteric progenitor cells are marked by the expression of phox2bb, a transcription factor required for the specification of enteric neural crest fate (Elworthy et al., 2005). To investigate whether Meis3 is required for the specification of enteric neural crest fate and/or localization of enteric progenitors along the rostral-caudal axis of the gut, we performed in situ hybridization against phox2bb. In control embryos, the phox2bb+ cell population had left the postotic vagal region by 52 hpf (Figure 3A, arrow) and migrated caudally, progressing along 40% of the length of the gut (Figure 3D), as evidenced by two migratory streams of phox2bb+ enteric precursors localized along the left and right sides of the gut tube at midgut level (Figure 3G; arrows, dorsolateral view; n = 30/30). In contrast, Meis3 morphants and pbcab-injected embryos possessed an expanded postotic vagal phox2bb+ domain at 52 hpf (Figure 3, B and C; arrows; n = 25/30 and 28/30, respectively). Along the gut of Meis3 morphants, phox2bb+ enteric precursors were located more rostrally (at foregut level) than control embryos and had only progressed 25% of the length of the gut (Figure 3, E and H; arrows, dorsolateral view). pbcab-injected embryos exhibited a more severe phenotype, with phox2bb+ enteric precursors located in the anterior foregut (Figure 3, F and I; arrow, dorsolateral view), only migrating 10% of the length of the gut. In transverse sections, although control embryos had enteric precursors localized on the left and right sides of the gut tube along the posterior foregut (Figure 3J, arrows), Meis3 morphants and pbcab-injected embryos had no detectable enteric precursors in a comparable location (Figure 3, K and L). These results indicate that Meis3 is required for the timely and efficient migration of enteric neural crest to and along the developing gut but that their specification occurs normally.
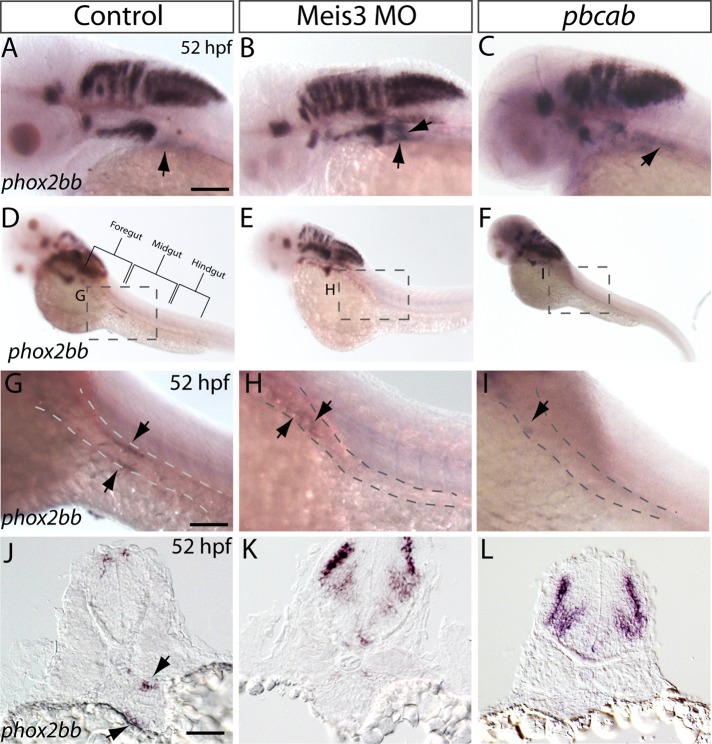
FIGURE 3:
Localization of phox2bb+ neural crest along the gut is altered after loss of Meis3. Lateral views of phox2bb localization within the cranial regions of (A) control, (B) Meis3 morphant, and (C) pbcab-injected embryos at 52 hpf reveals that phox2bb+ vagal neural crest cells have left the vagal region of control embryos (arrow) but persist in Meis3 morphants and pbcab-injected dominant-negative embryos (arrows) at the same location. phox2bb+ enteric neural crest cells are localized along the left and right sides of the midgut in (D, G) control embryos, whereas enteric crest cells localize rostrally within the foregut of (E, H) Meis3 morphants and are present in the most anterior region of the foregut in (F, I) pbcab-injected embryos. Sectioned views confirm that although phox2bb+ enteric crest cells localize around the gut tube in (J) control embryos (arrows), they are not detected at the comparable level in (K) Meis3 morphant and (L) pbcab-injected embryos. Scale bars, 50 μm (A–F), 40 μm (G–I).
To test further the hypothesis that loss of Meis3 function effects cell migration, we next used in vivo time-lapse confocal microscopy to examine directly enteric neural crest migration in control embryos compared with Meis3 morphants. To this end, we imaged the gut region of control and Meis3-morphant double-transgenic embryos positive for sox10:mRFP (membrane red fluorescent protein [RFP]+ neural crest) and sox17:GFP (GFP+ gut endoderm). Images were assumed laterally, as depicted in the schematic (Figure 4A), during the second day in development (∼54–57 hpf), when enteric neural crest cells are migrating along the developing gut tube. In control embryos, sox10:mRFP+ neural crest cells were observed migrating caudally as a chain along the sox17:GFP+ midgut at a rate of 20 μm/h (Figure 4C; arrows over time; n = 3; Supplemental Movie S1). In contrast, in Meis3 morphants, sox10:mRFP+ neural crest cells were observed at much more rostral locations along the sox17:GFP+ foregut at comparable times (Figure 4A) and exhibited a much slower migration rate of 10 μm/h (Figure 4B; arrows over time; n = 3; Supplemental Movie S2). Indeed, in agreement with phox2bb in situ analysis (Figure 3), these observations indicate that enteric neural crest migration is compromised after loss of Meis3 and suggest that Meis3 is required for the efficient migration of enteric neural crest along the developing gut.
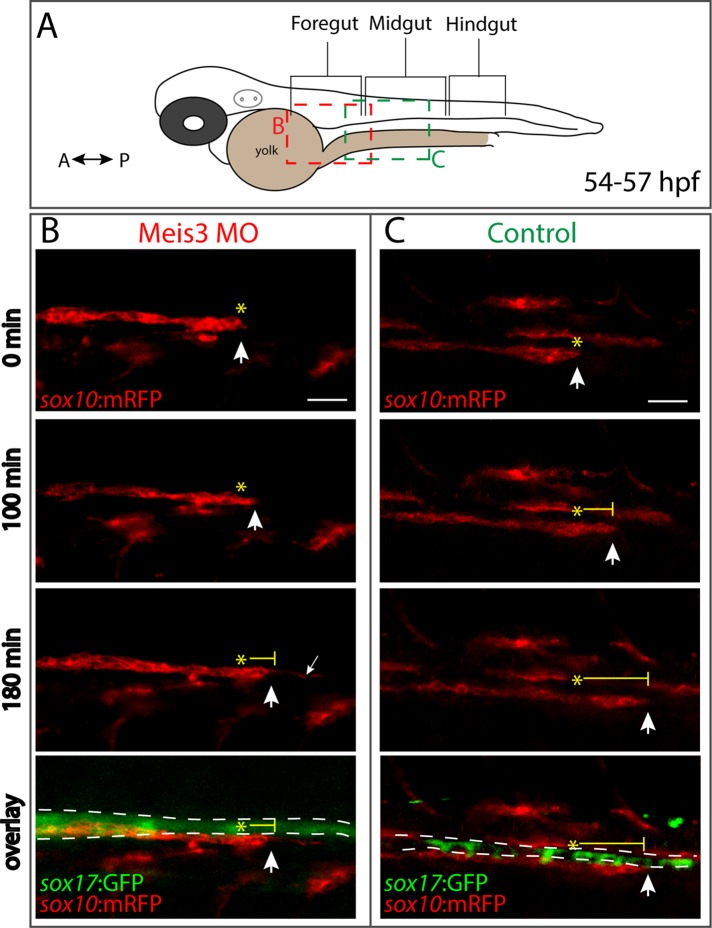
FIGURE 4:
Meis3 is required for efficient migration of enteric neural crest cells along the gut. (A) Schematic depicting where along the gut axis live time-lapse movies were acquired. Colored boxes depict where neural crest streams were observed in Meis3 morphant (red box) and Control embryos (green box). Lateral cropped view of time-lapse stills depicting sox10:mRFP+; sox17:GFP+ (B) Meis3 morphant and (C) control embryos over time during the second day in development (∼54–57 hpf). (B) In Meis3 morphant embryos, sox10:mRFP+ enteric neural crest chains were observed migrating at a rate of 10 μm/h along the posterior end of the foregut, where they migrated 30 μm within 3 h (arrow over time). In Meis3 morphants, the leading edge of the migratory front exhibited membrane protrusions caudally along the gut (small arrow). (C) In control embryos, sox10:mRFP+ enteric neural crest migratory chains were observed migrating at a rate of 23 μm/h along the midgut, where they migrated 70 μm within 3 h (arrow over time). Asterisks mark beginning of migratory front; bottom, merging shows location of sox10:mRFP+ enteric neural crest cells (red) along the sox17:GFP+ gut endoderm (green). Scale bar, 40 μm.
Loss of Meis3 results in colonic aganglionosis during ENS development
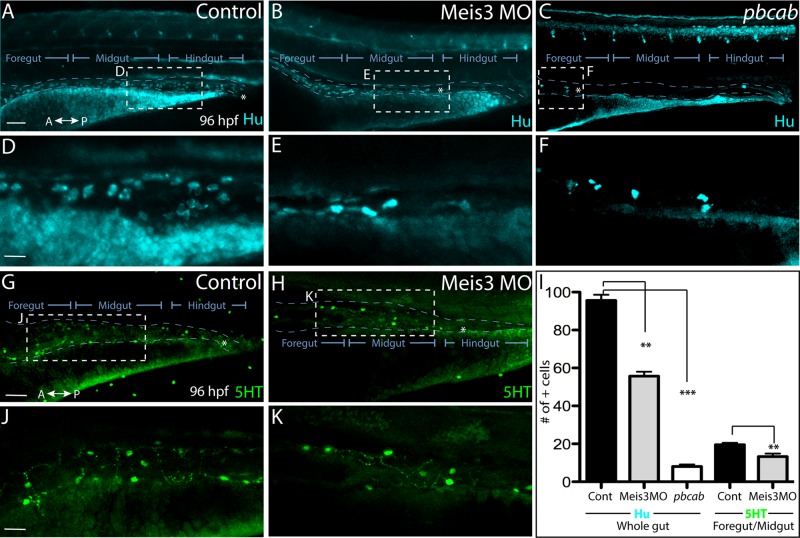
Because Meis3 loss results in perturbed enteric neural crest migration along the gut, it is possible that this may lead to defects in neuronal differentiation. During zebrafish enteric neuron differentiation, the first differentiating enteric neurons are detected along the anterior gut at ∼55 hpf (Elworthy et al., 2005; Olden et al., 2008), and by 74 hpf, Hu+ enteric neurons appear at the most caudal end of the gut, near the anus (Olden et al., 2008; Olsson et al., 2008). To examine neuronal differentiation, we analyzed control, Meis3 MO, and pbcab-injected embryos by whole- mount immunohistochemistry at 96 hpf, when enteric neurons encompass the entire length of the gut, using antibodies against the panneuronal marker Hu (Figure 5). In control embryos, Hu+ neurons were observed along the entire length of the gut, all the way to the anus (Figure 5, A and D; n = 12). In Meis3 morphants, Hu+ neurons were only present within the foregut and midgut regions, with no Hu+ neurons detected in the hindgut (Figure 5, B and E; n = 10/12), indicating the Meis3 loss results in colonic (hindgut) aganglionosis, a disorder in which the hindgut is devoid of neurons (Bergeron et al., 2013). Demonstrating a more severe phenotype, pbcab-injected embryos exhibited total intestinal aganglionosis, in which all but the foregut is devoid of neurons (Figure 5, C and F; n = 12/12). Compared with control embryos, which contained an average of 95.6 Hu+ neurons along the entire gut (Figure 5I, n = 8), Meis3-deficient embryos contained significantly less, with an average of 55.6 Hu+ neurons along the entire gut (Figure 5I, p = 0.0004, n = 8), whereas pbcab-injected embryos contained only eight (Figure 5I, p = 0.0001, n = 8).
FIGURE 5:
Loss of Meis3 leads to colonic aganglionosis during ENS development. Whole-mount immunohistochemistry against Hu at 96 hpf reveals enteric neurons present throughout the entire length of the gut of (A) control embryos, while they are absent from the hindgut region of (B) Meis3 morphants and present only within the foregut of (C) pbcab-injected embryos. (D) Zoomed-in view from A shows enteric neuron density and morphology in control embryos, whereas zoomed-in views from (E) Meis3 morphants in B and (F) pbcab-injected embryos in C reveal fewer neurons present after loss of Meis3. (G, J) Whole-mount immunohistochemistry against 5HT neurotransmitter in control embryos reveals the presence of terminally differentiated neurons throughout the gut. (H, K) In Meis3 morphants, the presence of 5HT+ terminally differentiated neurons was evident within the foregut and midgut but not the hindgut region, marked by asterisk. (I) Graph quantifying total Hu neurons in control-, Meis3 MO–, and pbcab-injected guts at 96 hpf, as well as total 5HT neurons within the foregut-midgut of control and Meis3 morphant guts. **p < 0.05; ***p < 0.01. Error bars indicate ± SEM. Scale bar, 80 μm (A– C, G, H), 30 μm (D–F, J, K).
We also examined the localization of 5-hydroxytryptamine (5HT) neurotransmitter as an indicator of terminal differentiation. In control embryos, 5HT was detected in the gut (Figure 5, G and J), with control foregut–midgut regions containing an average of 19.5 5HT+ neurons (Figure 5I; n = 8). In Meis3 morphants, 5HT+ neurons were detected, with an average of 13.25 5HT+ neurons in the foregut–midgut region (Figure 5, H, I, and K; p = 0.018, n = 6); however, no neurons were detected in the hindgut (Figure 5H, asterisk). Therefore, although Meis3-deficient enteric neural crest cells have the capacity to differentiate into enteric neurons, there are fewer of them compared with control and they are absent from the hindgut. Taken together, these data suggest that Meis3 is required for efficient colonization of the entire gut (Figures 3 and 4) such that its loss causes a deficit of enteric neurons within the hindgut region (Figure 5).
Loss of Meis3 results in a decrease in the number and proliferation of enteric neural crest cells around the gut
The observation that decreased numbers of enteric neurons form within the ENS in Meis3-deficient embryos (Figure 5I) suggests that Meis3 might play an important functional role in controlling the appropriate number of enteric neurons that are born during development. Enteric neural crest cells actively proliferate as they migrate (Pietsch et al., 2006; Olden et al., 2008). One current model for gut colonization is that neural crest cells undergo a proliferative expansion into new gut territory as they migrate, driving the neural crest front forward (Landman et al. 2007). Accordingly, the decreased efficiency of neural crest migration and diminished enteric neuron formation may reflect a decrease in precursor cell number and/or their proliferation during migration.
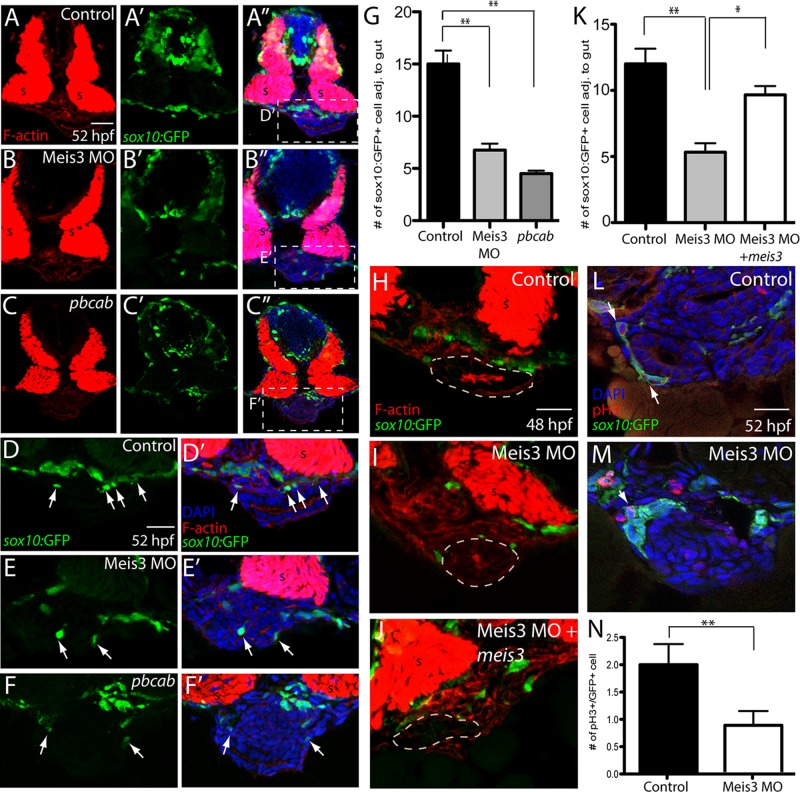
To test this possibility, we performed immunohistochemistry on serial transverse cryosections at caudal foregut levels at 52 hpf and quantified the number of neural crest cells near the gut (Figure 6). In control sections, sox10:GFP+ enteric neural crest cells were abundantly detected around the gut (Figure 6, A–A′′, D, and D′, arrows), with an average of 15 cells/section (Figure 5G, n = 6). In Meis3 morphants, the number of sox10:GFP+ neural crest cells was significantly reduced, with an average of 6.7 cells/section (Figure 6, B–B′′, E, E′, and G; p = 0.0012, n = 6). Similarly, pbcab-injected embryos exhibited only 4.5 cells/section (Figure 6, C–C′′, F, F′, and G; p = 0.0002, n = 6). To determine whether the cell number defect in Meis3 morphants could be rescued, we coinjected meis3 mRNA, which cannot be bound by Meis3 MO, with Meis3 MO into sox10:GFP embryos and analyzed it at 48 hpf by immunohistochemistry. Control embryos exhibited an average of 12 sox10:GFP+ enteric neural crest cells near the gut (Figure 6, H and K, n = 6), whereas Meis3 morphants contained significantly fewer, on average 5.3 (Figure 6, I and K; p = 0.0075, n = 6). Coinjection of embryos with Meis3 MO and meis3 resulted in a partial rescue in neural crest cell number around the gut, with embryos exhibiting an average of 9.6 sox10:GFP+ cells, a significant increase compared with Meis3 morphants (Figure 6, J and K; p = 0.01, n = 6).
FIGURE 6:
Meis3 function is required for the proper number of enteric neural crest cells around the gut and for their proliferation. By use of immunohistochemistry, enteric neural crest cell numbers were analyzed in (A–A′′) control-, (B–B′′) Meis3 MO–, and (C–C′′) pbcab-injected sox10:GFP+ embryos at 52 hpf. Zoomed-in views of control- (D, D′), Meis3 MO– (E, E′), and pbcab-injected embryos (F, F′) reveal fewer enteric neural crest cells near the gut after loss of Meis3 or Meis function; sox10:GFP (green), F-actin (red), and DAPI (blue). (G) Bar graph showing quantification of enteric neural crest near the gut in control-, Meis3 MO–, and pbcab-injected embryos at 52 hpf. (H–K) Rescue experiment immunohistochemistry and quantification showing that coinjection of meis3 mRNA, which cannot be bound by Meis3 MO, along with Meis3 MO (J) was sufficient to partially rescue enteric neural crest cell number near the gut at 48 hpf (K) when compared with injection of Meis3 MO alone (I, K); sox10:GFP (green), F-actin (red). (L) Control immunohistochemistry against the mitotic marker pH3 at 52 hpf and in (M) Meis3 MO–injected embryos; sox10:GFP (green), pH3 (red), and DAPI (blue). (N) Bar graph showing quantification of mitotic neural crest around the gut in control- and Meis3 MO–injected embryos. For all graphs, *p < 0.05; **p < 0.01. Error bars indicate ± SEM. Scale bars, 40 μm (A–C), 20 μm (D–F), 20 μm (H–M).
To test the hypothesis that reduced neural crest cell number in Meis3 morphants could be due to changes in their proliferation, we analyzed cell division in serial transverse sections using an antibody against phospho–histone H3, which marks mitotic cells in G2/M phase of the cell cycle. In control sections, mitotic sox10:GFP+ cells were present around the gut at an average of two cells/section (Figure 6, L and N, arrow; n = 6), whereas Meis3 MO–injected embryos exhibited fewer than an average of one mitotic cell (Figure 6, M and N, arrow; p = 0.02, n = 6). Collectively these quantitative data suggest that Meis3 is necessary for the correct numbers of enteric neural crest cells around the gut and for their proliferation, suggesting that the paucity of enteric neurons later in development results, at least partially, from reduction of the total migratory population.
Sonic Hedgehog pathway is disrupted after loss of Meis3
Shh signaling is required for proper ENS patterning and development in mice and zebrafish (Sukegawa et al., 2000; Ramalho-Santos et al., 2000; Reichenbach et al., 2008; Tobin et al., 2008) and for the proliferation and differentiation of enteric neural crest cells in explant culture (Fu et al., 2004) and zebrafish embryos (Reichenbach et al., 2008). Moreover, previous experiments showed that the dominant-negative Pbx4 construct pbcab leads to reduced endodermal shh expression domain (diIorio et al., 2007).
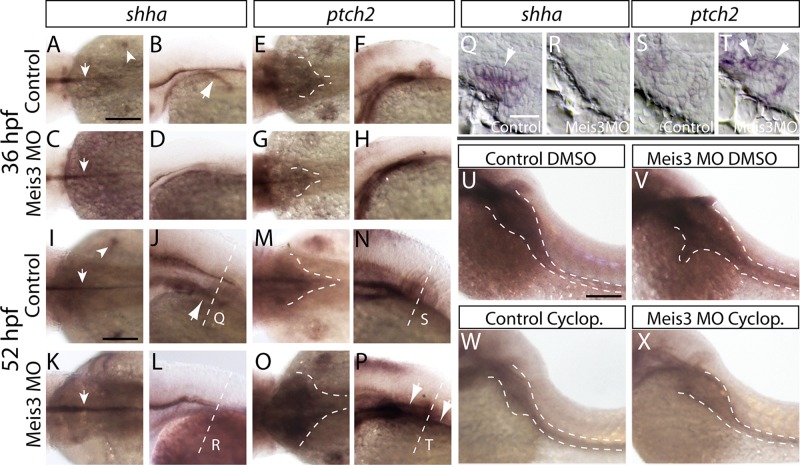
To determine whether Meis3 loss altered Shh pathway component expression during ENS phases of development, we performed in situ hybridization against shha, the zebrafish Shh orthologue, and ptch2 (zebrafish orthologue of Ptc1, formerly known as ptc-1) at 36 and 52 hpf in control and Meis3 MO–injected embryos. In control embryos at 36 hpf, shha transcript was observed in the notochord, fin buds, and endoderm (Figure 7, A and B; arrows, n = 30/30). In Meis3 MO–injected embryos, shha was detected in the notochord but not in the fin buds or the endoderm (Figure 7, C and D; arrow, n = 28/30). Accordingly, expression of the Shh receptor ptch2 was also reduced in the fin buds and diminished within the anterior foregut region in Meis3 MO–injected embryos (Figure 7, G and H; dashed lines; n = 26/30) compared with control (Figure 7, E and F; n = 30/30 normal). At 52 hpf, the lack of fin bud and endodermal shha expression was still pronounced in Meis3 MO–injected embryos (Figure 7, K and L; arrow, n = 28/30) compared with control embryos (Figure 7, I and J; n = 30/30 normal). By contrast, the foregut expression domain of ptch2 appeared expanded in Meis3 MO–injected embryos (Figure 7, O and P; dashed lines; n = 27/30) compared with controls (Figure 7, M and N; n = 30/30 normal). To obtain enhanced resolution of the shha and ptch2 expression territories around the gut, we imaged sections after in situ hybridization at 52 hpf. In control sections, expression of shha was detected in the gut endoderm (Figure 7Q, arrow), and ptch2 was observed in the surrounding mesenchyme (Figure 7S). By contrast, in Meis3 MO–injected embryos, shha was reduced (Figure 7R), whereas ptch2 was increased (Figure 7T, arrows).
FIGURE 7:
Meis3 loss is sufficient to alter the expression of Sonic Hedgehog pathway components during ENS phases of development. The expression of shha and ptch2 was examined by in situ hybridization at 36 hpf in control- (A, B and E, F, respectively) and Meis3 MO–injected embryos (C, D and G, H, respectively) and at 52 hpf in control- (I, J and M, N, respectively) and Meis3 MO–injected embryos (K, L and O, P, respectively). Dorsal views in A, E, C, and G at 36 hpf and I, M, K, and O at 52 hpf. Lateral views in B, F, D, and H at 36 hpf and J, N, L, and P at 52 hpf. At 52 hpf, transverse sections through the gut revealed shha expression in gut endoderm of control embryos (Q, arrow) and its absence in Meis3 MO–injected embryos (R), whereas ptch2 was expanded in Meis3 MO–injected embryos (T, arrows) compared with control embryos (S). The expression domain of ptch2 along the gut (dashed lines) in control embryos treated with DMSO (U) and in Meis3 MO–injected embryos (V). Cyclopamine treatment from 32 to 52 hpf was sufficient to reduce ptch2 gut expression (dashed lines) in control- (W) and Meis3 MO–injected embryos (X) compared with treatment of DMSO alone. Scale bars, 70 μm (A–P, U–X), 20 μm (Q–T).
Although Ptch1 is a negative regulator of the Hedgehog pathway, it is also up-regulated in response to Smoothened-dependent Hedgehog signaling activity (Concordet et al., 1996; Lewis et al., 1999; Johnson et al., 2000). To ascertain whether the expansion in ptch2 expression could be smoothened dependent, we treated control and Meis3 MO–injected embryos with 10 μM cyclopamine, a steroidal Hedgehog pathway inhibitor that acts by binding Smoothened (Chen et al., 2002), or dimethyl sulfoxide (DMSO) from 32 to 52 hpf and assayed the expression of ptch2 by in situ hybridization. In control embryos treated with DMSO, ptch2 was expressed broadly in the anterior foregut region and narrowed around the gut tube caudally (Figure 7U; dashed lines; n = 20/20). Meis3 MO–injected embryos treated with DMSO had laterally expanded ptch2 expression domains along the length of the gut (Figure 7V; dashed lines; n = 17/20). Meis3 MO–injected embryos treated with cyclopamine were largely “rescued” and exhibited reduced ptch2 expression fields (Figure 7X; n = 20/20), similar to control embryos (Figure 7W; n = 20/20). This notable rescue suggests that ptch2 up-regulation in Meis3-deficient embryos is due to elevated Hedgehog signaling.
DISCUSSION
Meis transcription factors are members of the TALE homeodomain subclass of protein involved in the intrinsic control of varied roles during embryonic development (for review, see Moens and Selleri, 2006; Cerdá-Esteban and Spagnoli, 2014). They are best known to function as part of a dimeric or trimeric complex, along with Pbx and/or Hox transcription factors, in order to regulate activation of target genes in a tissue-specific manner. For example, in zebrafish and mouse embryos, Meis1/2 and Hoxb1b bind the promoter of Hoxb1a to influence the magnitude of transcriptional output in a conserved and temporal manner during branchial arch development (Choe et al., 2014). In this study, we identified zebrafish Meis3 as a novel factor involved in ENS development. Meis3 loss was sufficient to perturb enteric neural crest proliferation and migration along the developing gut in zebrafish embryos. As a result, fewer enteric neurons were born in the ENS, leading to colonic aganglionosis—directly implicating Meis3 in ENS development and highlighting it as a novel candidate gene in Hirschsprung’s disease. Indeed, Meis3 loss resulted in the aberrant expression of Shh pathway components, which have known roles in the etiology of this disorder.
During ENS development, enteric neural crest precursors must proliferate in order to create sufficient numbers of enteric neurons and glial cells needed to populate the mature ENS. In humans, for instance, the total number of neurons rivals those found in the spinal cord, with numbers between 200 million and 600 million (Furness, 2006). Therefore enteric neural crest cells must maintain proliferation and prevent differentiation until the appropriate time, all while migrating through the gut. Our results demonstrate that Meis3 loss of function decreases the number of enteric precursors and their proliferation during zebrafish ENS development, contributing to ENS defects during later phases of development. This suggests that Meis3, in conjunction with yet-unknown cofactors, participates in controlling enteric precursor proliferation in order to ensure that the appropriate number of neurons is generated throughout the gut. Meis proteins have well-described roles in proliferation control in various tissues, making them logical candidates for proliferative regulators in neural crest cells. For example, Meis1 protein regulates the proliferative expansion of retinal progenitors during early phases of retinogenesis in zebrafish embryos by directly modulating the expression of ccnd1 (CyclinD1), a cell cycle progression factor (Bessa et al., 2008). Meis1 expression is associated with high levels of proliferation in neuroblastoma (Geerts et al., 2003), and its forced expression in immortalized myeloid progenitors leads to maintenance of self-renewal and blocks differentiation (Calvo et al., 2001). Meis2 is known as a player in the pathogenesis of cancer; its presence is essential for neuroblastoma proliferation via direct influence on progression of the M phase (Zha et al., 2014). In the frog hindbrain, using ectopic expression and knockdown of Meis3, Elkouby et al. (2012) showed that Meis3 is necessary and sufficient to induce the proliferation of neural progenitors while inhibiting their precocious differentiation downstream of Wnt signaling.
In addition to implicating Meis3 in regulation of enteric neural crest cell number, our experiments reveal a potential role for Meis proteins in migration of the neural crest. Although it has no effect on enteric neural crest specification, Meis3 loss causes a delay in the migration of vagal neural crest to the foregut and decreases the efficiency of migration of enteric neural crest along the gut. Meis3-deficient enteric neural crest migrated much more slowly along the gut than did control enteric crest, as assayed by monitoring the localization of enteric progenitors using the marker phox2bb (Figure 3) and live imaging of sox10:mRFP+ neural crest migration along the gut (Figure 4). The combined effects of decreased proliferation and slower migration contribute to diminished colonization of the caudal regions of the gut. We hypothesize that this leads to colonic aganglionosis. It is possible that defects in proliferation may attenuate the migratory progress of enteric crest to and along the gut as well, significantly contributing to lack of gut colonization. Indeed, enteric neural crest cells are highly proliferative during migration, and this proliferation is believed to drive the migratory front forward during gut colonization (Landman et al. 2007). Although Meis1 has been implicated in influencing early induction of neural crest cell fate in frog embryos (Maeda et al., 2002), there were no previously published reports of the Meis family playing a role in regulation of neural crest migration. Of interest, however, Meis protein expression has been described in diverse migratory cell types. For example, Meis1 is expressed in migratory cortical neurons of the embryonic mouse brain and neuroblastoma cancer cells (Friocourt and Parnavelas, 2011), and Meis 1/2/3 protein was detected within migratory and proliferative cerebellar neural stem cells of the adult brown ghost knifefish brain, implying functional roles for the Meis genes in proliferation and migration during adult homeostasis and regeneration (Sirbulescu et al., 2015). Here we describe meis3 expression within a subset of vagal neural crest cells migrating into the foregut (Figure 1). Of importance, all of these studies in varied model organisms suggest that Meis regulation may be a conserved method that cells use during tissue development, cell homeostasis, and cancer metastasis to regulate proliferation and migration.
Other transcription factors in addition to Meis3 have been shown to play roles in ENS development. Intrinsically, the transcription factor Phox2b is required for enteric cell fate specification in amniotes and zebrafish (Pattyn et al., 1999; Elworthy et al., 2005), and is required for Ret expression in the ENS (Pattyn et al., 1999). Highlighting its conserved and integral role in ENS development, zebrafish and mouse mutants for the transcription factor Sox10 completely lack ENS (Pingault et al., 1998; Kelsh and Eisen, 2000; Dutton et al., 2001; Carney et al., 2006), and Sox10 is required for the expression of Phox2b (Kim et al., 2003; Elworthy et al., 2005), indicating that it lies upstream of enteric neural crest specification. In addition, mutation or loss of Foxd3 (Montero-Balaguer et al., 2006; Stewart et al., 2006), Tfap2α (Arduini et al., 2009), Pax3 (Minchin and Hughes, 2008), Hoxb5 (Lui et al., 2008; Kam et al., 2014), and Hand2 (Reichenbach et al., 2008) all result in either a lack of enteric neural crest colonization of the gut or perturbed ENS development, highlighting the complexity of ENS development. Possible candidates for binding partners of Meis3 include pbx1a, which is expressed in the zebrafish vagal and foregut environment (Teoh et al., 2010), and hoxb3a and hoxb5b, which are expressed in the postotic vagal regions and gut mesoderm during enteric neural crest phases of development (Kudoh et al., 2001; Jarinova et al., 2008). In the mouse, both Hoxb3 and Hoxb5 have been implicated in vagal and enteric neural crest migration and development (Chan et al., 2005; Lui et al., 2008). In addition, we cannot rule out the possibility that other Meis proteins in addition to Meis3 are important for ENS development.
The Shh signaling pathway plays conserved roles in regulating the patterning, proliferation, and differentiation of various cell types (for review, see Choudhry et al., 2014). During ENS development in chicken embryos, temporally inhibiting Sonic hedgehog pathway activity with cylopamine leads to increased enteric neuron number, along with enhanced smooth muscle differentiation (Sukegawa et al., 2000). In mice, loss of Shh ectopically expands neuron distribution throughout the gut (Ramalho-Santos et al., 2000). In contrast, loss of shha, smoothened, or cilia function in zebrafish leads to complete loss of enteric neural crest cells in the gut at all phases of ENS development (Reichenbach et al., 2008; Tobin et al., 2008; Tu et al., 2012), whereas overexpression of shha creates an overabundance of enteric neural crest cells along the gut (Reichenbach et al., 2008). Thus the role of Shh in ENS development varies considerably among species.
Testing the hypothesis that the Shh signaling pathway was disrupted after Meis3 loss, we discovered that the expression of shha and its receptor ptch2 was decreased during early phases of enteric neural crest migration, at 36 hpf. During later phases of ENS development, at 52 hpf, however, we observed that although shha was diminished in the gut environment, the expression of ptch2 was expanded in a Smoothened-dependent manner, as incubation of Meis3 morphants in cyclopamine was sufficient to rescue the expansion of ptch2 at 52 hpf. These results suggest that the Hedgehog signaling pathway is disrupted after loss of Meis3. In mice, it was previously shown that Indian Hedgehog (Ihh) is expressed in the developing gut epithelium and its loss results in ENS defects, similar to what is seen in Hirschsprung’s disease (Ramalho-Santos et al., 2000; Mao et al., 2010). ihha is expressed during zebrafish intestinal development and required for the proper development of the gut and ENS (Winata et al., 2009; Korzh et al., 2011). One possibility is that ihha is up-regulated in Meis3 morphants, contributing to ENS defects during development. Alternatively, Meis3 may directly effect the expression of ptch2 to influence ENS development. Indeed, Ptch1 has recently been implicated as a novel factor involved in the pathogenesis of Hirschsprung’s disease; in humans and mice, single- nucleotide polymorphisms have been associated with high risk for this disorder, and conditional loss of Ptch1 within enteric neural crest cells causes a decreased ability to differentiate into enteric neurons in culture (Ngan et al., 2011). Collectively the results suggest a model in which loss of Meis3 effects Shh activity during early phases of vagal neural crest entry into the gut, leading to a delay in gut invasion, while also contributing to perturbed migration along the gut during later phases.
MATERIALS AND METHODS
Zebrafish maintenance
Zebrafish (Danio rerio) were maintained at 28.5°C on a 13-h light/11-h dark cycle. Animals were treated in accordance with California Institute of Technology Institutional Animal Care and Use Committee provisions.
Morpholino, mRNA injections, and mRNA rescue experiment
Meis3 MO (5′-ATCCATGCGATACGGAAGCCGAGCT-3′; Genetools, LLC, Philomath, OR; diIorio et al., 2007) directed against the 5′ untranslated region of meis3 was used in morpholino knockdown experiments. Standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was used as a negative control. Wild-type AB strain and Tg(-4.9sox10:GFP) (Carney et al., 2006) were injected with 2.3 nl of 0.75 mM Meis3 MO at the one-cell stage using a Nanoliter 2000 injector (World Precision Instruments, Sarasota FL). For pbcab injections, the pCS2-pbcab-myc construct (Choe et al., 2002), which encodes a truncated version of the pbx4 gene tagged with myc, was linearized with Not1, and capped mRNA was transcribed using the Sp6 RNA synthesis kit (Ambion, Waltham, MA). A 200-pg amount of pbcab was injected into one-cell-stage embryos for dominant-negative experiments. For mRNA rescue experiments, the meis3 coding sequence was PCR amplified from a 28-hpf zebrafish cDNA library starting from the start codon, thereby removing 19 base pairs of morpholino recognition sequence, with primers to add a 5′ EcoRI restriction site and a 3′ XbaI restriction site, respectively, as follows: forward, 5′-CGGAATTCCATGGATAAGAGG-3′; and reverse, 5′-GCTCTAGATCAGTGGGCATGT-3′. The resulting meis3 PCR fragment was then gel excised, digested with EcoRI and XbaI, directionally cloned into pCS2+, and sequence verified. pCS2-meis3 was linearized with NotI and used as a template to generate meis3 capped mRNA with the Sp6 RNA synthesis kit (Ambion). A 150-pg amount of meis3 mRNA was coinjected with Meis3 MO for rescue experiments at the one-cell stage and embryos fixed at 48 hpf for immunohistochemistry as described later or processed at 25 hpf for Western blot analysis.
Western blot
Control-injected and Meis3 MO–injected embryos were anesthetized using tricaine, deyolked manually using fine forceps, and transferred to 1.7-ml Eppendorf tubes on ice. Embryos were rinsed twice in 1× phosphate-buffered saline (PBS) and then immediately homogenized in protein lysis buffer (5 mM EDTA, 50 mM Tris, pH 8, 100 mM NaCl, 1% NP-40) supplemented with Complete protease inhibitor (Roche, Madison, WI). Protein homogenate was denatured using Invitrogen NuPAGE LDS sample buffer and reducing agent per manufacturer’s instructions, loaded, and run in a Novex Bis-Tris SDS–PAGE gel. Proteins were transferred onto nitrocellulose membrane, blocked in 5% milk/1× PBS/Tween-20 (PBST) for 1 h at room temperature and incubated in rabbit anti-Meis3 (1:1000; ab82761; Abcam, Cambridge, MA) overnight at 4°C. The blot was incubated in horseradish peroxidase–conjugated anti-rabbit secondary 1:13,000 for 2 h at room temperature and developed using the GE Healthcare ECL detection system.
Riboprobes and in situ hybridization
Hybridizations were performed essentially as described (Jowett and Lettice, 1994). All experiments were repeated using three biological replicates. The following cDNA constructs were used as templates to generate riboprobes for whole-mount in situ hybridization: meis3 (Rauch et al., 2003), crestin (Luo et al., 2001), foxa1 (Odenthal and Nusslein-Volhard, 1998), pdx1 (Biemar et al., 2001), and ptch2 (Lee et al., 2008). The following cDNAs were PCR amplified from a 28-hpf cDNA library, ligated into pGEM-Teasy vector, linearized, and used as templates to generate probe: phox2bb, forward, 5′-TTCCTTCTCCACTGACCCTT-3′, and reverse, 5′-CGTCGCTTCTTTTCTCCATC-3′; hand2, forward, 5′-TGGTTCGCCGTAGGGTATAG-3′, and reverse, 5′-TCTTGTCATTGCTGCTCCCT-3′; shha, forward, 5′-CTACGGCAGAAGAAGACAT-3′, and reverse, 5′-CTGGCCGCTATCATCAACAA-3′.
Immunohistochemistry
For immunohistochemistry processing, embryos were fixed overnight at 4°C in 4% paraformaldehyde (PFA), washed three times in 1× PBS, and incubated in 15% sucrose/1× PBS overnight at 4°C. Embryos were then incubated at 38°C in 7.5% gelatin/15% sucrose overnight, aligned in rubber molds, and snap frozen in liquid nitrogen. Cryoblocks were sectioned at 12 μm and mounted onto glass slides, and gelatin was removed by incubating slides in 1× PBS in a slide holder at 42°C for 20 min. Immunohistochemistry was then performed as previously described (Uribe and Gross, 2007). The following antibodies were used on sections: rabbit anti-GFP 1:500 (A-11122; Life Technologies, Waltham, MA), goat anti-GFP 1:250 (ab6673; Abcam), and rabbit anti-pH3 1:250 (EMD Millipore, Billerica, MA). The following secondary antibodies were used: Invitrogen Alexa Fluor goat anti-rabbit 488, donkey anti-goat 488, and goat anti-rabbit 568 were used at 1:700. Hoechst 3358 and/or Alexa Fluor 568 phalloidin were included in with secondary incubations. For whole-mount immunohistochemistry, embryos at 28, 36, or 96 hpf were fixed overnight at 4°C in 4% PFA, rinsed three times in 1× PBS, incubated in 100% methanol at −20°C for 1 h, and then rehydrated stepwise into 1× PBS at room temperature. Embryos were then incubated in 100% acetone at −20°C for 11 min, rinsed three times in 1× PBS, and then digested at room temperature in 10 μg/ml Proteinase K for 12 min (28 hpf), 16 min (36 hpf), or 45 min (96 hpf). Embryos were then rinsed three times in 1× PBS, fixed for 10 min in 4% PFA at room temperature, rinsed three times in 1× PBS, and then incubated in 5% normal goat serum block diluted in 1× PBST supplemented with 1% dimethyl sulfoxide (DMSO) for 3 h. Embryos were then incubated in rabbit anti-GFP 1:500 (A-11122; Life Technologies), mouse anti-HuC/D 1:200 (Invitrogen, Waltham, MA), or rabbit anti-5HT 1:1000 (Immunostar, Hudson, WI) overnight at 4°C. Embryos were then washed out of primary antibody in 1× PBST and then incubated at room temperature in the secondary antibody Alexa Fluor goat anti-rabbit 488 or goat anti-mouse 568 (1:700; Invitrogen) for 3 h at room temperature. Embryos were rinsed in 1× PBST and imaged in 75% glycerol/1× PBS on a Zeiss 710 two-photon confocal microscope at the Beckman Imaging Center, California Institute of Technology.
Time-lapse imaging
Embryos positive for Tg(sox10:mRFP) and Tg(sox17:GFP) were mounted laterally in 1% low-melt agarose dissolved in fish water supplemented with 1× N-phenylthiourea and tricaine anesthetic in an imaging chamber. Z-stacks of 10–13 μm were acquired every 5 min for ∼3 h using a 20× objective on a Zeiss LSM 710 microscope. Z-stacks were compiled and exported using Imaris Image Analysis software.
Quantification and statistics
For all experiments, three biological replicates were used, with n ≥ 6 for each experiment. For cell counts on sections, counts were performed on adjacent sections for each embryo and averaged. All graphs and calculations of statistical significance and SEM were performed using a two-parameter unpaired t test (GraphPad Prism).
Inhibitor experiments
A master stock of 10 mM cyclopamine (Sigma-Aldrich [St. Louis, MO]; C4116-1MG) was made in DMSO and stored at −80°C. Control- and Meis3 MO–injected embryos were incubated in 10 μM cyclopamine or DMSO in embryo medium between 32 and 52 hpf at 28.5°C and then immediately fixed and processed for in situ hybridization as described.
Supplementary Material
Acknowledgments
We thank Charles Sagerström for the pCS2-pbcab-myc construct, Tatiana Hochgreb-Hägele for foxa1 cDNA, Shuo Lin for pdx1 cDNA, and Jeff Gross for ptch2 cDNA. We thank the Caltech Beckman Institute Biological Imaging Center and Fish Facility, Marcos Simões-Costa for advice with double fluorescence in situ hybridization, and Martha Henderson and David Mayorga for fish care. Research was supported from grants from the National Institutes of Health (DE024157) to M.E.B. and National Institutes of Health F32 (HD080343) to R.A.U. and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Fellowship to R.A.U.
Abbreviation used:
- ENS
enteric nervous system.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-02-0112) on September 9, 2015.
REFERENCES
- Aamar E, Frank D. Xenopus Meis3 protein forms a hindbrain-inducing center by activating FGF/MAP kinase and PCP pathways. Development. 2004;131:153–163. doi: 10.1242/dev.00905. [DOI] [PubMed] [Google Scholar]
- Agoston Z, Heine P, Brill MS, Grebbin BM, Hau AC, Kallenborn-Gerhardt W, Schramm J, Gotz M, Schulte D. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 2014;141:28–38. doi: 10.1242/dev.097295. [DOI] [PubMed] [Google Scholar]
- Anderson RB, Stewart AL, Young HM. Phenotypes of neural-crest-derived cells in vagal and sacral pathways. Cell Tissue Res. 2006;323:11–25. doi: 10.1007/s00441-005-0047-6. [DOI] [PubMed] [Google Scholar]
- Arduini BL, Bosse KM, Henion PD. Genetic ablation of neural crest cell diversification. Development. 2009;136:1987–1994. doi: 10.1242/dev.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariki R, Morikawa S, Mabuchi Y, Suzuki S, Nakatake M, Yoshioka K, Hidano S, Nakauchi H, Matsuzaki Y, Nakamura T, Goitsuka R. Homeodomain transcription factor Meis1 is a critical regulator of adult bone marrow hematopoiesis. PLoS One. 2014;9:e87646. doi: 10.1371/journal.pone.0087646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BA, Liyanage VR, Zachariah RM, Olson CO, Bailey MA, Rastegar M. Dynamic expression of MEIS1 homeoprotein in E14.5 forebrain and differentiated forebrain-derived neural stem cells. Ann Anat. 2013;195:431–440. doi: 10.1016/j.aanat.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Bergeron KF, Silversides DW, Pilon N. The developmental genetics of Hirschsprung’s disease. Clin Genet. 2013;83:15–22. doi: 10.1111/cge.12032. [DOI] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tavares MJ, Santos J, Kikuta H, Laplante M, Becker TS, Gomez-Skarmeta JL, Casares F. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135:799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Burns AJ. Migration of neural crest-derived enteric nervous system precursor cells to and within the gastrointestinal tract. Int J Dev Biol. 2005;49:143–150. doi: 10.1387/ijdb.041935ab. [DOI] [PubMed] [Google Scholar]
- Burns AJ, Champeval D, Le Douarin NM. Sacral neural crest cells colonise aganglionic hindgut in vivo but fail to compensate for lack of enteric ganglia. Dev Biol. 2000;219:30–43. doi: 10.1006/dbio.1999.9592. [DOI] [PubMed] [Google Scholar]
- Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci USA. 2001;98:13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Tsukui T, Rodriquez Esteban C, Zappavigna V, Izpisua Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol Cell. 1999;4:839–849. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machín M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–30. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Cerdá-Esteban N, Spagnoli FM. Glimpse into Hox and tale regulation of cell differentiation and reprogramming. Dev Dyn. 2014;243:76–87. doi: 10.1002/dvdy.24075. [DOI] [PubMed] [Google Scholar]
- Chan KK, Chen YS, Yau TO, Fu M, Lui VC, Tam PK, Sham MH. Hoxb3 vagal neural crest-specific enhancer element for controlling enteric nervous system development. Dev Dyn. 2005;233:473–483. doi: 10.1002/dvdy.20347. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SK, Ladam F, Sagerstrom CG. TALE factors poise promoters for activation by Hox proteins. Dev Cell. 2014;28:203–211. doi: 10.1016/j.devcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SK, Lu P, Nakamura M, Lee J, Sagerstrom CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SK, Vlachakis N, Sagerstrom CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Choudhry Z, Rikani AA, Choudhry AM, Tariq S, Zakaria F, Asghar MW, Sarfraz MK, Haider K, Shafiq AA, Mobassarah NJ. Sonic hedgehog signalling pathway: a complex network. Ann Neurosci. 2014;21:28–31. doi: 10.5214/ans.0972.7531.210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, Scott MP, Ingham PW. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- Dibner C, Elias S, Frank D. XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development. 2001;128:3415–3426. doi: 10.1242/dev.128.18.3415. [DOI] [PubMed] [Google Scholar]
- diIorio P, Alexa K, Choe SK, Etheridge L, Sagerstrom CG. TALE-family homeodomain proteins regulate endodermal sonic hedgehog expression and pattern the anterior endoderm. Dev Biol. 2007;304:221–231. doi: 10.1016/j.ydbio.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. The pattern of neural crest advance in the cecum and colon. Dev Biol. 2005;287:125–133. doi: 10.1016/j.ydbio.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Elkouby YM, Polevoy H, Gutkovich YE, Michaelov A, Frank D. A hindbrain-repressive Wnt3a/Meis3/Tsh1 circuit promotes neuronal differentiation and coordinates tissue maturation. Development. 2012;139:1487–1497. doi: 10.1242/dev.072934. [DOI] [PubMed] [Google Scholar]
- Elworthy S, Pinto JP, Pettifer A, Cancela ML, Kelsh RN. Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mech Dev. 2005;122:659–669. doi: 10.1016/j.mod.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Epstein ML, Mikawa T, Brown AM, McFarlin DR. Mapping the origin of the avian enteric nervous system with a retroviral marker. Dev Dyn. 1994;201:236–244. doi: 10.1002/aja.1002010307. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Friocourt G, Parnavelas JG. Identification of Arx targets unveils new candidates for controlling cortical interneuron migration and differentiation. Front Cell Neurosci. 2011;5:28. doi: 10.3389/fncel.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Lui VC, Sham MH, Cheung AN, Tam PK. HOXB5 expression is spatially and temporarily regulated in human embryonic gut during neural crest cell colonization and differentiation of enteric neuroblasts. Dev Dyn. 2003;228:1–10. doi: 10.1002/dvdy.10350. [DOI] [PubMed] [Google Scholar]
- Fu M, Lui VC, Sham MH, Pachnis V, Tam PK. Sonic hedgehog regulates the proliferation, differentiation, and migration of enteric neural crest cells in gut. J Cell Biol. 2004;166:673–684. doi: 10.1083/jcb.200401077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB. The Enteric Nervous System. Oxford, UK: Blackwell; 2006. [Google Scholar]
- Geerts D, Schilderink N, Jorritsma G, Versteeg R. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 2003;197:87–92. doi: 10.1016/s0304-3835(03)00087-9. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Pachnis V. Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies. Nat Rev Neurosci. 2007;8:466–479. doi: 10.1038/nrn2137. [DOI] [PubMed] [Google Scholar]
- Heine P, Dohle E, Bumsted-O’Brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135:805–811. doi: 10.1242/dev.012088. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Asano Y, Iida H, Watanabe T, Nakamura T, Goitsuka R. Meis1 is required for the maintenance of postnatal thymic epithelial cells. PLoS One. 2014;9:e89885. doi: 10.1371/journal.pone.0089885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarinova O, Hatch G, Poitras L, Prudhomme C, Grzyb M, Aubin J, Berube-Simard FA, Jeannotte L, Ekker M. Functional resolution of duplicated hoxb5 genes in teleosts. Development. 2008;135:3543–3553. doi: 10.1242/dev.025817. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Milenkovic L, Scott MP. In vivo functions of the patched protein: requirement of the C terminus for target gene inactivation but not Hedgehog sequestration. Mol Cell. 2000;6:467–478. doi: 10.1016/s1097-2765(00)00045-9. [DOI] [PubMed] [Google Scholar]
- Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Kam MK, Cheung MC, Zhu JJ, Cheng WW, Sat EW, Tam PK, Lui VC. Perturbation of Hoxb5 signaling in vagal and trunk neural crest cells causes apoptosis and neurocristopathies in mice. Cell Death Differ. 2014;21:278–289. doi: 10.1038/cdd.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN, Eisen JS. The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development. 2000;127:515–525. doi: 10.1242/dev.127.3.515. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Korzh S, Winata CL, Zheng W, Yang S, Yin A, Ingham P, Korzh V, Gong Z. The interaction of epithelial Ihha and mesenchymal Fgf10 in zebrafish esophageal and swimbladder development. Dev Biol. 2011;359:262–276. doi: 10.1016/j.ydbio.2011.08.024. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, Clarke CJ, Kiang A, Schultz S, Epstein JA, Toyama R, Dawid IB. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- Kuo BR, Erickson CA. Vagal neural crest cell migratory behavior: a transition between the cranial and trunk crest. Dev Dyn. 2011;240:2084–2100. doi: 10.1002/dvdy.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman KA, Simpson MJ, Newgreen DF. Mathematical and experimental insights into the development of the enteric nervous system and Hirschsprung’s disease. Dev Growth Differ. 2007;49:277–286. doi: 10.1111/j.1440-169X.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Teillet MA. The migration of neural crest cells to the wall of the digestive tract in avian embryo. J Embryol Exp Morphol. 1973;30:31–48. [PubMed] [Google Scholar]
- Lee J, Willer JR, Willer GB, Smith K, Gregg RG, Gross JM. Zebrafish blowout provides genetic evidence for Patched1-mediated negative regulation of Hedgehog signaling within the proximal optic vesicle of the vertebrate eye. Dev Biol. 2008;319:10–22. doi: 10.1016/j.ydbio.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Concordet JP, Ingham PW. Characterisation of a second patched gene in the zebrafish Danio rerio and the differential response of patched genes to Hedgehog signalling. Dev Biol. 1999;208:14–29. doi: 10.1006/dbio.1998.9169. [DOI] [PubMed] [Google Scholar]
- Lui VC, Cheng WW, Leon TY, Lau DK, Garcia-Barcelo MM, Miao XP, Kam MK, So MT, Chen Y, Wall NA, et al. Perturbation of hoxb5 signaling in vagal neural crests down-regulates ret leading to intestinal hypoganglionosis in mice. Gastroenterology. 2008;134:1104–1115. doi: 10.1053/j.gastro.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Maeda R, Ishimura A, Mood K, Park EK, Buchberg AM, Daar IO. Xpbx1b and Xmeis1b play a collaborative role in hindbrain and neural crest gene expression in Xenopus embryos. Proc Natl Acad Sci USA. 2002;99:5448–5453. doi: 10.1073/pnas.082654899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann CJ, Voz ML, Martial JA, Peers B. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development. 2007;134:4011–4021. doi: 10.1242/dev.007823. [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Kim BM, Rajurkar M, Shivdasani RA, McMahon AP. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 2010;137:1721–1729. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin VS, Feng W, Hernandez-Lagunas L, Artinger KB, Williams T. A morpholino-based screen to identify novel genes involved in craniofacial morphogenesis. Dev Dyn. 2013;242:817–831. doi: 10.1002/dvdy.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin JE, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317:508–522. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulos AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev Dyn. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- Natarajan D, Marcos-Gutierrez C, Pachnis V, de Graaff E. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002;129:5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- Ngan ES, Garcia-Barcelo MM, Yip BH, Poon HC, Lau ST, Kwok CK, Sat E, Sham MH, Wong KK, Wainwright BJ, et al. Hedgehog/Notch-induced premature gliogenesis represents a new disease mechanism for Hirschsprung disease in mice and humans. J Clin Invest. 2011;121:3467–3478. doi: 10.1172/JCI43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Noisa P, Raivio T. Neural crest cells: from developmental biology to clinical interventions. Birth Defects Res C Embryo Today. 2014;102(3):263–274. doi: 10.1002/bdrc.21074. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Olden T, Akhtar T, Beckman SA, Wallace KN. Differentiation of the zebrafish enteric nervous system and intestinal smooth muscle. Genesis. 2008;46:484–498. doi: 10.1002/dvg.20429. [DOI] [PubMed] [Google Scholar]
- Olsson C, Holmberg A, Holmgren S. Development of enteric and vagal innervation of the zebrafish (Danio rerio) gut. J Comp Neurol. 2008;508:756–770. doi: 10.1002/cne.21705. [DOI] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Pietsch J, Delalande JM, Jakaitis B, Stensby JD, Dohle S, Talbot WS, Raible DW, Shepherd IT. lessen encodes a zebrafish trap100 required for enteric nervous system development. Development. 2006;133:395–406. doi: 10.1242/dev.02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;2:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Melton DA, McMahon A. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- Rauch GJ, Lyons DA, Middendorf I, Friedlander B, Arana N, Reyes T, Talbot WS. Submission and Curation of Gene Expression Data. 2003 ZFIN Direct Data Submission ( http://zfin.org) (accessed 2003) [Google Scholar]
- Reichenbach B, Delalande JM, Kolmogorova E, Prier A, Nguyen T, Smith CM, Holzschuh J, Shepherd IT. Endoderm-derived Sonic hedgehog and mesoderm Hand2 expression are required for enteric nervous system development in zebrafish. Dev Biol. 2008;318:52–64. doi: 10.1016/j.ydbio.2008.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Jayasena CS, Nie S, Bronner ME. Neural crest specification: tissues, signals, and transcription factors. Wiley Interdiscip Rev Dev Biol. 2012;1:52–68. doi: 10.1002/wdev.8. [DOI] [PubMed] [Google Scholar]
- Roy S, Qiao T, Wolff C, Ingham PW. Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr Biol. 2001;11:1358–1363. doi: 10.1016/s0960-9822(01)00402-x. [DOI] [PubMed] [Google Scholar]
- Shepherd IT, Beattie CE, Raible DW. Functional analysis of zebrafish GDNF. Dev Biol. 2001;231:420–435. doi: 10.1006/dbio.2000.0145. [DOI] [PubMed] [Google Scholar]
- Shepherd I, Eisen J. Development of the zebrafish enteric nervous system. Methods Cell Biol. 2011;101:143–160. doi: 10.1016/B978-0-12-387036-0.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd IT, Pietsch J, Elworthy S, Kelsh RN, Raible DW. Roles for GFRalpha1 receptors in zebrafish enteric nervous system development. Development. 2004;131:241–249. doi: 10.1242/dev.00912. [DOI] [PubMed] [Google Scholar]
- Sirbulescu RF, Ilies I, Vitalo AG, Trull K, Zhu J, Traniello IM, Zupanc GK. Adult stem cells in the knifefish cerebellum. Dev Neurobiol. 2015;75:39–65. doi: 10.1002/dneu.22210. [DOI] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Sukegawa A, Narita T, Kameda T, Saitoh K, Nohno T, Iba H, Yasugi S, Fukuda K. The concentric structure of the developing gut is regulated by Sonic hedgehog derived from endodermal epithelium. Development. 2000;127:1971–1980. doi: 10.1242/dev.127.9.1971. [DOI] [PubMed] [Google Scholar]
- Taraviras S, Marcos-Gutierrez CV, Durbec P, Jani H, Grigoriou M, Sukumaran M, Wang LC, Hynes M, Raisman G, Pachnis V. Signalling by the RET receptor tyrosine kinase and its role in the development of the mammalian enteric nervous system. Development. 1999;126:2785–2797. doi: 10.1242/dev.126.12.2785. [DOI] [PubMed] [Google Scholar]
- Teoh PH, Shu-Chien AC, Chan WK. Pbx1 is essential for growth of zebrafish swim bladder. Dev Dyn. 2010;239:865–874. doi: 10.1002/dvdy.22221. [DOI] [PubMed] [Google Scholar]
- Tobin JL, Di Franco M, Eichers E, May-Simera H, Garcia M, Yan J, Quinlan R, Justice MJ, Hennekam RC, Briscoe J, et al. Inhibition of neural crest migration underlies craniofacial dysmorphology and Hirschsprung’s disease in Bardet-Biedl syndrome. Proc Natl Acad Sci USA. 2008;105:6714–6719. doi: 10.1073/pnas.0707057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CT, Yang TC, Huang HY, Tsai HJ. Zebrafish arl6ip1 is required for neural crest development during embryogenesis. PLoS One. 2012;7:e32899. doi: 10.1371/journal.pone.0032899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe RA, Gross JM. Immunohistochemistry on cryosections from embryonic and adult zebrafish eyes. CSH Protoc. 2007 doi: 10.1101/pdb.prot4779. 2007, pdb.prot4779. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Choe SK, Sagerstrom CG. Meis3 synergizes with Pbx4 and Hoxb1b in promoting hindbrain fates in the zebrafish. Development. 2001;128:1299–1312. doi: 10.1242/dev.128.8.1299. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Ellstrom DR, Sagerstrom CG. A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev Dyn. 2000;217:109–119. doi: 10.1002/(SICI)1097-0177(200001)217:1<109::AID-DVDY10>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, Gong Z. Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol. 2009;331:222–236. doi: 10.1016/j.ydbio.2009.04.035. [DOI] [PubMed] [Google Scholar]
- Zha Y, Xia Y, Ding J, Choi JH, Yang L, Dong Z, Yan C, Huang S, Ding HF. MEIS2 is essential for neuroblastoma cell survival and proliferation by transcriptional control of M-phase progression. Cell Death Dis. 2014;5:e1417. doi: 10.1038/cddis.2014.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.