Dynamic changes in CCAN organization during progression of the cell cycle are examined in chicken DT40 cells. CENP-C166-324 is sufficient for interphase centromere localization through association with CENP-L-N, and CENP-C643-864 is essential for mitotic centromere localization through binding to CENP-A nucleosomes.
Abstract
The kinetochore is a crucial structure for faithful chromosome segregation during mitosis and is formed in the centromeric region of each chromosome. The 16-subunit protein complex known as the constitutive centromere-associated network (CCAN) forms the foundation for kinetochore assembly on the centromeric chromatin. Although the CCAN can be divided into several subcomplexes, it remains unclear how CCAN proteins are organized to form the functional kinetochore. In particular, this organization may vary as the cell cycle progresses. To address this, we analyzed the relationship of centromeric protein (CENP)-C with the CENP-H complex during progression of the cell cycle. We find that the middle portion of chicken CENP-C (CENP-C166–324) is sufficient for centromere localization during interphase, potentially through association with the CENP-L-N complex. The C-terminus of CENP-C (CENP-C601–864) is essential for centromere localization during mitosis, through binding to CENP-A nucleosomes, independent of the CENP-H complex. On the basis of these results, we propose that CCAN organization changes dynamically during progression of the cell cycle.
INTRODUCTION
The key objective of mitosis is equal distribution of genetic material to the daughter cells. In eukaryotes, this process is accomplished by attachment of the duplicated sister chromatids to a bipolar mitotic spindle and their subsequent separation and segregation to the daughter cells during mitosis. A large protein complex known as the kinetochore is formed at the centromeric region of each chromosome and mediates this attachment between centromeric chromatin and spindle microtubules for faithful chromosome segregation (Fukagawa and Earnshaw, 2014).
Electron microscopy observations have demonstrated that the kinetochore is composed of three layers (Rieder, 1982). The inner layer contains centromeric chromatin marked by the centromere-specific histone H3 variant centromeric protein (CENP)-A and consists of a 16-subunit protein complex called the constitutive centromere associated network (CCAN; Cheeseman and Desai, 2008; Perpelescu and Fukagawa, 2011). The outer layer consists of the Mis12 complex, the Ndc80 complex, and other intermediaries, which connect the inner kinetochore with the spindle microtubules (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009).
CENP-A (Palmer et al., 1987) specifies the centromeres through its deposition into the chromatin, and downstream kinetochore assembly subsequently takes place involving CCAN components (Black and Cleveland, 2011; Westhorpe and Straight, 2013). The CCAN components CENP-C and the CENP-T-W-S-X complex associate with the centromeric chromatin and the outer kinetochore components. These components are believed to function as a bridge between the inner and outer kinetochore (Gascoigne et al., 2011; Hori et al., 2013; Fukagawa and Earnshaw, 2014). The CENP-H-I-K-L-M-N (Okada et al., 2006; Basilico et al., 2014) and CENP-O-P-Q-R-U complexes (Hori et al., 2008b) may act as scaffolds or mediators for the establishment of centromeric chromatin.
CENP-C (Earnshaw and Rothfield, 1985) is the CCAN component closest to CENP-A (Wan et al., 2009), and it directly binds to CENP-A nucleosomes (Kato et al., 2013; Falk et al., 2015). CENP-C also binds to the Mis12 complex of the outer kinetochore (Petrovic et al., 2010; Przewloka et al., 2011), which is associated with the microtubule-binding Ndc80 complex (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). The CENP-T-W-S-X complex contains histone-fold domains and forms a nucleosome-like structure (Nishino et al., 2012; Takeuchi et al., 2014). Whereas CENP-T-W-S-X is located close to the centromeric chromatin, the N-terminal region of CENP-T binds directly to the Ndc80 complex of the outer kinetochore through phosphorylation of CENP-T (Gascoigne et al., 2011; Nishino et al., 2013). On the basis of artificial tethering experiments performed with CENP-C and CENP-T, we proposed two parallel pathways (CENP-C and CENP-T pathways) for Ndc80 recruitment at kinetochores (Hori et al., 2013; Fukagawa and Earnshaw, 2014).
Although these two pathways for Ndc80 binding are distinct, centromeric chromatin is complicated, and it is unclear how these two pathways organize the centromeric chromatin. In particular, it is unclear how the CENP H-I-K-L-M-N complex (CENP-H complex) is involved in these two pathways. Although centromeric localization of the CENP-H complex depends on CENP-T (Hori et al., 2008a), CENP-H–related proteins are present in CENP-C–deficient chicken DT40 cells (Kwon et al., 2007). In addition, although CENP-C localization in interphase is abolished in CENP-H– or CENP-K–deficient DT40 cells, mitotic CENP-C localization is not altered in these knockout cells (Fukagawa et al., 2001; Kwon et al., 2007). Moreover, centromeric localization of CENP-H–associated proteins depends on CENP-C in human cells (Basilico et al., 2014; Klare et al., 2015). Therefore it is possible that the CENP-C and CENP-T pathways interact during organization of centromeric chromatin. In addition, the relationship may vary during the cell cycle (Kwon et al., 2007; Hemmerich et al., 2008).
To address how these two pathways are organized during progression of the cell cycle, we analyzed various CENP-C domains using chicken DT40 cells and recombinant proteins. We found that different regions of CENP-C are responsible for its centromeric localization during progression of the cell cycle. The middle portion of chicken CENP-C (amino acids [aa] 166–324) is sufficient for centromeric localization during interphase, potentially through association with CENP-L-N. The C-terminus of CENP-C (aa 643–864) is essential for centromeric localization during mitosis, through binding to CENP-A nucleosomes, independent of the CENP-H complex.
RESULTS
The middle portion and C-terminus of CENP-C are responsible for localization of interphase and mitotic centromeres, respectively
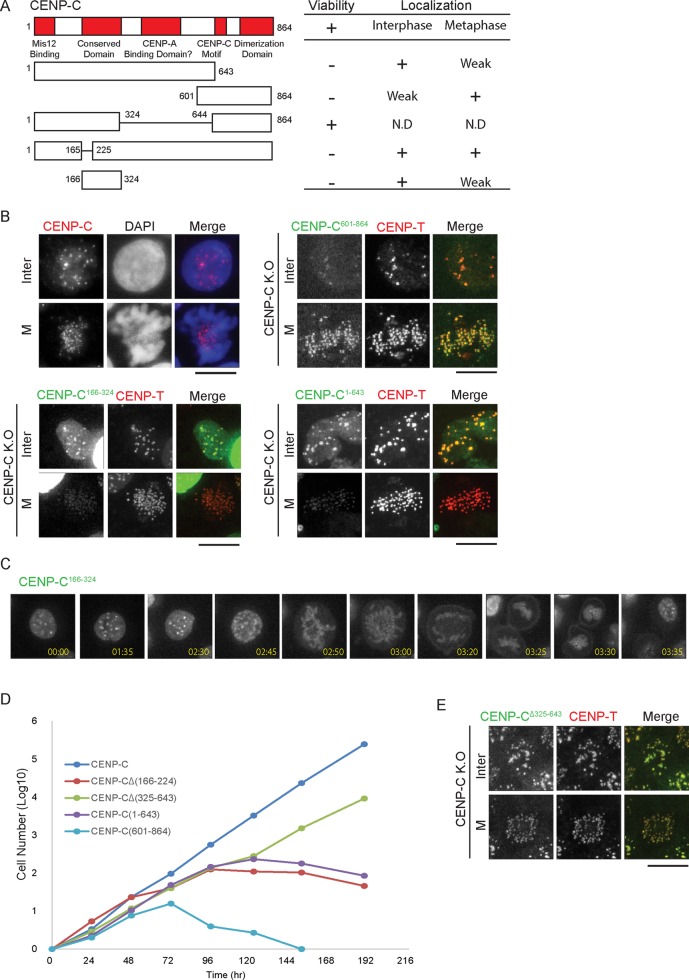
We previously showed that depletion of CENP-H or CENP-K in DT40 cells results in loss of CENP-C localization in interphase nuclei (Fukagawa et al., 2001; Kwon et al., 2007) but not in mitotic chromosomes. This suggests that CENP-C has different modes of localization to the kinetochore at different stages in the cell cycle. To test this hypothesis, we prepared various chicken CENP-C–truncated mutants fused with green fluorescent protein (GFP; Figure 1A), introduced them into CENP-C–deficient cells, and examined the localization of each mutant CENP-C in both interphase and mitosis (Figure 1B). We noted that numbers of centromere foci in interphase nuclei are less than those in mitotic cells because centromere foci were clustered in interphase nuclei.
FIGURE 1:
CENP-C166-324 and CENP-C601-864 localize to interphase and mitotic centromeres, respectively. (A) Schematic showing CENP-C domains (red) and the constructs used, along with their ability to rescue CENP-C depletion and localization throughout the cell cycle. (B) Representative images showing wild-type CENP-C (using an anti–ggCENP-C antibody), GFP-CENP-C601-864, GFP-CENP-C166-324, and GFP-CENP-C1-643 localization during interphase and metaphase. Kinetochores in CENP-C conditional-knockout cells expressing GFP constructs were visualized by CENP-T immunostaining at 36 h after addition of tetracycline. Bars, 10 μm. (C) Live-cell imaging of cells expressing GFP-CENP-C166-324. (D) Growth curves for CENP-C–deficient cells expressing the indicated CENP-C constructs. Tetracycline was added at t = 0, and the number of live cells was counted. (E) Localization of CENP-C∆325–643 and CENP-T in CENP-C–deficient cells expressing CENP-C∆325–643. Bars, 10 μm.
CENP-C contains various functional domains (Basilico et al., 2014; Klare et al., 2015). The N-terminal end is required for Mis12 binding (Petrovic et al., 2010; Przewloka et al., 2011), whereas the C-terminal end forms a dimer and is crucial for CENP-A binding (Hori et al., 2013; Kato et al., 2013). These two regions are relatively conserved. Another CENP-A–binding region (the middle of CENP-C) is not conserved in chicken CENP-C (Figure 1A). However, although the aa 166–324 region of chicken CENP-C is relatively conserved, its function is unclear.
We found that CENP-C601–864 preferentially localized to centromeres in mitotic cells but that these CENP-C signals were diffused in interphase nuclei (Figure 1B). In contrast, CENP-C1–643, which lacks a CENP-A–binding dimerization domain, preferentially localized to centromeres in interphase cells, but its centromeric localization in mitotic cells was lower than CENP-C localization in wild-type cells (Figure 1B). These data suggest that the entire N-terminal portion of CENP-C (aa 1–643) is primarily responsible for centromere localization in interphase nuclei, and the C-terminal dimerization domain of CENP-C is critical for centromeric localization during mitosis. We further analyzed the N-terminal CENP-C region responsible for centromere localization in interphase nuclei and found that CENP-C166–324 is sufficient for centromeric localization in interphase nuclei (Figure 1B). We also confirmed that clear punctate signals of CENP-C166–324–GFP in interphase were lost in mitosis and were visible again in next G1 cells by live-cell imaging (Figure 1C). On the basis of these results, we conclude that the middle portion of CENP-C (aa 166–324) is potentially responsible for interphase centromeric localization and that the C-terminus of CENP-C (aa 601–864) is critical for mitotic centromeric localization.
Another putative CENP-A–binding region of chicken CENP-C is dispensable for CENP-C function
Human CENP-C contains two CENP-A binding regions (Figure 1A; Kato et al., 2013). The first is the C-terminal dimerization domain, which is highly conserved among species. The second is the middle of human CENP-C (aa 444–537 in human CENP-C); no clearly homologous region is present in chicken CENP-C. To determine the function of the corresponding region in chicken CENP-C (aa 325–643), we introduced a CENP-C∆325–643 mutant into CENP-C–deficient cells and investigated cell viability. As shown in Figure 1D, CENP-C∆325–643 rescued the CENP-C deficiency, suggesting that the middle region of chicken CENP-C (aa 325–643) is dispensable for its function and that this region may not be required for centromeric localization of CENP-C. We confirmed that CENP-C∆325–643 and CENP-T showed clear centromere localization in CENP-C–deficient cells expressing CENP-C∆325–643 (Figure 1E). In contrast, CENP-C1–643 did not rescue the CENP-C deficiency (Figure 1D), because the C-terminus of CENP-C is responsible for CENP-A binding. On the basis of these results, we suggest that the putative CENP-A– binding region of chicken CENP-C (aa 325–643) is dispensable for cell viability and may not be involved in CENP-A binding in chicken cells.
Centromeric localization of CENP-C166–324 depends on the CENP-H complex
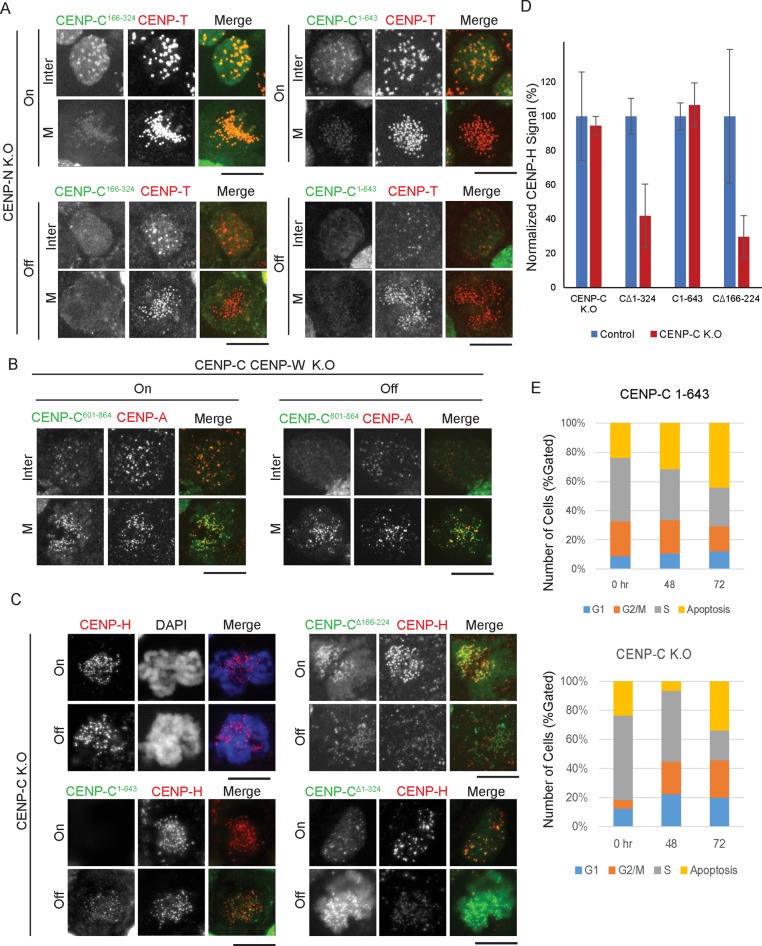
CENP-C166–324 preferentially localizes to interphase centromeres (Figure 1, B and C). We previously showed that interphase CENP-C localization depends on the CENP-H complex (Fukagawa et al., 2001; Kwon et al., 2007), and, therefore, interphase centromeric localization of CENP-C166–324 may depend on the CENP-H complex. To test this possibility, we expressed CENP-C166–324-GFP in CENP-N conditional-knockout cells (Supplemental Figure S1), which exhibit complete loss of CENP-H, -I, -K, -L, and -M (unpublished data) and a reduction of CENP-T levels (Supplemental Figure S1), similar to CENP-H– or CENP-K–knockout cells (Fukagawa et al., 2001; Okada et al., 2006; Kwon et al., 2007; Hori et al., 2008a). Although CENP-C166–324–GFP localized to interphase centromeres in the presence of CENP-N, this localization was abolished in the absence of CENP-N (Figure 2A). We also confirmed that interphase centromere localization of CENP-C1–643 depends on CENP-N (Figure 2A). On the basis of these results, we conclude that interphase centromeric localization of CENP-C166–324 depends on the CENP-H complex.
FIGURE 2:
Centromere localization of CENP-C166-324 depends on CENP-N during interphase and that of CENP-C601-864 depends on CENP-A during mitosis. (A) Representative images showing loss of centromere localization of CENP-C N-terminal constructs (green) during the cell cycle after CENP-N depletion (36 h after addition of tetracycline). Kinetochores were visualized by CENP-T immunostaining (red). Bars, 10 μm. (B) CENP-C601-864 (green) localizes to the centromeres in mitotic cells after CENP-C and CENP-W double depletion. Bars, 10 μm. (C) Immunostaining for CENP-H (red) in CENP-C–deficient cells expressing various GFP–CENP-C constructs showing their dependence on the N-terminus of CENP-C for localization to the kinetochore. Images were captured at 36 h after addition of tetracycline. (D) Quantification of signal intensities for CENP-H in DT40 cells expressing the indicated GFP–CENP-C constructs in control and CENP-C–deficient cells. Results are plotted as an average of the signal intensities of 20 kinetochores per cell (n > 7 cells). Error bars indicate SD. (E) Cell cycle distribution of CENP-C–deficient cells and these cells expressing CENP-C1-643 at 0, 48, and 72 h after tetracycline addition.
Given that CENP-C601–864 localizes to mitotic centromeres and mitotic CENP-C localization does not depend on the CENP-H complex (Kwon et al., 2007), it is possible that CENP-C601–864 localizes to the mitotic centromere after depletion of CENP-H-related proteins. To test this hypothesis, we expressed mutant CENP-C601–864 in CENP-C and CENP-W double-knockout cells to avoid dimerization of CENP-C601–864 and wild-type CENP-C. We previously demonstrated that CENP-H–complex proteins do not localize to centromeres in CENP-C and CENP-W double-knockout cells (Hori et al., 2008a). In control cells (in the presence of endogenous CENP-C and CENP-W), we observed centromere localization of CENP-C601–864 in both interphase and mitotic cells (Figure 2B, left). When expression of both genes was turned off, interphase localization CENP-C601–864 was abolished, but mitotic localization was not altered (Figure 2B, right). This indicates that mitotic localization of CENP-C601–864 does not depend on the CENP-H complex. Because only CENP-A is present in the centromeric region in CENP-C and CENP-W double-knockout cells, it is possible that CENP-C601–864 directly recognizes CENP-A.
Finally, we tested whether CENP-H depends on each mutant CENP-C. We performed these experiments using CENP-C conditional-knockout cells at 36 h after addition of tetracycline (Kwon et al., 2007), which does not cause significant CENP-H reduction (Figure 2, C and D). When we expressed CENP-C∆1–324 or CENP-C∆166–224 under these conditions, CENP-H was reduced to 30–40% of its previous levels (Figure 2, C and D, and Supplemental Figure S2), suggesting a dominant-negative effect on CENP-H localization. We also observed a 40–50% reduction in CENP-T, consistent with loss of CENP-H (Supplemental Figures S1E and S2B). Consistent with this, CENP-C∆166–224 did not rescue CENP-C deficiency (Figure 1C), and strong mitotic defects were observed. We noted that whereas CENP-C166–324 is sufficient for centromere localization in interphase cells, CENP-C∆166–224 still localizes to interphase centromeres (Supplemental Figure S2), suggesting that an additional N-terminal region of CENP-C (aa 225–324) may be responsible for centromere localization in interphase cells. On the other hand, in CENP-C–deficient cells expressing CENP-C1–643, CENP-H and CENP-T levels remained the same as in control cells (Figures 1B and 2, C and D, and Supplemental Figure S2). Consistent with this, we also observed Mis12 at kinetochores in these cells (unpublished data). However, CENP-C–deficient cells expressing CENP-C1–643 are not viable, showing similar mitotic index as that of CENP-C–deficient cells (Figure 2E). This suggests that proper kinetochore structure is not formed even if CENP-H localizes to centromeres, because CENP-C1–643 does not localize to centromeres during mitosis.
On the basis of these data, we conclude that the CENP-H complex is involved in centromeric localization of the middle portion of CENP-C (aa 166–324) and that the C-terminus of CENP-C (aa 601–864) is not related to the CENP-H complex but may be related to CENP-A.
The middle portion of CENP-C directly binds the CENP-L-N complex
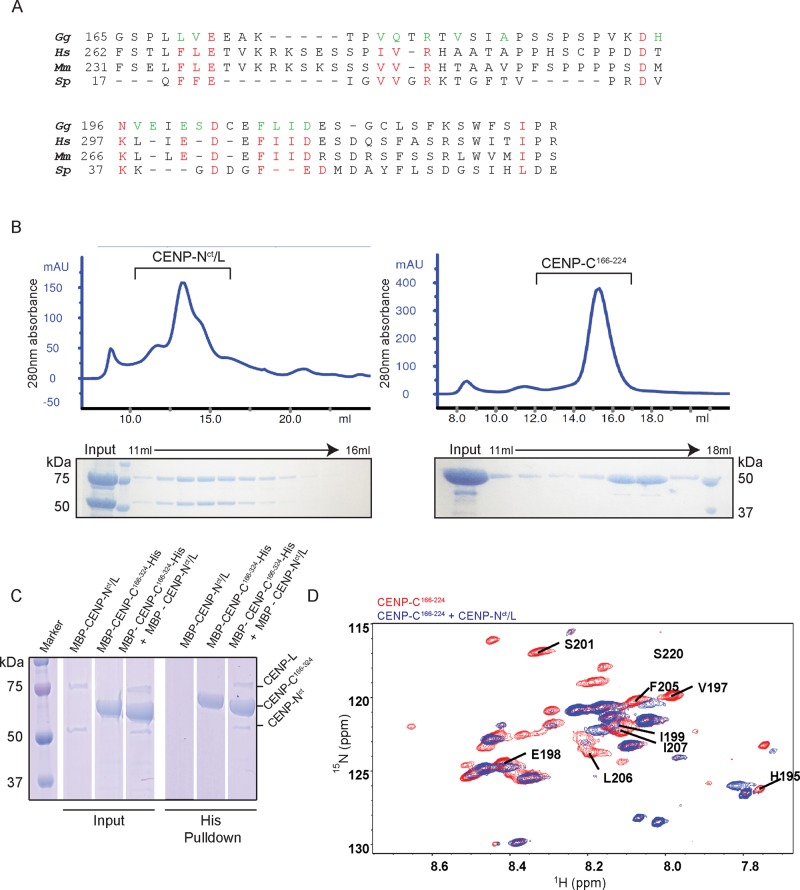
Given that the middle portion of CENP-C (aa 166–324) is related to the CENP-H complex, we next analyzed direct interactions between the middle portion of CENP-C– and CENP-H–complex proteins. The yeast homologue of CENP-C, Mif2, directly interacts with Iml3-Chl4 dimers, the counterpart of the CENP-L-N complex (Hinshaw and Harrison, 2013). Therefore we focused on the CENP-L-N complex among the CENP-H complex proteins. Because the 166–224 aa region of chicken CENP-C is relatively conserved, conserved residues are shown in red in Figure 3A.
FIGURE 3:
Middle portion of CENP-C directly binds to the CENP-N-L complex in vitro. (A) Red letters show residues conserved between Gallus gallus (Gg), Homo sapiens (Hs), Mus musculus (Mm), and Schizosaccharomyces pombe (Sp). Green letters indicate residues possibly involved in the interaction between ggCENP-C166-324 and ggCENP-Nct/L (see D and Supplemental Figure S3). (B) Gel-filtration profiles for the CENP-Nct/L complex and CENP-C166-224. Peak fractions were visualized using SDS–PAGE. The CENP-Nct/L complex was prepared from MBP-CENP-Nct (54 kDa) and MBP-CENP-L (80 kDa). (C) Histidine pull down of CENP-C166-324 with the CENP-Nct/L complex. (D) 1H15N HSQC spectrum of free CENP-C166-224 (red) overlaid with that of CENP-C166-224 in complex with CENP-Nct/L (blue). Residues that showed shifted or abolished peaks are labeled.
We first expressed and purified a complex of full-length CENP-L and the C-terminal domain of CENP-N fused with the maltose binding protein (MBP; MBP-CENP-Nct/L; Figure 3B and Supplemental Figure S3). We also prepared recombinant CENP-C166–224 or CENP-C166–324 fused with MBP and histidine (His; MBP-CENP-C166–224-His or MBP-CENP-C166–324-His; Figure 3B). To test the direct interaction of CENP-C with the CENP-L-N complex, we immobilized MBP-CENP-C166–324-His on Ni-Sepharose beads and used it as a bait to pull down MBP-CENP-Nct/L. We observed reproducible pull down of MBP-CENP-Nct/L (Figure 3C). We also observed that MBP-CENP-Nct/L-His interacted with MBP-CENP-C166–324 (unpublished data). We also tested for interactions between the CENP-L-N complex and other domains of CENP-C and found that other domains, including the C-terminus of CENP-C, did not bind the CENP-L-N complex (Supplemental Figure S3).
To verify the interaction and identify residues responsible, we performed nuclear magnetic resonance (NMR) experiments with CENP-C166–324 and found that it was disordered when alone but showed peak shifts on addition of CENP-Nct/L. Because CENP-C166–324 was slightly aggregated in the NMR sample tube, we repeated the experiment using CENP-C166–224, which showed a much clearer peak shift, indicating a clear interaction between CENP-C166–224 and CENP-Nct/L (Figure 3D and Supplemental Figure S3). We highlighted the CENP-C residues that showed a clear peak shift upon addition of CENP-Nct/L (Figure 3, A and D, and Supplemental Figure S3).
On the basis of biochemical and NMR analyses, we conclude that the middle portion of CENP-C (aa 166–324) directly binds the CENP-L-N complex.
The C-terminus of CENP-C, but not the middle portion of CENP-C, directly binds to the CENP-A nucleosome
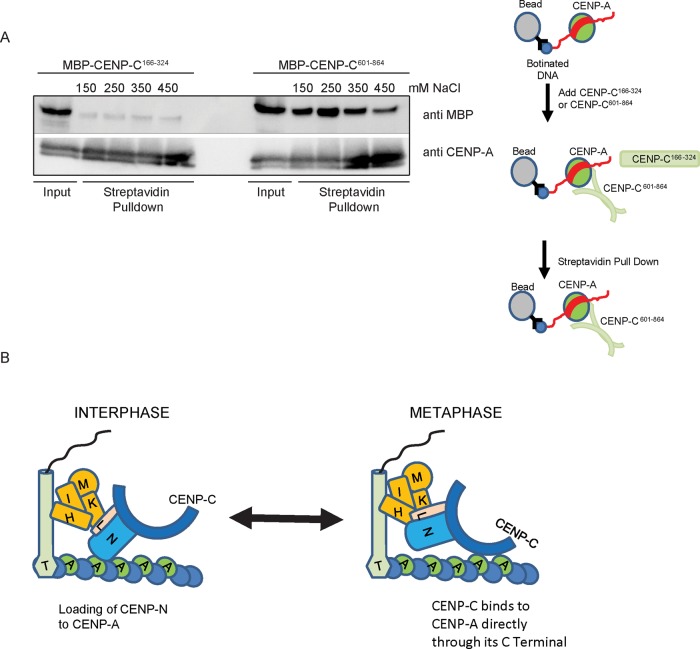
Human CENP-C binds to CENP-A (Kato et al., 2013; Falk et al., 2015). We previously showed that the C-terminal dimerization domain has activity to recruit CENP-A to a noncentromeric locus, using LacO-LacI experiments (Hori et al., 2013). In addition, microscopic observations in this study suggest that mitotic centromeric localization of CENP-C601–864 depends on CENP-A (Figure 2). We directly tested whether CENP-C fragments bind to reconstituted CENP-A nucleosomes in vitro. We prepared a biotinylated CENP-A nucleosome with MBP-fused CENP-C166–324 or CENP-C601–864 and performed a pull-down experiment using streptavidin beads. As shown in Figure 4A, CENP-C601–864 binds to the CENP-A nucleosome, but CENP-C166–324 does not. On the basis of these results, we conclude that the C-terminus of CENP-C, but not the middle portion of CENP-C, directly binds to the CENP-A nucleosome.
FIGURE 4:
CENP-C601-864 directly binds to the CENP-A nucleosome in vitro. (A) Analysis of MBP-ggCENP-C601-864 and MBP-ggCENP-C166-324 proteins by Western blot after streptavidin pull down using a biotinylated CENP-A nucleosome. The pull-down experiment was performed at the various salt concentrations indicated. CENP-C601–864 clearly bound to CENP-A, with the strongest interaction at 250 mM NaCl (gel on the right). (B) Model showing cell cycle–dependent kinetochore organization. During interphase, the N-terminus of CENP-N preferentially binds to CENP-A nucleosomes (Fang et al., 2015), whereas the C-terminus of CENP-N and CENP-L binds to the middle portion of CENP-C. The CENP-C–CENP-A interaction is probably weak during interphase. In contrast, CENP-C binds to CENP-A nucleosomes via its C-terminal dimerization domain during mitosis. CENP-N may not associate with CENP-A nucleosomes, because CENP-N does not bind to compact chromatin (Fang et al., 2015).
DISCUSSION
In recent years, multiple centromeric proteins have been identified (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009; Fukagawa and Earnshaw, 2014). Another important question concerns how these proteins are organized to form a functional kinetochore. Although we previously proposed two distinct pathways (CENP-C and CENP-T pathways; Hori et al., 2013; Fukagawa and Earnshaw, 2014) for binding of the Ndc80 complex, a crucial microtubule protein complex in kinetochores, it was unclear how these two pathways are organized in centromeric chromatin. In particular, the relationship between the CENP H-I-K-L-M-N complex (CENP-H complex) and CENP-C was unclear. Although we previously suggested that the localization dependence of CENP-C on the CENP-H complex varies during the progression of the cell cycle (Kwon et al., 2007), the region of CENP-C responsible for this dependence was unknown.
We conclude that the middle portion of chicken CENP-C (CENP-C166–324) is sufficient for centromeric localization in interphase through association with CENP-L-N. The C-terminus of CENP-C (CENP-C601–864) is essential for centromeric localization during mitosis, through binding to CENP-A nucleosomes. On the basis of these findings and other recent reports, we propose a model for CCAN organization during progression of the cell cycle (Figure 4B). During interphase, the N-terminus of CENP-N preferentially binds to CENP-A nucleosomes (Fang et al., 2015), and the complex of the C-terminus of CENP-N and CENP-L binds to the middle portion of CENP-C (Figure 3). During interphase, the CENP-C–CENP-A interaction must be weak, because centromeric localization of CENP-C during interphase depends on CENP-N, CENP-H, and CENP-K, whereas the C-terminus of CENP-C does not localize to interphase centromeres (Figure 1). However, during mitosis, CENP-C binds to CENP-A nucleosomes via its C-terminal dimerization domain. Because the middle portion of CENP-C does not efficiently localize to mitotic centromeres, this middle portion is not required for mitotic CENP-C localization. In this case, CENP-N may not associate with CENP-A nucleosomes, because it does not bind to compact chromatin (Fang et al., 2015). In fact, live-cell imaging of CENP-N suggests that CENP-N levels at mitotic kinetochores are low (Hellwig et al., 2011). In addition, CENP-N knockout does not cause a CENP-C reduction during mitosis, because CENP-C binds tightly to CENP-A nucleosomes, consistent with our previous observation (Kwon et al., 2007). In this model, we propose that CENP-T associates with centromeric DNA, but this association must be facilitated by the CENP-H complex, because CENP-H knockout causes reduction of CENP-T (Hori et al., 2008a; Supplemental Figure S2).
Recent reports proposed that CENP-C is upstream of other CCAN proteins in human cells (Basilico et al., 2014; Klare et al., 2015). In our model, CENP-C may largely contribute to localization of the CENP-H complex to the mitotic centromere through association of CENP-C to CENP-A (Figure 4B). In addition, although we did not demonstrate CENP-H–CENP-C interactions using chicken proteins, it is possible that, in addition to CENP-L-N, CENP-H-I-K-M may bind directly to CENP-C, as suggested for human proteins (Klare et al., 2015). Klare et al. (2015) demonstrated that mutation to alanine (3A mutation) of hydrophobic residues in human CENP-C, corresponding to residues in chicken CENP-C responsible for binding to CENP-L-N (based on our NMR studies), caused disruption of interaction with the CENP-H complex. However, we observed ∼60% CENP-H at mitotic chromosomes in CENP-C–deficient chicken DT40 cells (Kwon et al., 2007; Hori et al., 2008a). We believe that chromatin association of CENP-T largely contributes to CCAN assembly in chicken cells. Therefore CENP-C deficiency may not cause a strong reduction in CENP-H proteins in chicken cells because CENP-T tightly binds chromatin even in CENP-C–deficient cells.
We still do not know why protein dependence in CCAN is slightly different between chicken and human cells. Of course, we cannot conclude that chicken and human systems are entirely conserved without completing all analyses using both chicken and human cells. However, we believe that essential architecture of the kinetochore is similar between chicken and human cells. However, there are subtle differences as well. For example, we demonstrated that the second CENP-A–binding domain of CENP-C (aa 444–537 in human CENP-C, which corresponds to aa 325–643 in chicken CENP-C) is not essential in chicken cells, but this region is required for CENP-A binding in human cells (Kato et al., 2013; Falk et al., 2015).
Our model suggests that during interphase, CENP-C localization occurs downstream of CENP-H-I-K-L-M-N, which is inconsistent with the reported model in which CENP-C is upstream of other CCAN proteins (Basilico et al., 2014; Klare et al., 2015). Although CENP-C dependence has not been analyzed in human cells, the architecture of CCAN organization must be altered during human cell cycle progression. We propose that these dynamic changes in CCAN organization are essential for formation of a plastic kinetochore structure.
MATERIALS AND METHODS
Molecular biology, cell culture, and transfections
For expression of CENP-C deletion mutants in DT40 cells, Ecogpt gene or blasticidin S resistance cassette under control of the chicken β-actin promoter was inserted as selection marker. A Gene Pulser II electroporator (Bio-Rad, Tokyo, Japan) was used for all transfections into DT40 cells. All molecular biology experiments, including Southern and Western blot analyses, were followed by standard methods. All chicken DT40 cells were cultured at 38.5°C in DMEM supplemented with 10% fetal bovine serum, 1% chicken serum, β-mercaptoethanol, penicillin, and streptomycin.
Immunofluorescence
Chicken DT40 cells were prepared by the cytospin method and fixed in 3% paraformaldehyde for 10 min at room temperature. Immunofluorescence staining of DT40 cells was performed using anti–CENP-T (a rabbit antibody against recombinant full-length chicken CENP-T), anti–CENP-C (a rabbit antibody against recombinant chicken CENP-C 1–330 aa), and anti–CENP-H (a rabbit antibody against recombinant chicken CENP-H). Several cell lines expressing GFP or monomeric red fluorescent protein fusions were also used to examine protein localization. Images were collected with a cooled charge-coupled device camera (CoolSnap HQ; Roper Scientific, Tokyo, Japan) mounted on an inverted microscope (IX71; Olympus, Tokyo, Japan) with a 100× objective lens together with a filter wheel. All subsequent analysis and processing of images were performed using MetaMorph software (Molecular Devices, Sunnyvale, CA). High-resolution images were also collected using a confocal scanner box (Cell Voyager CV1000; Yokogawa, Tokyo, Japan) with an oil immersion objective lens (100×).
Protein preparation
Chicken MBP-CENP-C1-165-His, MBP-CENP-C166-324-His, MBP-CENP-C166-224-His, and MBP-CENP-C601-864-His fragments and chicken MBP-CENP-Nct/L were cloned into pMal coexpression vector. Proteins were expressed in BL21(DE3)Star-pRARE2LysS by addition of 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) for 12 h at 16°C. The proteins were purified using Ni-Sepharose and Amylose resin and a Superdex 200 column. Isotope-labeled proteins were prepared by growing cultures in M9 medium containing [15N]ammonium chloride and [15N]ammonium sulfate (Cambridge Isotope Laboratory, Tewksbury, MA) and expressed by addition of 0.2 mM IPTG for 14 h at 16°C. Cell pellets were resuspended in lysis buffer (25 mM Tris-HCl, pH 7.5, 2 mM EDTA, 100 μg/ml lysozymes) and lysed by sonication. The samples were cleared by centrifugation at 18,000 rpm at 4°C for 30 min. The cleared supernatant was filtered through a 0.8-μm filter to remove residual debris and large aggregates. The lysate was then passed through either a 5-ml His-Trap FF (GE Healthcare, Tokyo, Japan) or 5-ml MBP-Trap (GE Healthcare) preequilibrated with His-running buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM dithiothreitol [DTT], 20 mM imidazole) or MBP-running buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT), respectively, depending on requirement. The samples were eluted in appropriate running buffer supplemented with 500 mM imidazole or 10 mM maltose. Fractions containing CENP-C or CENP-Nct/L were concentrated and dialyzed into MBP-running buffer and loaded onto a Superdex 200 10/300 or HiLoad Superdex 200 16/600 preequilibrated with MBP-running buffer. Fractions containing protein were concentrated and stored at 4°C. For NMR, samples were treated with TEV protease to cleave MBP, which was removed by passing through a Ni-Sepharose column. Because chicken CENP-C tended to be aggregated during storage, all samples were used within 1 wk from purification.
His-affinity pull-down assay
Purified MBP-CENP-C1-165-His, MBP-CENP-C166-324-His, MBP-CENP-C166-224-His, or MBP-CENP-C601-864-His fragments and MBP-CENP-Nct/L were mixed in 1:1 M ratio and incubated at 4°C for 2 h in wash buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM DTT, 20 mM imidazole). The mixture of each CENP-C fragment and CENP-Nct/L was added to 30 μl of Ni-Affinity beads (Roche, Tokyo, Japan), made up to 100 μl with wash buffer, and incubated for 1 h at 4°C with rotating. Beads were washed three times with 1 ml of wash buffer and then eluted with 40 μl of elution buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM DTT, 500 mM imidazole). The eluate was collected and boiled with SDS sample buffer and run on 10–20% SDS–PAGE gel. Bands were visualized with Coomassie brilliant blue staining.
NMR analysis
NMR experiments were performed on AVANCE DRX600 (Bruker BioSpin, Billerica, MA) at 298 K using samples dissolved in 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, and 5% D2O. Sequential assignments of the backbone 1H, 13C, and 15N chemical shifts of CENP-C166-224 were obtained from the standard triple-resonance NMR spectra HNCO, HN(CA)CO, HNCA, HN(CO)CA, CBCACONH, and HNCACB (Ikura et al., 1990). Data were processed and analyzed using the program NMRPipe (Delaglio et al., 1995) and the program Kujira (Kobayashi et al., 2007). For the chemical shift perturbation experiment, CENP-Nct/L was added to the CENP-C166-224 at a protein ratio of 1:1, and a 1H-15N HSQC spectrum of the complex was measured.
CENP-A streptavidin pull-down assay
Dynabeads M280 Streptavidin (Invitrogen, Tokyo, Japan) were washed in the nucleosome binding buffer (20 mM Tris-HCl, pH 7.5, 1 mM DTT, 250 mM KCl). CENP-A nucleosomes were prepared as described previously (Tachiwana et al., 2011; Arimura et al., 2012). A 100 μl amount of biotinylated CENP-A nucleosome (0.1 μg/μl) was added to 100 μl of washed beads and incubated for 2 h at 4°C while under rotation. Beads were washed three times in binding buffer (20 mM (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 20% glycerol, 1 mM DTT, 0.2 mM EDTA, 150–450 mM NaCl). A 30 μl amount of beads was incubated with purified MBP-CENP-C166-324-His or MBP-CENP-C601-864-His for 1 h at 4°C with rotation. The supernatant was removed, and beads were washed three times with binding buffer. Beads were subsequently boiled in SDS sample buffer and run on a 10–20% SDS–PAGE gel. The gel was transferred to a polyvinylidene fluoride membrane using a Semi-Dry transfer system (Bio-Rad). The bands were visualized by Western blot analysis with anti-MBP antibody (NEB, Tokyo, Japan) and anti-CENP-A antibody.
Supplementary Material
Acknowledgments
We are very grateful to Mayumi Takahashi, Kaeko Nakaguchi, Reika Fukuoka, and Yuko Fukagawa for technical assistance and Tatsuya Nishino for help with protein purification. This work was supported by Grants-in-Aid for Scientific Research (S) and for Scientific Research on Innovative Areas (Chromosome OS) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.F.
Abbreviations used:
- CCAN
constitutive centromere-associated network
- CENP
centromere protein
- GFP
green fluorescent protein
- NMR
nuclear magnetic resonance
- MBP
maltose-binding protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0531) on September 9, 2015.
REFERENCES
- Arimura Y, Tachiwana H, Oda T, Sato M, Kurumizaka H. Structural analysis of the hexasome, lacking one histone H2A/H2B dimer from the conventional nucleosome. Biochemistry. 2012;51:3302–3309. doi: 10.1021/bi300129b. [DOI] [PubMed] [Google Scholar]
- Basilico F, Maffini S, Weir JR, Prumbaum D, Rojas AM, Zimniak T, De Antoni A, Jeganathan S, Voss B, van Gerwen S, et al. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 2014;3:e02978. doi: 10.7554/eLife.02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LE, Van Duyne GD, Vinogradov SA, et al. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L, Teng Y, Yao T, You Q, Ruan H, et al. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev. 2015;29:1058–1073. doi: 10.1101/gad.259432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, et al. Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci. 2011;124:3871–3883. doi: 10.1242/jcs.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw SM, Harrison SC. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep. 2013;5:29–36. doi: 10.1016/j.celrep.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008a;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008b;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Guntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- Kwon MS, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Rieder CL ( The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Nishino T, Mayanagi K, Horikoshi N, Osakabe A, Tachiwana H, Hori T, Kurumizaka H, Fukagawa T. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 2014;42:1644–1655. doi: 10.1093/nar/gkt1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.