Microvilli are actin-based protrusions that amplify plasma membrane area and mediate interactions with the extracellular environment. We found that the multifunctional actin regulator cordon bleu promotes the growth of intestinal brush border microvilli. These results provide a new framework for investigating brush border biogenesis.
Abstract
Microvilli are actin-based protrusions found on the surface of diverse cell types, where they amplify membrane area and mediate interactions with the external environment. In the intestinal tract, these protrusions play central roles in nutrient absorption and host defense and are therefore essential for maintaining homeostasis. However, the mechanisms controlling microvillar assembly remain poorly understood. Here we report that the multifunctional actin regulator cordon bleu (COBL) promotes the growth of brush border (BB) microvilli. COBL localizes to the base of BB microvilli via a mechanism that requires its proline-rich N-terminus. Knockdown and overexpression studies show that COBL is needed for BB assembly and sufficient to induce microvillar growth using a mechanism that requires functional WH2 domains. We also find that COBL acts downstream of the F-BAR protein syndapin-2, which drives COBL targeting to the apical domain. These results provide insight into a mechanism that regulates microvillar growth during epithelial differentiation and have significant implications for understanding the maintenance of intestinal homeostasis.
INTRODUCTION
Found on the surface of numerous cell types, microvilli are finger-like membrane protrusions supported by parallel bundles of F-actin. One of the most elaborate arrays of microvilli, known as the brush border (BB), projects from the apex of the intestinal epithelial cell (IEC), where it functions in nutrient processing and host defense (Crawley et al., 2014). Loss of these protrusions due to genetic causes such as microvillus inclusion disease or infections with enteric pathogens such as enteropathogenic Escherichia coli leads to nutrient malabsorption and osmotic imbalances that cause dramatic dehydration and, in susceptible populations, death (Davidson et al., 1978; Vallance et al., 2002). However, molecular mechanisms controlling the growth of microvilli on the surface of transporting epithelia have yet to be defined.
Given that microvilli are supported by polarized actin bundles, understanding how these bundles form during differentiation and are stabilized thereafter will be critical for understanding BB assembly and function. Actin-based cytoskeletal structures in general are assembled through the coordinated action of factors that mediate initial filament formation (i.e., nucleation), elongation, capping, bundling, and cross-linking to membranes or other cytoskeletal systems (Chhabra and Higgs, 2007). Factors that bundle actin filaments within microvilli (villin, fimbrin, espin, and EPS8; Bretscher and Weber, 1979, 1980; Bartles et al., 1998; Hertzog et al., 2010) or link the resulting core bundles to the overlying plasma membrane (e.g., myosin-1a, ezrin; Bretscher, 1983; Howe and Mooseker, 1983) have been the subject of extensive investigation. However, molecules that control upstream events in microvillar formation, such as actin filament nucleation and elongation, remain poorly understood. Studies on other types of protrusions, such as filopodia, which extend from the leading edge of crawling cells (Faix et al., 2009), might provide mechanistic clues. The parallel actin bundles that support filopodia are formed by at least two distinct mechanisms. In the first case, bundle formation is driven by elongation and bundling of Arp 2/3–nucleated filaments that originate in the dendritic array of the lamellipodium (Svitkina et al., 2003). In the second, filopodial actin bundle formation is initiated at the membrane by barbed end–associated factors such as formins, which are capable of both nucleating and elongating filaments in linear bundles (Schirenbeck et al., 2005; Steffen et al., 2006). Of interest, recent studies show that mice lacking Arp C3, an essential component of Arp 2/3 complex, still assemble well-organized BBs (Zhou et al., 2015), suggesting that branched nucleation is unlikely to play a role in the formation of epithelial microvilli. However, proteomic analyses of isolated BBs also identified formin DIAPH1 and the Wiskott–Aldrich homology 2 (WH2) domain–containing factor cordon bleu (COBL) as potential regulators of microvillar formation (McConnell et al., 2011; Revenu et al., 2012), although the extent to which these molecules contribute to BB assembly remains unclear.
The objective of the present study was to identify factors that promote the growth of IEC BB microvilli. Our results indicate that COBL plays an important role in promoting the growth of microvillar actin bundles during intestinal epithelial differentiation. Moreover, a COBL interacting partner, syndapin-2, targets this factor to the apical domain during this process. Of importance, these studies provide a molecular framework for future investigations into the early steps of BB biogenesis. These findings also hold implications for understanding apical morphogenesis in other epithelial systems, such as the kidney, lung, and inner ear.
RESULTS
BB assembly requires dynamic turnover of the actin cytoskeleton
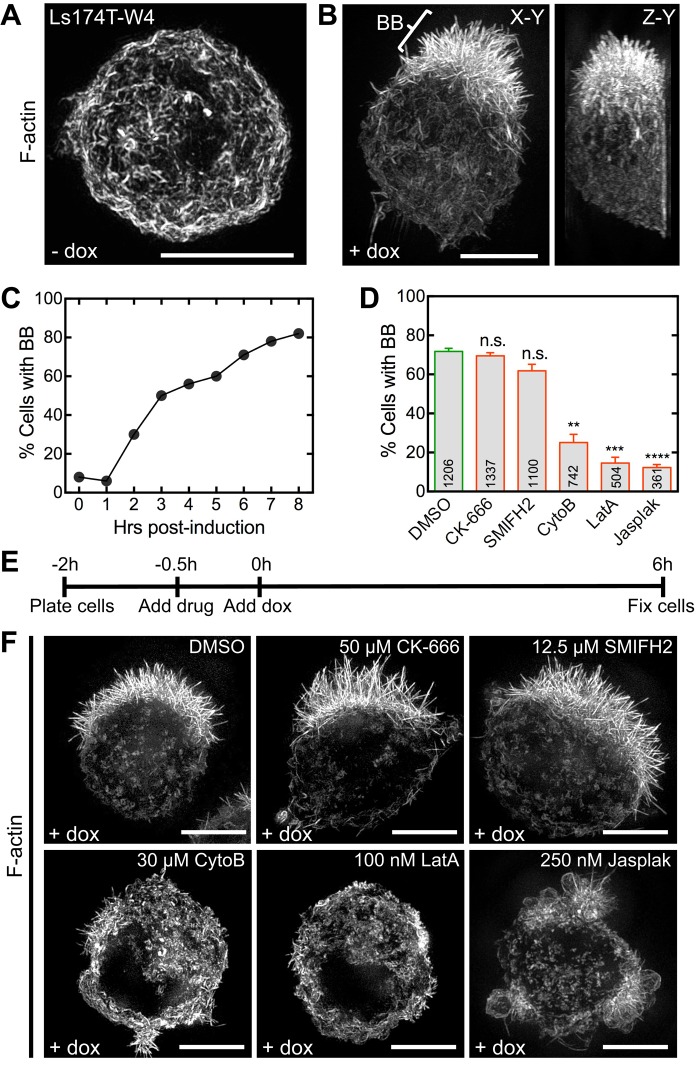
To explore the molecular basis of microvillar assembly, we used a cell culture model that allowed switch-like control over epithelial differentiation. Ls174T-W4 cells are human IECs that stably express myc-LKB1 and an inducible FLAG-STRADα. LKB1 is a serine/threonine kinase and master regulator of cell polarity (homologous to Par-4 in Caenorhabditis elegans and Drosophila melanogaster; Martin-Belmonte and Perez-Moreno, 2012). STRADα is an LKB1-specific adaptor protein that activates and translocates LKB1 from the nucleus to the cytoplasm (Baas et al., 2003). Exposing Ls174T-W4 cells to doxycycline induces STRADα expression, thereby activating LKB1 and initiating apical/basal polarity establishment and downstream events such as BB formation (Baas et al., 2004). Uninduced Ls174T-W4 cells exhibit an unpolarized morphology with cortical F-actin labeling (Figure 1A and Supplemental Movie S1). After induction with 1 μg/ml doxycycline for 8 h, ∼80% of Ls174T-W4 cells polarize and display a prominent BB on one side of the cell (Figure 1, B and C, and Supplemental Movie S2). Although the Ls174T-W4 model has been used to study polarity signaling upstream of BB assembly (ten Klooster et al., 2009; Gloerich et al., 2012), we took advantage of the switchable nature of this system to dissect the mechanism of microvillar growth.
FIGURE 1:
Actin dynamics is required for BB assembly. (A) SIM projection of an uninduced Ls174T-W4 cell stained with phalloidin to visualize F-actin (white). (B) En face (left) and lateral (right) SIM projections of an induced Ls174T-W4 cell; bracket highlights the BB. (C) Percentage of BB-positive cells at time points indicated. (D) Percentage of BB-positive cells from various drug treatments. Mean ± SEM from three separate experiments; number per condition listed within the bars (Student’s t test, **p < 0.005, ***p < 0.0005, ****p < 0.0001). (E) Schematic of experimental design and (F) SIM projections of Ls174T-W4 cells treated with drugs during induction. Scale bars, 5 μm.
We first sought to determine whether Arp 2/3 complex or formins functioned in assembling microvilli. BB assembly was induced in Ls174T-W4 cells in the presence of CK-666, which inhibits Arp 2/3 complex (Nolen et al., 2009), or SMIFH2 (Rizvi et al., 2009), which directly inhibits the FH2 domains of numerous formins from diverse species and specifically inhibits mouse DIAPH1/mDia1 (Rizvi et al., 2009), the only formin detected in BB proteomic studies (McConnell et al., 2011; Revenu et al., 2012; Figure 1, D–F, and Supplemental Figure S1). These compounds had little effect on the number of BB-positive cells after 6 h of induction (CK-666, 69.5 ± 1.8%; SMIFH2, 61.8 ± 3.3%; dimethyl sulfoxide [DMSO] control, 71.7 ± 1.6%). We did note that microvilli in CK-666–treated cells were slightly but significantly longer than control cells (2.40 ± 0.03 vs. 2.21 ± 0.03 μm); one possible explanation is that Arp 2/3 inhibition increases the concentration of actin monomers available for microvillar elongation (Burke et al., 2014; Rotty et al., 2015; Suarez et al., 2015). In contrast, the fraction of BB-positive cells was significantly reduced when barbed ends of actin filaments were capped using cytochalasin B (25.1 ± 4.2%), actin monomers were sequestered with latrunculin A (14.6 ± 3.0%), or filaments were stabilized with jasplakinolide (12.3 ± 1.5%; Figure 1, D–F). Although the results of small-molecule inhibition studies should be interpreted with caution, these data are consistent with previous reports showing that mice lacking functional Arp 2/3 complex continue to build normal BBs (Zhou et al., 2015) and lymphocytes lacking Arp 2 or upstream activator WASP still assemble microvilli (Majstoravich et al., 2004; Nicholson-Dykstra and Higgs, 2008). Together these results suggest that microvillar growth is driven by the assembly of new actin filaments that elongate from their barbed ends and that Arp 2/3 and formins are not needed for this process.
COBL—a multifunctional actin regulator that localizes to the base of microvilli
Another actin regulator identified in BB proteomic studies is the multi-WH2 domain–containing molecule COBL (McConnell et al., 2011; Revenu et al., 2012). Individual WH2 domains are G-actin–binding modules capable of mediating filament nucleation, severing, and a variety of other activities when linked in tandem (Dominguez, 2009). Since its initial identification as a nucleator (Ahuja et al., 2007), further studies revealed profilin-like binding to ATP-actin, sequestration of ADP-actin, and filament severing (Husson et al., 2011). How the multiple activities demonstrated by COBL WH2 domains in vitro contribute to COBL activity in cells remains unclear. However, COBL has been implicated in neural tube formation (Carroll et al., 2003), neurite outgrowth (Ahuja et al., 2007), assembly of motile cilia in Kupffer’s vesicle (Ravanelli and Klingensmith, 2011), and differentiation of sensory cells (Schuler et al., 2013).
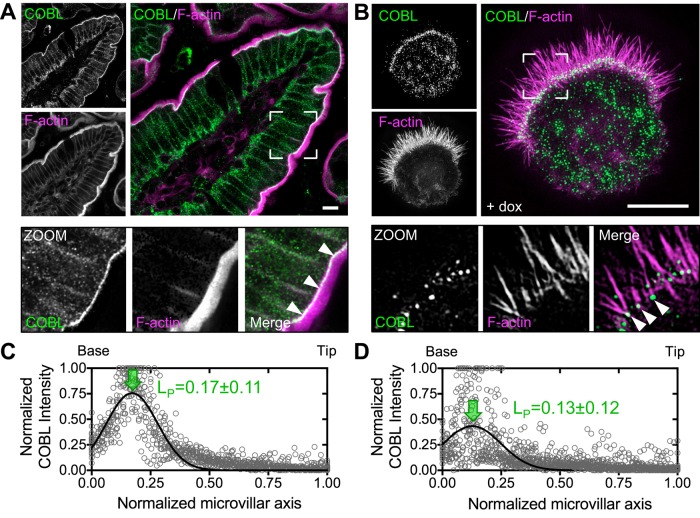
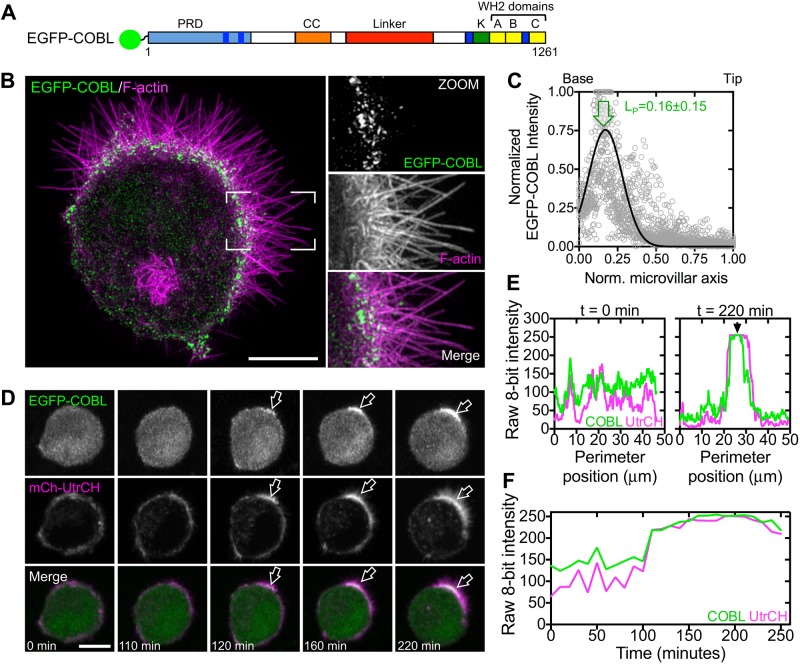
Immunostaining of mouse intestinal tissue revealed that COBL was strongly enriched at the base of the BB (Figure 2A, zoom), where the pointed ends of microvillar actin bundles are anchored (Hirokawa et al., 1982). Structured illumination microscopy (SIM) of endogenous COBL labeling in Ls174T-W4 cells showed a striking enrichment of puncta near or at the base of individual microvillar actin bundles (Figure 2B, zoom). This localization is consistent with a previous report on the targeting of enhanced green fluorescent protein (EGFP)–tagged COBL in the JEG-3 placental epithelial cell line (Wayt and Bretscher, 2014). Line scans revealed peak COBL signal at 0.17 ± 0.11 (tissue, Figure 2C) and 0.13 ± 0.12 (Ls174T-W4 endogenous, Figure 2D) when plotted as a function of normalized microvillar length (0 = base, 1 = tip). Overexpressed EGFP-tagged COBL exhibited localization similar to endogenous COBL, with peak localization at 0.16 ± 0.15 (Figure 3, A–C). Moreover, time-lapse imaging of EGFP-COBL in polarizing Ls174T-W4 cells revealed tight temporal and spatial overlap with F-actin, as visualized using mCherry-tagged UtrCH (Burkel et al., 2007), during BB assembly (Figure 3, D–F, and Supplemental Movie S3). Thus COBL enriches at the nascent apical domain during polarity establishment and localizes near or at the pointed ends of microvillar actin bundles, the expected site of action for a WH2 domain–containing nucleator.
FIGURE 2:
COBL localizes to the base of BB microvilli. (A) Endogenous COBL (green) and F-actin (magenta) labeling in a single villus from mouse small intestine. Boxed corners indicate zoom region, which highlights terminal web localization of COBL. (B) SIM projection of an induced Ls174T-W4 cell showing localization of endogenous COBL (green) and F-actin (magenta). Boxed corners indicate zoom region. (C, D) Line scans (n = 25) parallel to the microvillar axis show the distribution of endogenous COBL in mouse small intestine and Ls174T-W4 cells, respectively. Arrows highlight the peak localization (LP, mean ± SE) along the normalized microvillar axis. Scale bars, 5 μm.
FIGURE 3:
COBL localization at the base of the BB coincides with microvillar formation. (A) Schematic of full-length COBL domain organization; A–C refer to COBL WH2 domains; CC, coiled-coil; K, lysine-rich region; PRD, proline-rich domain. (B) SIM projection of an induced Ls174T-W4 cell overexpressing EGFP-COBL (green) and stained for F-actin (magenta). Boxed corners indicate zoom region. (C) Line scans (n = 25) parallel to the microvillar axis show the distribution of EGFP-COBL in Ls174T-W4 cells. Arrow highlights the peak localization. (D) Time series montage of a single Ls174T-W4 cell coexpressing EGFP-COBL and mCherry-UtrCH undergoing cell polarization and BB assembly. (E) Perimeter line-scan analysis of the cell shown in C at t = 0 and 220 min; black arrow in right-hand plot marks position of the BB. (F) Intensity profile over time of a 2.5-μm2 region of interest at the base of the forming BB in C. Scale bars, 5 μm.
Overexpression of COBL induces microvillar growth
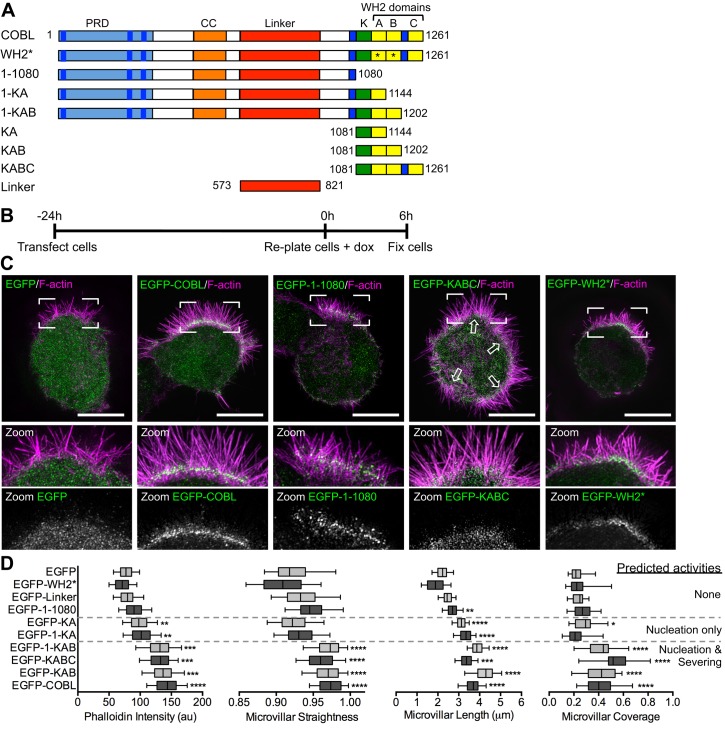
COBL comprises an N-terminal proline-rich domain (PRD), three C-terminal WH2 domains, and intervening predicted coiled-coil (CC) and linker motifs (Figure 4A). In Ls174T-W4 cells overexpressing EGFP-COBL, we noted that microvillar actin bundles appeared more numerous, longer, and straighter relative to EGFP-transfected controls (Figures 3B and 4C). Scoring of SIM images revealed that microvillar length, straightness, phalloidin intensity (proportional to F-actin content), and fraction of cell surface covered with microvilli were all significantly increased in cells overexpressing EGFP-COBL (Figure 4D). Truncation of COBL from the C-terminus, and thus elimination of WH2 domains, had a graded effect on microvillar morphology. Constructs with no WH2 domains showed little effect (1–1080, linker). Constructs with a single WH2 domain, which nucleate actin in vitro (1-KA, KA; Husson et al., 2011), showed a modest effect, whereas constructs with two or three WH2 domains, which nucleate and sever in vitro (Husson et al., 2011), elicited the most significant microvillar induction (1-KAB, KAB, KABC, COBL; Figure 4, C and D, and Supplemental Figure S2A). However, a full-length COBL construct with inactivating mutations in the first two WH2 domains (Chen et al., 2013) did not significantly affect microvillar morphology (WH2*, Figure 4, C and D). These results suggest that COBL promotes the growth of microvilli using a mechanism that takes advantage of multiple functional WH2 domains.
FIGURE 4:
Overexpression of COBL promotes microvillar growth. (A) Schematic of full-length COBL domain organization and truncated/mutated COBL constructs used in this study; CC, coiled-coil; K, lysine-rich region; PRD, proline-rich domain; *point mutations (L1113A and L1166A) that inactivate G-actin binding ability of COBL WH2 domains. (B) Schematic of procedure for overexpression experiments followed by (C) SIM projections of Ls174T-W4 cells overexpressing COBL constructs (green) and stained with phalloidin (magenta). Boxed corners indicate zoom region. White arrows in the EGFP-KABC image highlight multiple sites of microvillar assembly. (D) Quantification of microvillar phalloidin intensity, straightness, length, and surface coverage for cells overexpressing COBL constructs. Predicted activities listed to the right of the quantification are based on biochemical experiments published in Husson et al. (2011). Student’s t test (*p < 0.05, **p < 0.005, ***p < 0.0007) was used to determine significance for phalloidin intensity. For straightness, length, and coverage measurements, Mann–Whitney U tests (*p < 0.05, **p < 0.005, ***p < 0.0004, ****p < 0.0001) were used to determine significance. For all graphs, the boxes represent the 25th and 75th percentiles around the median, and the whiskers represent the 5th and 95th percentiles; 25 cells/condition, 10 microvilli/cell. Scale bars, 5 μm.
A recent study investigating COBL function in the JEG-3 placental cell line reported that overexpression reduced microvillar length (Wayt and Bretscher, 2014). Using SIM, we observed that JEG-3 cells overexpressing COBL assembled more microvilli, with higher levels of F-actin in individual protrusions (Supplemental Figure S2, B and C), findings that are consistent with our studies in IECs. We did observe shorter protrusions, however, suggesting that in the JEG-3 context, COBL overexpression and the subsequent assembly of numerous microvilli taxes the limited actin pool available for protrusion elongation (Burke et al., 2014). The same study suggested that the linker domain of COBL (Figure 4A and Supplemental Figure S2A, far right) was necessary and sufficient to target this molecule to the base of microvilli on the surface of JEG-3 placental epithelial cells (Wayt and Bretscher, 2014). When we overexpressed this domain in Ls174T-W4 cells, it enriched in the BB, but its localization was diffuse throughout the microvillus. Our truncation analysis and SIM imaging revealed that only fragments containing amino acids (aa) 1–1080 were enriched at the base of the BB, in a manner similar to endogenous COBL. Therefore, in addition to the linker domain, other motifs within the N-terminus are likely required for proper targeting.
Loss of COBL function impairs BB assembly
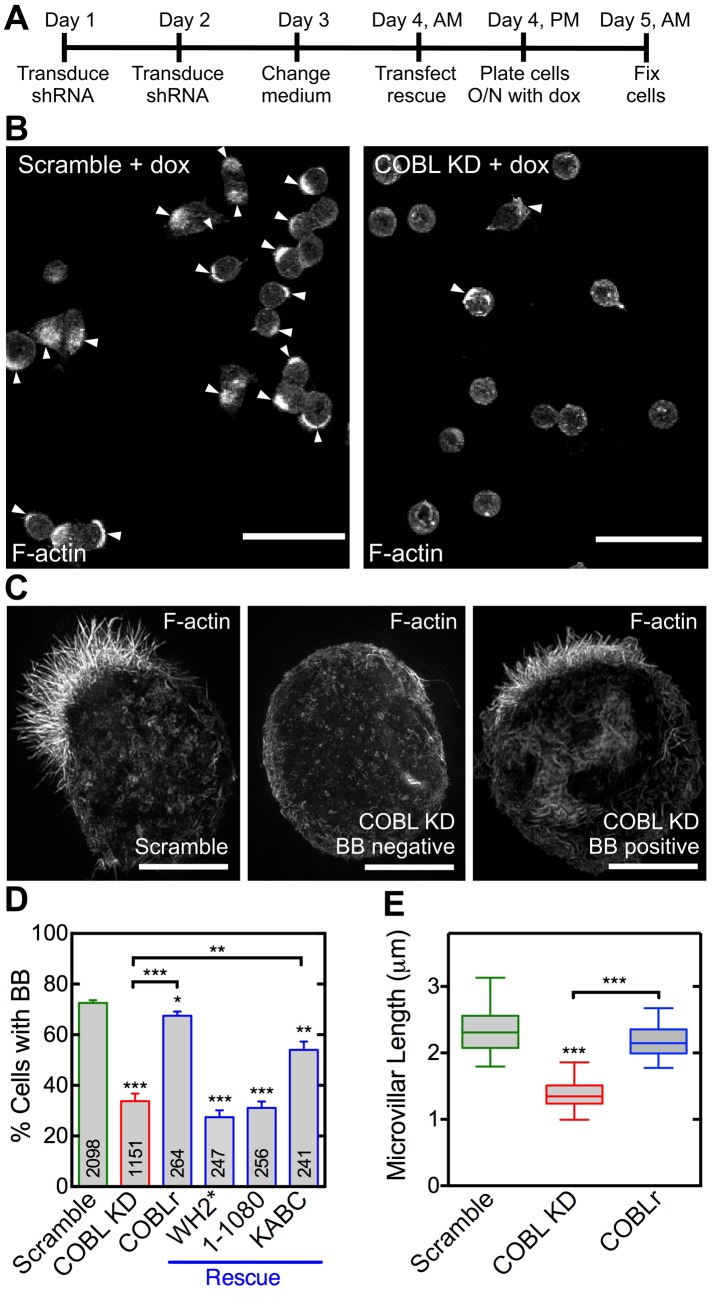
To determine how loss of COBL affects the formation of BB microvilli, we used short hairpin RNA (shRNA) to knock down expression in Ls174T-W4 cells (Figure 5 and Supplemental Figure S3). COBL knockdown (KD) significantly reduced the fraction of cells able to assemble BBs (33.8 ± 3.0% of COBL KD vs. 72.6 ± 1.0% of scramble control cells; Figure 5, A–D). This perturbation was specific to loss of COBL, as reexpression of a full-length COBL construct refractory to KD (COBLr) rescued BB formation (67.5 ± 1.6%; Figure 5D and Supplemental Figure S4A). Moreover, functional WH2 domains were required for rescue, as a full-length COBL construct with inactivated WH2 domains (WH2*) or a construct lacking WH2 domains (1–1080) failed to improve the KD phenotype (27.3 ± 2.7 and 31.4 ± 2.7%, respectively; Figure 5D and Supplemental Figure S4, B and C). Reexpression of the KABC fragment containing all three functional WH2 domains yielded a partial rescue (54.0 ± 3.1%; Figure 5D and Supplemental Figure S4D).
FIGURE 5:
COBL is required for microvillar growth. (A) Schematic of experimental design. (B) Images of scramble control or COBL KD cells scored for BB-positive cells. Arrowheads highlight BB-positive cells. Scale bar, 40 μm. (C) SIM projection images of scramble control cell, COBL KD cell without a BB, and COBL KD cell with a BB stained with phalloidin. Scale bars, 5 μm. (D) Percentage of BB-positive cells in KD and rescue experiments. Mean ± SEM from four separate experiments; number per condition is listed within bars (Student’s t test, *p < 0.05, **p ≤ 0.008, ***p ≤ 0.0004). (E) Quantification of microvillar length from scramble, COBL KD, and rescue cells with COBLr that were scored as BB-positive cells in D. Boxes represent the 25th and 75th percentiles around the median, and the whiskers represent the 5th and 95th percentiles; 30 cells/condition (Mann–Whitney U test, ***p < 0.0005).
COBL KD did not completely abolish BB assembly in Ls174T-W4 cells, despite >95% reduction in protein levels (Supplemental Figure S3B), suggesting that other molecules probably contribute to microvillar formation. This would be consistent with the partial penetrance observed in studies with knockout mouse models lacking major microvillar structural proteins (Saotome et al., 2004; Tyska et al., 2005; Revenu et al., 2012). However, when we used SIM to more closely examine COBL KD cells that were scored as “BB positive” in low-magnification fields (Figure 5B), their microvilli appeared significantly shorter than those assembled by scramble control cells (1.39 ± 0.05 vs. 2.35 ± 0.07 μm; Figure 5, C and E). Shorter length was rescued by expression of a full-length COBL construct refractory to KD (COBLr; 2.17 ± 0.05 μm; Figure 5E). These loss-of-function and rescue experiments indicate that COBL is required for normal microvillar formation, and functional WH2 domains are needed for proper function in this context.
COBL targeting to the apical domain requires syndapin-2
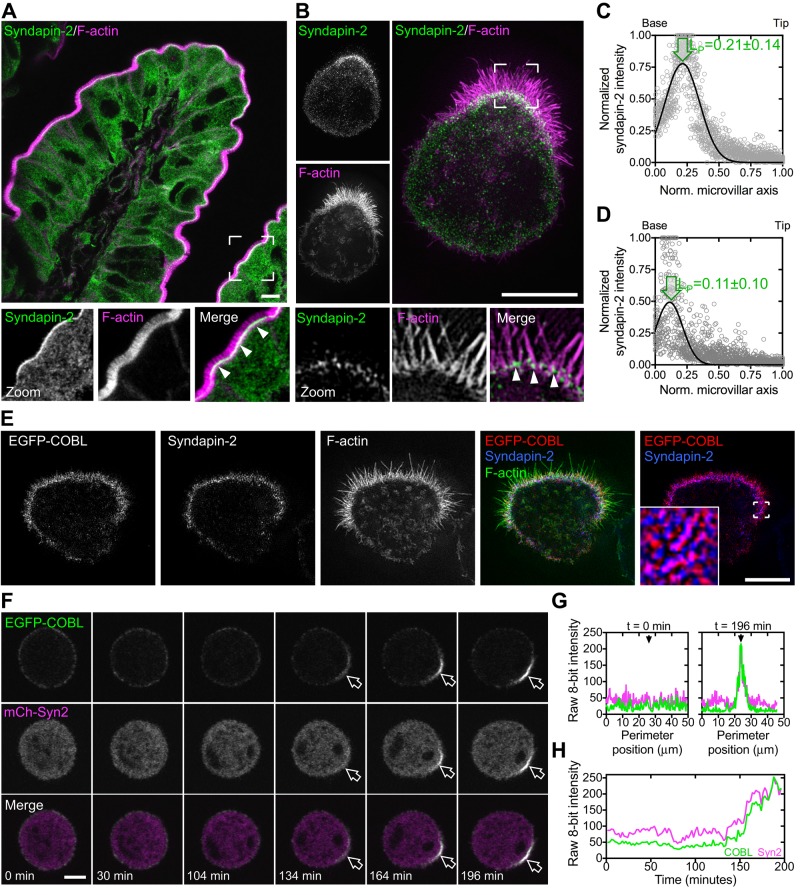
The F-BAR–domain protein syndapin-1 has been shown to target COBL to sites of neurite outgrowth in neurons (Schwintzer et al., 2011). In that context, targeting is mediated by direct binding between the N-terminal PRD of COBL and the C-terminal SH3 domain of syndapin-1. Whereas syndapin-1 is specific to the CNS and syndapin-3 is only expressed in skeletal muscle and heart tissue, syndapin-2 is ubiquitously expressed (Quan and Robinson, 2013). Of importance, COBL has been shown to directly bind all three syndapins in vitro (Schwintzer et al., 2011). However, the functional implications of interactions between COBL and syndapin-2 or -3 have yet to be investigated in cells. To determine whether syndapin-2 plays a role in targeting COBL to the apical surface of IECs, we first examined the expression and localization of syndapin-2 in small intestinal tissue and the Ls174T-W4 model system. Syndapin-2 was enriched at the base of microvilli in human small intestine (Figure 6A) and in Ls174T-W4 cells (Figure 6B). Line scans showed that, similar to COBL, syndapin-2 localizes near the pointed ends of microvillar actin bundles, with peak localization at 0.21 ± 0.14 (tissue, Figure 6C) and 0.11 ± 0.10 (Ls174T-W4 cells, Figure 6D) when plotted as a function of microvillar length. Furthermore, endogenous syndapin-2 shows striking colocalization with EGFP-COBL at the base of microvilli in Ls174T-W4 cells (Figure 6E). Time-lapse imaging of EGFP-COBL and mCherry–syndapin-2 in polarizing Ls174T-W4 cells also revealed spatial and temporal colocalization at the base of forming BBs (Figure 6, F–H, and Supplemental Movie S4). Thus syndapin-2 is well positioned to interact with COBL and facilitate its targeting to the apical membrane during epithelial differentiation.
FIGURE 6:
Syndapin-2, similar to COBL, localizes to the base of BB microvilli. (A) Endogenous syndapin-2 localization (green) and F-actin signal (magenta) in a villus from human small intestine. Boxed corners indicate zoom region; arrowheads highlight syndapin-2 localization near the base of the BB. (B) SIM projection of endogenous syndapin-2 (green) in an Ls174T-W4 cell reveals enrichment at the base of the BB. Boxed corners indicate zoom region. Arrowheads highlight syndapin-2 localization at the base of the BB. (C, D) Line scans parallel to the microvillar axis (n = 25) show the distribution of endogenous syndapin-2 in human small intestine and Ls174T-W4 cells, respectively. Arrows highlight peak localization. (E) SIM projection of EGFP-COBL (red), endogenous syndapin-2 (blue), and F-actin (green) shows colocalization of EGFP-COBL and syndapin-2 at the base of the BB. Boxed corners in the red/blue channel overlay indicate the zoom region. (F) Time series montage of an Ls174T-W4 cell coexpressing EGFP-COBL and mCherry–syndapin-2 undergoing cell polarization and BB assembly. (G) Perimeter line-scan analysis of the cell shown in C at t = 0 and 196 min; black arrow in right-hand plot marks the position of the BB. (H) Intensity profile over time of a 2.5-μm2 region of interest at the base of the forming BB in F. Scale bars, 5 μm.
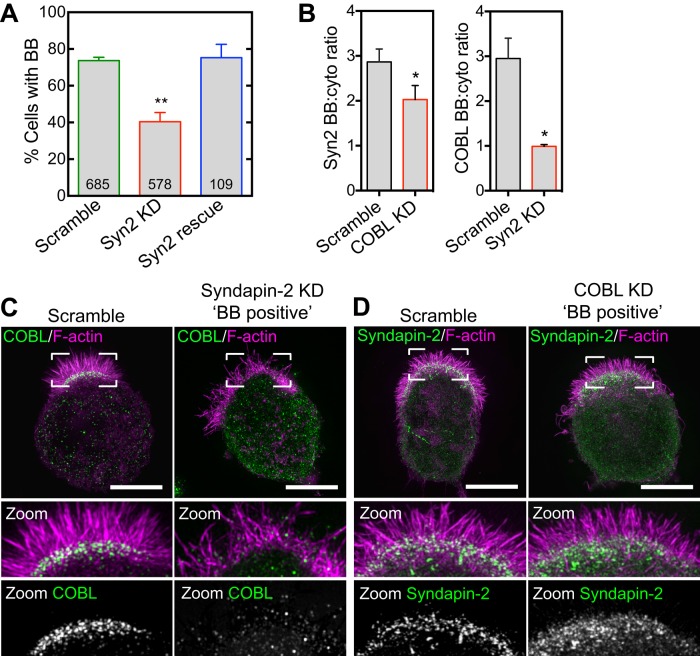
We next sought to determine the effect of syndapin-2 KD on BB assembly and COBL targeting in Ls174T-W4 cells. Similar to COBL KD, syndapin-2 KD resulted in a significant decrease in the percentage of cells capable of forming BBs (40.7 ± 2.4%) relative to controls (73.8 ± 1.2%; Figure 7A and Supplemental Figure S5, A–C). This phenotype was rescued by reintroducing a mouse syndapin-2 construct refractory to KD (75.3 ± 4.2%; Figure 7A and Supplemental Figure S5D). Moreover, inspection of BB-positive syndapin-2 KD cells revealed a significant loss of COBL localization from the base of microvilli (Figure 7, B and C). In the reciprocal experiment, COBL KD had only a minor effect on syndapin-2 signal at the apical domain (Figure 7, B and D). Thus these data suggest that in the context of IECs, syndapin-2 functions upstream of COBL and is needed for targeting of this factor to the apical membrane.
FIGURE 7:
Syndapin-2 targets COBL to the base of the BB. (A) BB formation in response to syndapin-2 KD and rescue. Mean ± SEM from three separate experiments; number per condition is listed within the bars (Student’s t test, **p < 0.004). (B) Left, syndapin-2 BB enrichment in scramble and COBL KD cells. Right, COBL BB enrichment in scramble and syndapin-2 KD cells. Data are displayed as mean ± SEM (Student’s t test, *p < 0.05); 30 cells/condition. (C) SIM projections of COBL immunostaining (green) and F-actin (magenta) in control and syndapin-2 KD cells. Boxed corners indicate zoom region. (D) SIM projections of syndapin-2 immunostaining (green) and F-actin (magenta) in control and COBL KD cells. Boxed corners indicate zoom region. Scale bars, 5 μm.
A recent study examining the localization of overexpressed proteins in JEG-3 cells concluded that COBL is responsible for targeting syndapin-2 to the membrane (Wayt and Bretscher, 2014). This conflict with our observations might suggest that the function of COBL–syndapin-2 complex is context specific. However, our results are consistent with studies performed in neurons, which revealed that syndapin-1 targets COBL to sites of F-actin assembly during dendritogenesis (Schwintzer et al., 2011).
DISCUSSION
Effect of COBL on microvillar morphology
Our findings indicate that COBL plays a significant role in promoting microvillar growth and BB assembly during the differentiation of cultured IECs. In vitro biochemical studies with COBL fragments revealed a range of activities, including nucleation, severing of ADP–F-actin, sequestration of ADP–G-actin, and profilin-like binding to ATP-actin (Husson et al., 2011). In our assays, overexpression of COBL and COBL fragments containing multiple active WH2 domains induced striking effects on microvillar growth. Cells transfected with longer fragments of the COBL C-terminus (1-KAB, KAB, KABC), which are predicted to nucleate and sever (Husson et al., 2011), significantly increased microvillar surface coverage (Figure 4D). One possible interpretation of this effect is that generating large numbers of microvilli during BB assembly requires cooperation between COBL nucleation and severing activities, both of which would increase the number of elongation-competent filaments.
COBL overexpression also increased phalloidin intensity along the microvillar axis, suggesting more filaments per core bundle (Figure 4D). Microvilli were also longer and straighter, which might represent secondary effects of having more filaments per core (Figure 4D). Thicker bundles would exhibit a higher flexural rigidity (Gittes et al., 1993), which could explain their straighter appearance in SIM images. Bundles containing more filaments would also have more force-generating barbed ends pushing against the apical membrane, which could explain the increased length of COBL-induced microvilli. However, we cannot rule out other potential mechanisms that might influence microvillar length. For example, COBL might recruit elongation/anticapping factors such as Ena/VASP family members or formins (Chesarone and Goode, 2009) to the apical domain, although our experiments would argue against significant involvement of formins in BB assembly (Figure 1, D and F).
COBL KD completely inhibited microvillar formation in a large fraction of cells and stunted microvillar growth in the remaining cells (Figure 5). Rescue experiments also revealed that functional WH2 domains, which mediate nucleation and severing (Ahuja et al., 2007; Husson et al., 2011), are needed for these effects. Although COBL KD was >95% effective based on Western blots (Supplemental Figure S3), ∼35% of COBL KD cells still formed rudimentary BBs, suggesting that other pathways compensate for the loss of COBL in this model. Such compensatory mechanisms are expected when we consider that BBs still form in mouse models lacking key structural proteins, including myosin-1a (Tyska et al., 2005), ezrin (Saotome et al., 2004), or villin, espin, and plastin-1 (Revenu et al., 2012).
Significance of COBL localization
Superresolution analysis of COBL localization in native intestinal tissue and cultured IECs revealed enrichment near, but not precisely at, the pointed ends of microvillar actin bundles. Population averages show that COBL enrichment exhibits a peak at ∼15% of total bundle length from the pointed end (Figures 2, C and D, and 3B). Because our SIM images are limited to a lateral resolution of ∼110 nm (approximate diameter of a core bundle), we are unable to determine whether COBL is integrated within mature microvillar actin bundles, binds to the sides of core bundles, or is merely anchored near bundle rootlets. However, a truncated form of COBL lacking C-terminal WH2 domains targeted to the apical domain but failed to demonstrate the focused enrichment of full-length COBL near the pointed ends (Figure 4C). Thus direct binding to actin contributes to the precise distribution of COBL observed in our SIM images.
Is COBL localization consistent with a role in generating microvillar actin bundles? Because WH2 domain–containing proteins generate new filaments by creating nuclei that elongate primarily from their barbed ends, these factors are expected to localize to filament pointed ends (Quinlan et al., 2005; Ahuja et al., 2007; Zuchero et al., 2009). However, the actin nucleator APC remains bound to the filament it nucleates without inhibiting elongation of the pointed end (Breitsprecher et al., 2012). As elongation proceeds, APC is left near but not precisely at the pointed end. A similar mechanism might help explain the localization of COBL: after formation of a COBL-actin nucleus, elongation might proceed from both ends of the nucleus, leaving COBL behind at the original site of nucleation. This model is consistent with a recent structural study that proposed that COBL-actin nuclei initially elongate from the pointed end before barbed-end elongation begins (Chen et al., 2013). Biochemical studies have also shown that both ends of microvillar core bundles are uncapped and remain competent for elongation (Mooseker et al., 1982). Enrichment of COBL near the base of microvillar F-actin bundles would also be consistent with a role in binding and severing ADP–F-actin. Although it is appealing to interpret COBL localization data only in terms of nucleation and severing activities (Ahuja et al., 2007; Husson et al., 2011; Haag et al., 2012), our experiments do not allow us to rule out other contributions to microvillar formation, such as anchoring core bundles in the terminal web.
Apical targeting of COBL requires syndapin-2
For COBL to function in microvillar growth, it must be targeted to the apical plasma membrane during differentiation. BAR domain–containing proteins are now recognized as membrane-targeting factors for actin polymerization machinery (Aspenstrom, 2009). BAR domains are small helix bundles that form dimers capable of sensing and inducing membrane curvature (Peter et al., 2004). Our studies indicate that syndapin-2, which contains an N-terminal F-BAR domain, is required for COBL targeting to the apical domain (Figure 7). Because the N-terminal polyproline motifs in COBL interact with the SH3 domains of all three syndapin family members (Schwintzer et al., 2011), this requirement probably reflects direct binding between syndapin-2 and COBL. Consistent with this, the N-terminal region of COBL is sufficient for apical targeting (Figure 4). Thus syndapin-2 likely provides COBL with a physical link to the apical plasma membrane. Of interest, syndapin-family F-BAR domains bind membranes with high positive (i.e. inward) curvature (Quan and Robinson, 2013), such as the plasma membrane found between adjacent microvilli. The intermicrovillar region is also an established site of endocytosis, which is intriguing, given that syndapins have been implicated in membrane internalization (Qualmann and Kelly, 2000).
Model and conclusions
Our studies lead us to the following model for microvillar growth during BB assembly (Figure 8). We propose that COBL targets to the nascent apical domain of differentiating IECs using the membrane-binding capacity of its binding partner, syndapin-2. There COBL contributes to microvillar formation using one or more of its actin-regulating activities, including nucleation, severing, and/or anchoring. After filament formation, new filaments elongate and are bundled by factors such as villin, fimbrin, espin, and EPS8 to generate a core bundle of mature length (∼1–3 μm; Crawley et al., 2014). Although minor elongation may take place off the pointed ends, the barbed ends grow much faster. Dynamics at the barbed ends will be controlled by capping and anticapping factors, which are likely components of the electron-dense “tip complex” that appears to serve as a membrane attachment site for filaments in both growing and mature microvilli (Tilney and Cardell, 1970; Burgess and Grey, 1974; Mooseker et al., 1982).
FIGURE 8:
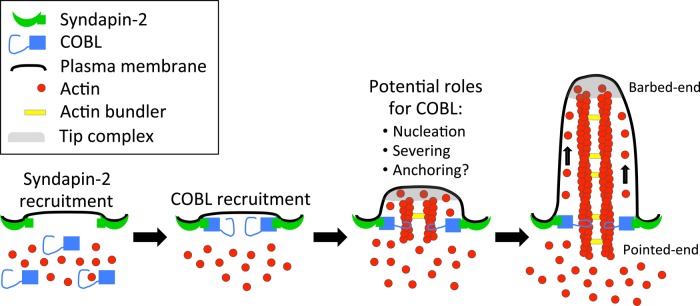
Model depicting a role for COBL in the formation of brush border microvilli.
In conclusion, our results provide new insight on a COBL-dependent mechanism of microvillar growth during BB assembly. We expect future studies to focus on identifying elongation and capping factors that function in microvillar formation and determining how syndapin-2–dependent targeting of COBL is controlled by upstream factors that regulate polarity establishment (Gloerich et al., 2012). Future work must also elaborate on the findings presented here by exploring roles for COBL in intestinal diseases characterized by perturbations of BB microvilli (Davidson et al., 1978; Vallance et al., 2002).
MATERIALS AND METHODS
Cell culture
Ls174T-W4 cells (a generous gift of Hans Clevers, Hubrecht Institute for Developmental Biology and Stem Cell Research, Royal Dutch Academy of Sciences, and UMC Utrecht, The Netherlands) were cultured in DMEM with high glucose and 2 mM l-glutamine supplemented with 10% tetracycline-free fetal bovine serum (FBS), G418 (1 mg/ml), blasticidin (10 μg/ml), and phleomycin (20 μg/ml). JEG-3 and B16F1 cells (American Type Culture Collection) were cultured in DMEM with high glucose and 2 mM l-glutamine supplemented with 10% FBS. All cells were grown at 37°C and 5% CO2.
Transfections and lentivirus production
Transient transfections were performed using Lipofectamine 2000 (Invitrogen, Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. Lentivirus particles were generated by cotransfecting HEK293FT cells (T75 flasks at 80% confluency) with 6 μg of pLKO.1 shRNA KD plasmids (Open Biosystems; COBL, TRCN0000005530, TRCN0000005531; syndapin-2, TRCN0000037982), 4 μg of psPAX2 packaging plasmid, and 0.8 μg of pMD2.G envelope plasmid using FuGENE transfection reagent (Roche, Basel, Switzerland). Cells were incubated for 2 d to allow for lentiviral production. Media containing lentivirus was collected and particles concentrated with the addition of Lenti-X concentrator reagent (Clontech, Mountain View, CA). For COBL KD, separate viruses were prepared and concentrated for each shRNA KD plasmid and then pooled together before transduction to achieve a suitable KD. Lentivirus transduction of Ls174T-W4 cells and rescue of KD was performed as described previously (Gloerich et al., 2012). Briefly, cells were infected with lentiviral shRNAs targeting human COBL on two consecutive days. Two days after the second infection, cells were seeded overnight in the absence or presence of doxycycline. Cells were then prepared for microscopy or for SDS–PAGE and immunoblotting. For rescue experiments, cells were transiently transfected 2 d after the second infection using Lipofectamine 2000 (Invitrogen), induced with 1 μg/ml doxycycline overnight, and fixed the following morning for immunofluorescence.
Immunofluorescence
Cells were first washed once with warm phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde in PBS for 15 min at 37°C. After fixation, cells were washed three times with PBS and permeabilized with 0.1% Triton X-100/PBS for 15 min at room temperature. Cells were then washed three time with PBS and blocked for 1 h at 37°C in 5% bovine serum albumin (BSA)/PBS. Primary antibodies anti-COBL (1:100, HPA019033; Sigma-Aldrich, St. Louis, MO), anti–syndapin-2 (1:200, HPA049854; Sigma-Aldrich), or anti-GFP (1:200, GFP-1020; Aves Labs) were diluted in PBS and incubated with cells at 37°C for 2 h, followed by four washes with PBS. Cells were then incubated for 1 h with goat anti-rabbit Alexa Fluor 488 (A11008; Invitrogen) and Alexa Fluor 568–phalloidin (A12380; Invitrogen) at room temperature (each at 1:200 in PBS). Coverslips were then washed four times and mounted in ProLong Gold Antifade Mountant (P36930; Invitrogen). Cells or tissue samples were imaged at low magnification using a 40× objective on a TCS SP5 laser-scanning confocal microscope (Leica) at the Vanderbilt Cell and Developmental Biology Core, and SIM images were acquired using an Applied Precision DeltaVision OMX (GE Healthcare, Pittsburgh, PA) located in the Vanderbilt Cell Imaging Shared Resource and processed using softWoRx software (GE Healthcare). High-magnification confocal images of Ls174T-W4 cells were acquired on a Nikon A1R confocal microscope equipped with 488- and 561-nm excitation lasers and a 100×/1.49 numerical aperture (NA) Apo total internal reflection fluorescence (TIRF) objective.
OCT-embedded mouse (anti-COBL) or human (anti–syndapin-2) small intestinal tissue was cut into 5-μm-thick sections onto Superfrost plus–coated slides (Fisher, Waltham, MA) and stored at −80°C. For immunohistochemistry, slides were warmed to room temperature, and OCT was removed with three washes for 5 min each with PBS supplemented with 50 mM ethylene glycol tetraacetic acid (EGTA)-K; this buffer was used for all subsequent steps. Tissue was then permeabilized with 0.3% Triton X-100 in PBS/EGTA for 5 min at room temperature. After three washes, sections were blocked using 10% BSA in PBS/EGTA for 40 min at room temperature. Block was removed with one wash, and sections were incubated in anti-COBL (1:50, HPA019167) or anti–syndapin-2 (1:100, HPA049854) antibodies in PBS/EGTA overnight at 4°C. The next day, sections were washed three times with PBS/EGTA and incubated for 1 h with goat anti-rabbit Alexa Fluor 488 and Alexa Fluor 568–phalloidin at room temperature (each at 1:200 in supplemented PBS). Slides were then washed three times and mounted in ProLong Gold Antifade Mountant and imaged using a Leica TCS SP5 laser-scanning confocal microscope.
Drug treatments
Ls174T-W4 cells were plated on coverslips and allowed to adhere; 30 min before induction with 1 μg/ml doxycycline, cells were incubated with 100 nM latrunculin A (L5163; Sigma-Aldrich), 30 μM cytochalasin B (C6762; Sigma-Aldrich), 250 nM jasplakinolide (J4580; Sigma-Aldrich), 50 μM CK-666 (SML0006; Sigma-Aldrich), or 12.5 μM SMIFH2 (S4826; Sigma-Aldrich). After 6 h of incubation with doxycycline and inhibitors, cells were fixed, washed, and stained with Alexa Fluor 488–phalloidin (A12379; Invitrogen), washed four times with PBS, and mounted on slides using Prolong Gold Antifade Mountant. Cells were imaged as described earlier.
Microscopy
SIM was performed using an Applied Precision DeltaVision OMX equipped with a 60× Plan-Apochromat N/1.42 NA objective and processed using softWoRx software (GE Healthcare). Confocal microscopy was performed using either a Leica TCS SP5 laser-scanning confocal microscope (fixed cells) or a Nikon A1R laser-scanning confocal microscope (fixed and live cells). For live-cell imaging of Ls174T-W4 cells, 1 d before imaging, cells were transfected in a T25 flask at 80% confluency using Lipofectamine 2000 as described by the manufacturer (11668; Invitrogen). The next day, cells were plated on glass-bottom dishes and allowed to adhere for 6 h. Imaging was performed on a Nikon A1R confocal microscope equipped with 488- and 561-nm excitation lasers and a 100×/1.49 NA Apo TIRF objective. Doxycycline was added to the media at 1 μg/ml just before image acquisition began. Cells were maintained in a humid environment at 37°C and 5% CO2 using a stage-top incubation system (Tokai Hit). Image acquisition was controlled with Nikon Elements software.
Western blots
Samples were diluted with Laemmli sample buffer and heated at 95°C for 5 min, and equal volumes were loaded on a 4–12% Nu-Page gradient gel (Invitrogen). Proteins were transferred in Towbin buffer, pH 8.3, to nitrocellulose at 10 V overnight, which was subsequently blocked for 1 h in 5% milk-PBS. Primary antibodies against COBL (1:250, HPA019167; Sigma-Aldrich), syndapin-2 (1:250, HPA049854; Sigma-Aldrich) and glyceraldehyde 3-phosphate dehydrogenase (1:1000; Cell Signaling) were diluted in PBS containing 0.1% Tween-20 (PBS-T) and incubated with the membranes overnight at 4°C. Membranes were washed four times with PBS-T and then incubated with donkey anti-rabbit 800 IRdye (1:5000, 926-32213; Li-Cor) for 30 min. Membranes were again washed four times with PBS-T and imaged using a Li-Cor Odyssey infrared imaging system. Images of membrane scans were cropped and contrast adjusted using ImageJ (National Institutes of Health, Bethesda, MD).
Plasmids and molecular biology
pENTR(tm)221 COBL (IOH536; Invitrogen) corresponding to full-length human COBL was purchased and shuttled into a pEGFP-C1 vector (Clontech) adapted for Gateway cloning using the Gateway conversion kit (Invitrogen) to generate pEGFPC1-COBL and verified by sequencing. Using full-length COBL as template, PCR was used to create the truncated fragment KABC (aa 1081–1261) with 5′ XhoI and 3′ BamHI sites, which allowed ligation into linearized pEGFP-C1 to create pEGFP-KABC. To generate pEGFP-WH2*, mutations L1113A and L1166A were introduced into pEGFP-COBL using QuikChange site-directed mutagenesis (Agilent, Santa Clara, CA). All other EGFP-tagged truncation constructs, 1–1080, 1-KA (aa 1–1144), 1-KAB (aa 1–1202), KA (aa 1081–1144), KAB (aa 1081–1202), and linker (aa 573-821), were generated using PCR with the full-length COBL construct described earlier as template. PCR products were first shuttled into pDONR/Zeo or pCR8/GW/TOPO, and then into a Gateway-adapted pEGFP-C1 vector using Gateway cloning (Invitrogen). To generate a COBL construct refractory to KD, two silent mutations per shRNA target were introduced into full-length COBL using site-directed mutagenesis. For COBL shRNA TRCN0000005530, which targets COBL nucleotides (nts) 1840–1860, nt 1845 (t → c) and nt 1851 (c → t) were mutated. For COBL shRNA TRCN0000005531, which targets COBL nts 3208–3228, nt 3213 (a → g) and nt 3219 (c → t) were mutated. These silent mutations were also introduced into pEGFP-1-1080 and pEGFP-WH2*. Neither shRNA targeted the KABC sequence. pmCherry–syndapin-2 (mouse) was purchased from Addgene (27681; deposited by C. Merrifield; Taylor et al., 2011), as were EGFP- and mCherry-tagged variants of UtrCH (26737 and 26740, deposited by W. Bement; Burkel et al., 2007). Syndapin-2 was then excised from pmCherry–syndapin-2 using EcoRI and BamHI and ligated into pEGFP-C1 to generate pEGFP–syndapin-2. All newly generated plasmids were verified by sequencing.
Image analysis and quantification
Excluding line-scan analysis, all quantitative data are from at least three experiments. For length, straightness, phalloidin intensity, and microvillar coverage, the D’Agostino and Pearson omnibus normality test was performed to determine whether the data were normally distributed. Normally distributed data were statistically analyzed using the unpaired Student’s t test. If the data were not normal, the Mann–Whitney U test was used to determine significance. Statistical analyses performed are stated in the figure legends. All graphs were generated and statistical analyses performed using Prism, version 6 (GraphPad, La Jolla, CA). All image analysis was performed using ImageJ. For quantification of percentage of cells with BB, cells were scored as BB positive if they displayed polarized F-actin accumulation as visualized using a 40× objective on a Leica TCS SP5 confocal microscope. To perform line-scan analysis, a line was drawn along the microvillar axis using F-actin signal (visualized with phalloidin staining) as a reference; the intensity of the COBL or syndapin-2 signal along that line was then recorded. Intensities from individual line-scans were normalized such that the maximum value was equal to 1. The length axis from individual scans was also normalized such that the base of the microvillus was set equal to 0 and the distal tip to 1. Normalized line-scans were then plotted together and fit to a single Gaussian using nonlinear regression (Prism, version 6); the resulting fit parameters revealed the position of peak localization (LP) along the microvillar axis. For perimeter analysis of live differentiating Ls174T-W4 cells, a line beginning on the opposite side of the cell from where the BB was formed was drawn along the perimeter of the cell, and the COBL and UtrCH intensities were plotted along that line for each time point indicated. To measure intensity over time in differentiating Ls174T-W4 cells, a 2.5-μm2 region of interest was drawn at the center of the base where the BB formed, and the average intensity of that area for COBL and UtrCH was plotted over time. Microvillar length measurements were performed on projected SIM images by tracing individual microvillar actin bundles using ImageJ. Microvillar straightness was determined as the ratio between a straight line drawn directly from the base to the tip of a microvillus and the actual length of the protrusion. Microvillar phalloidin intensity was measured in unprocessed images by drawing a line along the length of single protrusion and averaging the intensity along that line. For analyses in which individual microvilli were measured, 10 microvillar actin bundles were scored per cell. Microvillar coverage, that is, fraction of the cell perimeter occupied by microvilli, was determined from projected SIM images using ImageJ. In JEG-3 cells, microvillar number was quantified in ImageJ by tracing an area of a cell, counting the number of microvilli in that area for COBL-overexpressing and untransfected cells, and then normalizing the value to microvilli per 100 μm2. For measuring enrichment of COBL and syndapin-2 at the base of the BB in Ls174T-W4 cells, the base of the BB as determined by phalloidin staining was traced with an eight-pixel-wide line, and the mean intensity of both signals along that line was determined. The same line was then moved to a region ∼2 μm below the base of the BB (that did not include the nucleus), and the mean intensity of COBL and syndapin-2 along that line was determined. BB:cytosol enrichment was defined as the ratio of these two mean intensities.
Supplementary Material
Acknowledgments
We thank all members of the Tyska laboratory, Vanderbilt Microtubule and Motors Club, and Vanderbilt Epithelial Biology Center for advice and support. We thank Anne Powell for providing human intestinal tissue samples. Acquisition of the OMX superresolution microscope was supported by 1S10-OD012324 (M.J.T.). This work was also supported by a Vanderbilt University Medical Center Cellular, Biochemical, and Molecular Sciences training grant (N.G.L.), an American Heart Association Predoctoral Fellowship (N.G.L.), an American Heart Association Postdoctoral Fellowship (S.W.C.), and National Institutes of Health Grants R01-DK075555 and R01-DK095811 (M.J.T.).
Abbreviations used:
- Arp 2/3
actin-related protein 2/3
- BB
brush border
- COBL
cordon bleu
- EGFP
enhanced green fluorescent protein
- F-BAR
Fes and Cip4 homology Bin-Amphiphysin-Rvs
- FH2
formin homology 2 domain
- IEC
intestinal epithelial cell
- PRD
proline-rich domain
- SIM
structured illumination microscopy
- WH2
Wiskott–Aldrich homology 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0443) on September 9, 2015.
REFERENCES
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenstrom P. Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol. 2009;272:1–31. doi: 10.1016/S1937-6448(08)01601-8. [DOI] [PubMed] [Google Scholar]
- Baas AF, Boudeau J, Sapkota GP, Smit L, Medema R, Morrice NA, Alessi DR, Clevers HC. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–466. doi: 10.1016/s0092-8674(04)00114-x. [DOI] [PubMed] [Google Scholar]
- Bartles JR, Zheng L, Li A, Wierda A, Chen B. Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol. 1998;143:107–119. doi: 10.1083/jcb.143.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol. 1983;97:425–432. doi: 10.1083/jcb.97.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K. Villin: the major microfilament-associated protein of the intestinal microvillus. Proc Natl Acad Sci USA. 1979;76:2321–2325. doi: 10.1073/pnas.76.5.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DR, Grey RD. Alterations in morphology of developing microvilli elicited by cytochalasin B. Studies of embryonic chick intestine in organ culture. J Cell Biol. 1974;62:566–574. doi: 10.1083/jcb.62.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EA, Gerrelli D, Gasca S, Berg E, Beier DR, Copp AJ, Klingensmith J. Cordon-bleu is a conserved gene involved in neural tube formation. Dev Biol. 2003;262:16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- Chen X, Ni F, Tian X, Kondrashkina E, Wang Q, Ma J. Structural basis of actin filament nucleation by tandem W domains. Cell Rep. 2013;3:1910–1920. doi: 10.1016/j.celrep.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Crawley SW, Mooseker MS, Tyska MJ. Shaping the intestinal brush border. J Cell Biol. 2014;207:441–451. doi: 10.1083/jcb.201407015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson GP, Cutz E, Hamilton JR, Gall DG. Familial enteropathy: a syndrome of protracted diarrhea from birth, failure to thrive, and hypoplastic villus atrophy. Gastroenterology. 1978;75:783–790. [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors—structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloerich M, Ten Klooster JP, Vliem MJ, Koorman T, Zwartkruis FJ, Clevers H, Bos JL. Rap2A links intestinal cell polarity to brush border formation. Nat Cell Biol. 2012;14:793–801. doi: 10.1038/ncb2537. [DOI] [PubMed] [Google Scholar]
- Haag N, Schwintzer L, Ahuja R, Koch N, Grimm J, Heuer H, Qualmann B, Kessels MM. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J Neurosci. 2012;32:17842–17856. doi: 10.1523/JNEUROSCI.0843-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, Milanesi F, Hazelwood L, Disanza A, Liu H, Perlade E, Malabarba MG, Pasqualato S, Maiolica A, Confalonieri S, et al. Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PLoS Biol. 2010;8:e1000387. doi: 10.1371/journal.pbio.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Tilney LG, Fujiwara K, Heuser JE. Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J Cell Biol. 1982;94:425–443. doi: 10.1083/jcb.94.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CL, Mooseker MS. Characterization of the 110-kdalton actin-calmodulin-, and membrane-binding protein from microvilli of intestinal epithelial cells. J Cell Biol. 1983;97:974–985. doi: 10.1083/jcb.97.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson C, Renault L, Didry D, Pantaloni D, Carlier MF. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell. 2011;43:464–477. doi: 10.1016/j.molcel.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Majstoravich S, Zhang J, Nicholson-Dykstra S, Linder S, Friedrich W, Siminovitch KA, Higgs HN. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require Wiskott-Aldrich syndrome protein (WASp) for their morphology. Blood. 2004;104:1396–1403. doi: 10.1182/blood-2004-02-0437. [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer. 2012;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- McConnell RE, Benesh AE, Mao S, Tabb DL, Tyska MJ. Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol. 2011;300:G914–G926. doi: 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, Pollard TD, Wharton KA. Nucleated polymerization of actin from the membrane-associated ends of microvillar filaments in the intestinal brush border. J Cell Biol. 1982;95:223–233. doi: 10.1083/jcb.95.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan A, Robinson PJ. Syndapin—a membrane remodelling and endocytic F-BAR protein. FEBS J. 2013;280:5198–5212. doi: 10.1111/febs.12343. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Ravanelli AM, Klingensmith J. The actin nucleator Cordon-bleu is required for development of motile cilia in zebrafish. Dev Biol. 2011;350:101–111. doi: 10.1016/j.ydbio.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C, Ubelmann F, Hurbain I, El-Marjou F, Dingli F, Loew D, Delacour D, Gilet J, Brot-Laroche E, Rivero F, et al. A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol Biol Cell. 2012;23:324–336. doi: 10.1091/mbc.E11-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell. 2015;32:54–67. doi: 10.1016/j.devcel.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome I, Curto M, McClatchey AI. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev Cell. 2004;6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Schuler S, Hauptmann J, Perner B, Kessels MM, Englert C, Qualmann B. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator Cobl. J Cell Sci. 2013;126:196–208. doi: 10.1242/jcs.111674. [DOI] [PubMed] [Google Scholar]
- Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011;30:3147–3159. doi: 10.1038/emboj.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 2015;32:43–53. doi: 10.1016/j.devcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster JP, Jansen M, Yuan J, Oorschot V, Begthel H, Di Giacomo V, Colland F, de Koning J, Maurice MM, Hornbeck P, et al. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev Cell. 2009;16:551–562. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Cardell RR. Factors controlling the reassembly of the microvillous border of the small intestine of the salamander. J Cell Biol. 1970;47:408–422. doi: 10.1083/jcb.47.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Mackey AT, Huang JD, Copeland NG, Jenkins NA, Mooseker MS. Myosin-1a is critical for normal brush border structure and composition. Mol Biol Cell. 2005;16:2443–2457. doi: 10.1091/mbc.E04-12-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance BA, Chan C, Robertson ML, Finlay BB. Enteropathogenic and enterohemorrhagic Escherichia coli infections: emerging themes in pathogenesis and prevention. Can J Gastroenterol. 2002;16:771–778. doi: 10.1155/2002/410980. [DOI] [PubMed] [Google Scholar]
- Wayt J, Bretscher A. Cordon Bleu serves as a platform at the basal region of microvilli, where it regulates microvillar length through its WH2 domains. Mol Biol Cell. 2014;25:2817–2827. doi: 10.1091/mbc.E14-06-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Sumigray KD, Lechler T. The Arp2/3 complex has essential roles in vesicle trafficking and transcytosis in the mammalian small intestine. Mol Biol Cell. 2015;26:1995–2004. doi: 10.1091/mbc.E14-10-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.