The actin-binding protein Mena regulates RTK signaling after growth factor stimulation in tumor cells by a novel mechanism. The alternatively spliced MenaINV isoform disrupts this attenuation to drive sensitivity to growth factors, resistance to targeted inhibitors, and ultimately tumor invasion and metastasis.
Abstract
During breast cancer progression, alternative mRNA splicing produces functionally distinct isoforms of Mena, an actin regulator with roles in cell migration and metastasis. Aggressive tumor cell subpopulations express MenaINV, which promotes tumor cell invasion by potentiating EGF responses. However, the mechanism by which this occurs is unknown. Here we report that Mena associates constitutively with the tyrosine phosphatase PTP1B and mediates a novel negative feedback mechanism that attenuates receptor tyrosine kinase signaling. On EGF stimulation, complexes containing Mena and PTP1B are recruited to the EGFR, causing receptor dephosphorylation and leading to decreased motility responses. Mena also interacts with the 5′ inositol phosphatase SHIP2, which is important for the recruitment of the Mena-PTP1B complex to the EGFR. When MenaINV is expressed, PTP1B recruitment to the EGFR is impaired, providing a mechanism for growth factor sensitization to EGF, as well as HGF and IGF, and increased resistance to EGFR and Met inhibitors in signaling and motility assays. In sum, we demonstrate that Mena plays an important role in regulating growth factor–induced signaling. Disruption of this attenuation by MenaINV sensitizes tumor cells to low–growth factor concentrations, thereby increasing the migration and invasion responses that contribute to aggressive, malignant cell phenotypes.
INTRODUCTION
Tumor initiation, growth, and malignant progression are governed by interactions between cancer cells and their microenvironment (Hanahan and Weinberg, 2011). Aggressive, invasive cancer cells exit the primary tumor in response to growth factors, extracellular matrix (ECM) proteins, and other signals that cause them to invade surrounding tissue. After extravasation, invasive cells migrate to and enter blood or lymphatic vessels and are transported to sites of metastasis (Joyce and Pollard, 2009). Invading cells encounter numerous signals that trigger multiple intracellular pathways, whose activity is integrated to evoke appropriate, spatiotemporally coordinated responses. Tumor cell migration within this complex microenvironment requires continuous, coordinated cytoskeletal remodeling, which matches corresponding dynamic changes in cell–matrix and cell–cell adhesion (Bear and Haugh, 2014). Although distinct tumor cell migration modalities have been described (Petrie and Yamada, 2012), motility is typically initiated by rapid actin polymerization-driven membrane protrusion in response to acute activation of epidermal growth factor receptor (EGFR) and other receptor tyrosine kinases (RTKs; Nürnberg et al., 2011; Roussos et al., 2011b). Many aspects of RTK-regulated actin remodeling have been established (Bear and Haugh, 2014), but little is known about whether, or how, actin networks provide feedback to RTKs.
The Mena protein acts via multiple processes that are important for tumor cell invasion and metastasis: actin polymerization, adhesion, and EGF-elicited motility responses (Gertler and Condeelis, 2011). Mena and the related vasodilator-stimulated phosphoprotein (VASP) and EVL proteins are members of the Ena/VASP family, which increase F-actin elongation rates and delay the termination of filament growth by capping proteins (Bear and Gertler, 2009; Hansen and Mullins, 2010; Breitsprecher et al., 2011). Ena/VASP proteins localize primarily to lamellipodia and filopodia and sites of cell–matrix and cell–cell adhesion (Pula and Krause, 2008). They contain two highly conserved regions—an N-terminal Ena/VASP homology (EVH) 1 domain and a C-terminal EVH2 domain—as well as a central polyproline-rich core. The EVH1 domain mediates protein–protein interactions and typically binds to molecules via a conserved proline-rich motif (Ball et al., 2002). The EVH2 domain harbors actin-binding motifs and a coiled-coil that mediates formation of stable Ena/VASP tetramers (Pula and Krause, 2008). Mena also contains an LERER repeat region not found in other Ena/VASP proteins, and multiple functionally distinct Mena isoforms are produced from alternatively spliced Mena mRNAs (Gertler and Condeelis, 2011).
Mena is up-regulated in breast cancers and other solid tumor types (Gertler and Condeelis, 2011). The aggressive mortality, morbidity, and metastatic burden associated with the polyoma middle T antigen–mouse mammary tumor virus transgenic mouse breast cancer model is almost fully eliminated by genetic ablation of Mena; Mena deficiency does not affect tumor formation or growth but does slow progression and decreases tumor cell invasion, intravasation, and metastasis (Roussos et al., 2010). Highly migratory and invasive tumor cell subpopulations produce Mena mRNAs that contain a 57-nucleotide, alternately included exon (designated INV) to produce MenaINV (Gertler and Condeelis, 2011). Further, MenaINV mRNA levels in biopsies correlate with the density of Tumor Microenvironment of Metastasis (Roussos et al., 2011c; Pignatelli et al., 2014), a prognostic indicator of metastatic risk that consists of a Mena-expressing carcinoma cell, an endothelial cell, and a macrophage that are all in contact (Robinson et al., 2009; Rohan et al., 2014). MenaINV has effects on tumor cell behavior that are potent but differ from those of Mena in magnitude, type, or both (Gertler and Condeelis, 2011). Ectopic MenaINV sensitizes carcinoma cells to EGF and allows them to extend lamellipodia, chemotax, or invade in response to significantly lower concentrations of EGF than required for similar responses by control cells (Philippar et al., 2008; Roussos et al., 2011a). Ectopic MenaINV also increases invasion, intravasation, and lung metastasis from xenograft mammary tumors. The mechanisms underlying MenaINV-driven increases in metastatic potential and tumor cell sensitivity to EGF are unknown.
Here we show that Mena participates in a mechanism that attenuates RTK signaling by interacting with the tyrosine phosphatase PTP1B and the 5′ inositol phosphatase SHIP2. Expression of MenaINV disrupts this regulation and results in a prometastatic phenotype characterized by increased RTK/activation signaling from low ligand stimulation and decreased sensitivity to targeted RTK inhibitors. Overall our findings explain why MenaINV-expressing tumor cells display enhanced sensitivity to several growth factors both in vitro and in vivo.
RESULTS
MenaINV increases sensitivity to ligands for specific RTKs and confers resistance to targeted kinase inhibitors
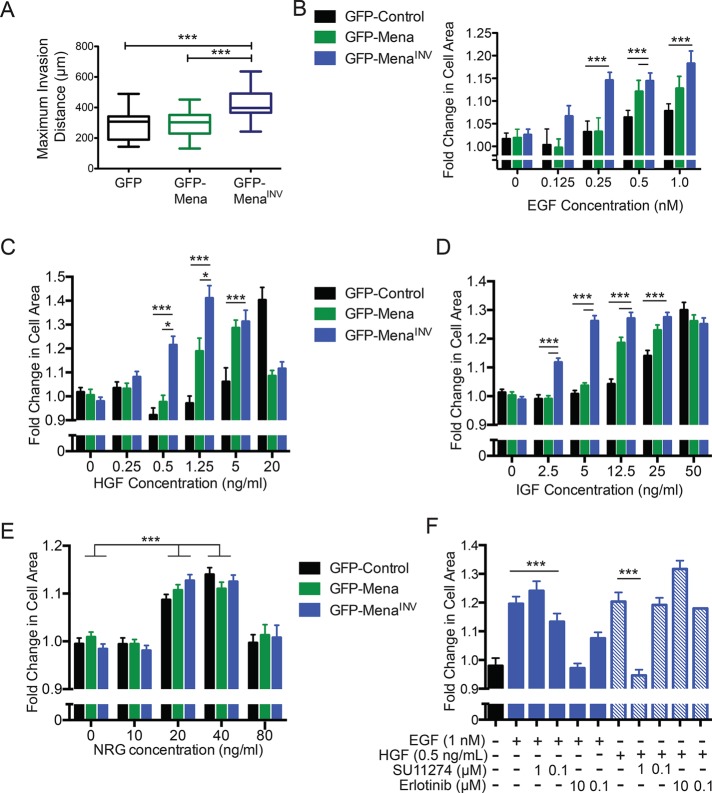
Although MenaINV is expressed robustly in aggressive tumor cell populations harvested from rodent mammary carcinomas, and in biopsies of human breast cancer patients, we have been unable to identify immortalized breast cancer cell lines that express more than trace amounts of the MenaINV mRNA or protein. Thus, to facilitate analysis of the mechanism of MenaINV-dependent growth factor sensitization, we used human cell lines that ectopically express green fluorescent protein (GFP)–Mena, GFP-MenaINV, or GFP alone. Mena was found to be upregulated ∼10-fold at the RNA level in invasive tumor cells collected from primary xenograft tumors, and therefore we engineered our cells to express levels roughly comparable to those seen in invasive cells from primary tumors (Wang et al., 2004; Supplemental Figure S1). As expected, MenaINV-expressing cells invaded further into three-dimensional (3D) collagen gels (Figure 1A). When looking at lamellipodial protrusion, an assay that has been described as accurately predicting 3D invasion responses (Meyer et al., 2012), we found that MenaINV-expressing cells exhibited increased lamellipodial protrusion in response to fourfold-lower EGF concentrations than required to elicit similar responses by GFP control cells (Figure 1B). Supplemental Figure S2 shows representative images of cells treated with EGF in this assay. Our engineered human cell lines thus faithfully replicate our previously published results demonstrating that ectopic expression of MenaINV in rat MTLn3 mammary adenocarcinoma cells enhanced protrusion responses elicited by stimulation with low EGF concentrations (Philippar et al., 2008).
FIGURE 1:
MenaINV expression confers sensitivity to select growth factors. (A) Invasion distance into 2 mg/ml collagen gel in full serum medium, using high-throughput 3D collagen invasion Iuvo platform (Bellbrook Labs) of MDA-MB231 cells expressing different Mena isoforms. ***p < 0.001 by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test. (B) Dose-response of lamellipodial protrusion in MDA-MB231 cells, reflected by fold change in cell area 8 min after addition of EGF. Results shown as mean ± SEM. Asterisks indicate significant difference by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test, with comparisons indicated by lines above; 35–158 cells/condition. (C–E) Dose-response of lamellipodial protrusion in MDA-MB231 cells, reflected by fold change in cell area 8 min after addition of IGF (C), HGF (D), or NRG (E). Results shown as mean ± SEM. Asterisks indicate significant difference by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test, with comparisons indicated by lines above; 35–158 cells/condition. (F) Lamellipodial protrusion dose-response 8 min after costimulation with 1 nM EGF or 0.5 ng/ml HGF and SU11274 or erlotinib, respectively. Significance measured by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test; *p < 0.05, ***p < 0.001 as indicated. See also Supplemental Figures S1 and S2.
In addition to EGF, other growth factors can drive tumor invasion and metastasis. We thus asked whether MenaINV also increased sensitivity to three other breast cancer–relevant growth factors—insulin-like growth factor (IGF), hepatic growth factor (HGF), and neuregulin 1 (NRG1; Friedl and Alexander, 2011)—using the lamellipodial protrusion assay. Similar to the sensitization seen with EGF, ectopic GFP-MenaINV enabled MDA-MB231 cells to extend lamellipodia when stimulated with significantly lower IGF and HGF concentrations (20- and 5-fold, respectively) than required for similar responses by control cells (Figure 1, C and D). GFP-Mena–expressing cells exhibited approximately twofold increase in response to IGF and HGF (as well as EGF); however, these effects were modest compared with those induced by GFP-MenaINV (Figure 1, A–D). Thus MenaINV expression evokes enhanced tumor cell responses, which are mediated by at least three RTKs relevant to breast cancer progression: EGFR, IGFR, and Met (receptors for EGF, IGF, and HGF, respectively; Alexander and Friedl, 2012). Note, however, that the response to NRG1, a ligand for HER3 and HER4, was unaffected by ectopic GFP-Mena or GFP-MenaINV (Figure 1E), indicating that the potentiating effects of MenaINV on RTK ligand sensitivity specifically affect only a subset of RTK signaling pathways. Because activation of Met by HGF decreases the efficacy of EGFR TKIs (Suda et al., 2010; Gusenbauer et al., 2013), we asked whether the MenaINV-mediated sensitivity to EGF and HGF involved cross-talk between the two receptors Inhibition of EGFR using the EGFR tyrosine kinase inhibitor (TKI) erlotinib or of Met using the Met-TKI inhibitor SU11274 in MenaINV-expressing cells had no effect on lamellipodial protrusion in response to HGF or EGF, respectively, indicating that the increased sensitivity to EGF and HGF observed in GFP-MenaINV–expressing cells was receptor specific (Figure 1F).
To analyze the effects of MenaINV on targeted RTK inhibitors, we performed dose-response experiments using the EGFR TKI erlotinib and the Met-TKI SU11274. Cells expressing GFP-MenaINV were resistant to at least 10-fold-greater inhibitor than required to block lamellipodial protrusion in GFP control cells (Figure 2, A and B). Collectively these data indicate that MenaINV confers increases in ligand sensitivity for responses by EGFR, Met, and IGFR, as well as in resistance to EGFR- and Met- targeted TKIs.
FIGURE 2:
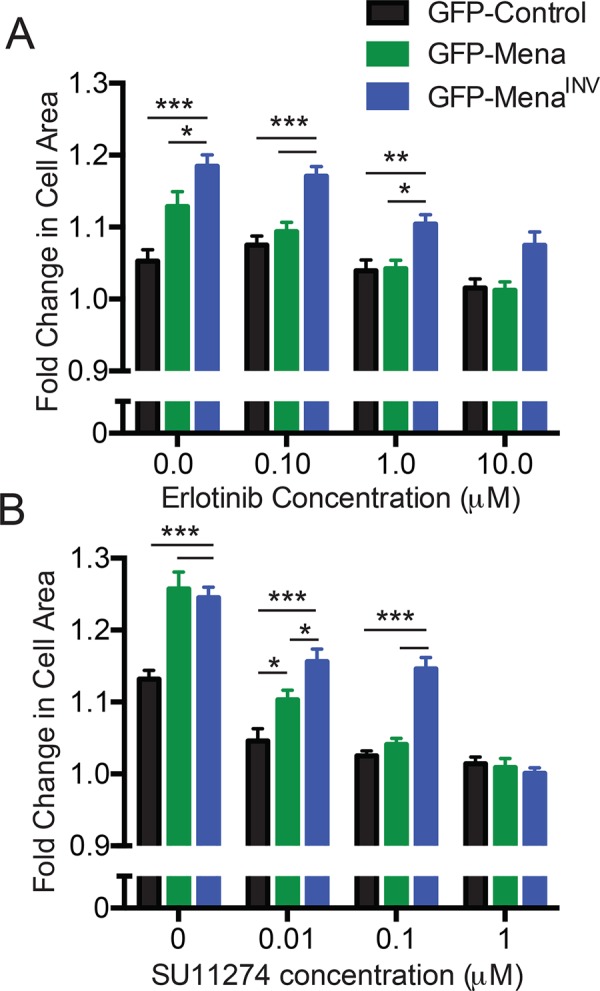
MenaINV expression confers resistance to targeted kinase inhibitors. (A) Lamellipodial protrusion response after stimulation with 1 nM EGF and increasing erlotinib doses. Fold change in cell area calculated 8 min poststimulation. Results shown as mean ± SEM; 16–48 cells/condition. Asterisks indicate significant difference by two-way analysis of variance (ANOVA). (B) Lamellipodial protrusion dose-response after costimulation with 0.5 ng/ml HGF and increasing SU11274. Fold change in cell area calculated 8 min poststimulation. Results shown as shown as mean ± SEM; 39–71 cells/condition. Asterisks indicate significant difference by two-way ANOVA; *p < 0.05, **p < 0.01, and ***p < 0.001 as indicated.
MenaINV dysregulates tyrosine kinase signaling
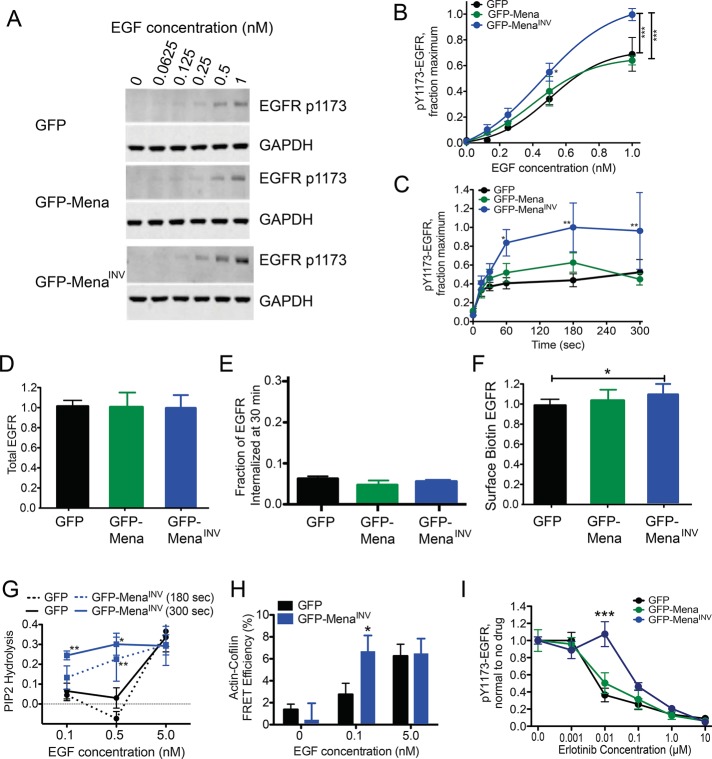
To address whether cell behaviors influenced by the MenaINV-induced increase in EGF sensitivity and resistance to erlotinib arise from increased actin polymerization (a process that is directly modulated by Mena), by a specific effect on EGFR pathway function, or both, we used anti-pY1173 antibodies to measure EGFR phosphorylation levels. We observed increased phosphorylation in cells expressing MenaINV compared with those expressing equivalent levels of ectopic Mena or GFP (Figure 3, A and B). Similarly, expression of MenaINV in a second triple-negative breast cancer cell line, BT-549, also significantly increased EGFR phosphorylation at Y1173 (Supplemental Figure S3). Despite the significant differences in magnitude, the relative kinetics of EGFR phosphorylation were similar across the cell lines stimulated with 0.25 nM EGF (Figure 3C) and mirrored the kinetics of membrane protrusion in MDA-MB-231 cells expressing MenaINV. Measurement of EGFR abundance and internalization indicated that the MenaINV--driven increase in EGFR phosphorylation was not attributable to increased total EGFR protein level or altered receptor internalization (Figure 3, D and E). Whereas MenaINV expression induced a small (<10%) increase in surface levels of EGFR (Figure 3F), this minor increase is unlikely to account for the observed MenaINV-dependent enhanced response to EGF, given that an ∼10-fold increase in total EGFR level is required to increase lamellipodial protrusion detectably at the EGF concentrations tested (Bailly et al., 2000). Furthermore, no apparent changes in EGFR clustering or spacing were evident in immuno–electron microscopy analyses (unpublished data). Overall these data indicate that the increased sensitivity to EGF conferred by MenaINV expression is unlikely to arise through changes in EGFR abundance or distribution.
FIGURE 3:
MenaINV expression does not affect EGFR presentation but increases signaling pathway activation at low levels of EGF. (A) Representative Western blot of MDA-MB231 cells expressing GFP, GFP-Mena, or GFP-MenaINV stimulated after bolus stimulation with EGF for 3 min. (B) Quantification of EGFR phosphorylation at Y1173 shown in A by densitometry. Results are mean ± SD; three experiments. Asterisks indicate significant difference by ANOVA with Tukey multiple-comparison test (*p < 0.05 vs. MDA-MB231-GFP cells, ***p < 0.001). (C) Time course of EGFR phosphorylation at Y1173 in response to 0.25 nM EGF. Asterisks indicate significant difference by ANOVA with Tukey multiple-comparison test (*p < 0.05 vs. MDA-MB231-GFP cells at 0.5 nM or **p < 0.01 vs. conditions indicated by bar). (D) Total EGFR protein normalized to GFP control measured by ELISA. n > 10 for each bar in MDA-MB231 cells. Data shown as mean ± SD. (E) Fraction of total EGFR internalized at basal (no EGF) conditions in serum-free medium after 30 min at 37C. Data shown as mean ± range; n = 2. (F) Membrane level of EGFR measured by biotin labeling of surface proteins, EGFR capture ELISA, and detection of protein by HRP-labeled streptavidin in MDA-MB231 cells. Data normalized to GFP control cells in each experiment. Data shown as mean ± SD; n > 10 for each bar. (G) Fraction of PI(4,5)P2 lost from the cell membrane at specified time after EGF stimulation, measured using PLCδ-PH domain FRET assay in MTLn3 cells. *p < 0.05 and **p < 0.01 vs. GFP at respective time point using ANOVA with Tukey posttest. (H) Cofilin activity, reflected by the amount of cofilin bound to actin quantified by an antibody FRET assay 300 s after EGF stimulation in MTLn3 cells. *p < 0.05 vs. GFP at respective EGF concentration using two-tailed t test. (I) EGFR phosphorylation dose-response 3 min poststimulation with 1 nM EGF and increasing erlotinib in MDA-MB231 cells. Data shown as mean ± SEM; ***p < 0.001 vs. MDA-MB231-GFP and GFP-Mena by two-way ANOVA. See Supplemental Figure S3.
We reasoned that the ligand-dependent RTK activation that leads to MenaINV-induced increases in membrane protrusion should be reflected in increased activity of downstream signaling pathways that mediate the protrusion. Analysis of several canonical signaling components downstream of EGFR (such as Erk and Akt) revealed no obvious effect of MenaINV on their activities despite increased activation of the receptor itself (Philippar et al., 2008). In breast cancer cells, the actin polymerization needed for EGF-elicited protrusion is initiated by the phospholipase C γ (PLCγ)–cofilin pathway (van Rheenen et al., 2007); therefore we asked whether EGFR-dependent activation of PLCγ enzymatic activity is altered by MenaINV. PLCγ activity can be quantified in living cells by measuring the abundance of its substrate, phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), using monomeric red fluorescent protein (mRFP)–PLCδ-PH, a reporter that is localized to the plasma membrane by binding to PI(4,5)P2 and is released into the cytosol upon its hydrolysis (van Rheenen et al., 2007). MDA-MB231 cells were unsuitable for this assay due to sensitivity to expression of mRFP-PLCδ-PH; thus we performed the assay in MTLn3 mammary adenocarcinoma cell lines, which we used for our initial characterization of MenaINV-dependent effects on metastasis (Philippar et al., 2008). MenaINV expression increased both PI(4,5)P2 hydrolysis (Figure 3G) and cofilin activity (Figure 3H) in response to significantly less EGF than required to trigger similar responses in GFP control cells. Consistent with these data, MenaINV-expressing cells required 10-fold more erlotinib than controls to decrease ligand-induced EGFR phosphorylation significantly (Figure 3I). We conclude that MenaINV potentiates EGF-elicited responses by increasing the activity of EGFR-mediated signaling pathways that are upstream of, and are required for, the initiation of actin polymerization.
Mena may associate with the tyrosine phosphatase PTP1B
Because abundance, surface levels, or internalization of EGFR did not account for the MenaINV-dependent increases in receptor phosphorylation and downstream signaling, we investigated other mechanisms that could increase RTK phosphorylation at a low ligand concentration. Activated RTKs cycle rapidly between phosphorylation and dephosphorylation; the half-life of EGFR tyrosine phosphorylation is ∼10–30 s (Kleiman et al., 2011). Changes in the balance of this rapid cycling are believed to control receptor sensitivity to ligand by altering net kinase activity. We hypothesized that MenaINV-dependent increases in ligand sensitivity could arise via increased net receptor phosphorylation due to dysregulation of a phosphatase. The tyrosine phosphatase PTP1B (PTPN1) regulates EGFR, IGFR, and MET-mediated signaling responses (Haj et al., 2003; Sangwan et al., 2011), and, given that MenaINV sensitizes responses to ligands of all three RTKs (Figure 1), we asked whether Mena is involved in PTP1B regulation.
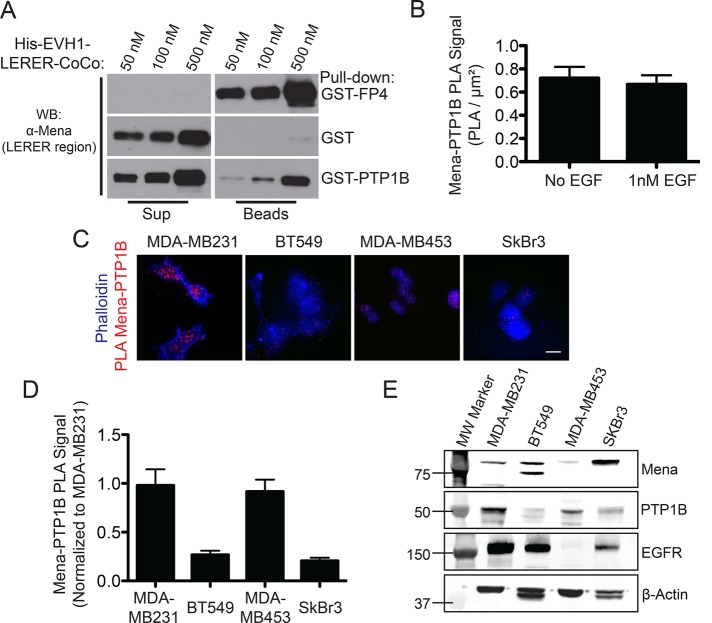
PTP1B contains the sequence LEPPPEHIPPPP, with similarities to the consensus EVH1-binding motif ([F/L/W/PXφP]; X = any residue, φ = any hydrophobic residue; Ball et al., 2002; Pula and Krause, 2008), raising the possibility that PTP1B binds to Mena through its EVH1 domain and thereby allows Mena/MenaINV to modulate RTK dephosphorylation. We used purified, recombinant glutathione S-transferase (GST)–PTP1B and His-Mini-Mena proteins to test whether PTP1B can bind Mena directly. Immobilized GST-PTP1B, but not the immobilized control GST protein, bound to soluble, purified His-Mini-Mena in vitro (Figure 4A). To determine whether endogenous Mena and PTP1B are present in protein complexes in situ, we used proximity ligation assays (PLAs). To confirm the specificity of the assay, we performed control PLAs using only one antibody or in Mena−/− cells (Supplemental Figure S4, A and B), which significantly reduced the detected PLA signal. Furthermore, transient knockdown of PTP1B expression in MDA-MB231 cells also reduced the Mena-PTP1B PLA signal, further supporting the specificity of this assay for Mena/PTP1B-containing complexes (Supplemental Figure S4, C–E). Significant PLA signal between Mena and PTP1B was observed in MDA-MB231 cells and was unchanged by EGF stimulation, indicating that Mena and PTP1B are in complex independent of EGFR signaling (Figure 4B). Quantification of Mena-PTP1B PLA signal abundance across four human breast cancer cell lines revealed that the amount of Mena-PTP1B PLA signal was correlated with PTP1B expression levels (Figure 4, C–E). The extremely low levels of MenaINV normally expressed in cultured cell lines precluded use of PLA to determine whether endogenous MenaINV is present in complexes with PTP1B. These results, however, demonstrate that PTP1B and Mena can directly bind each other in vitro and that complexes containing endogenously expressed Mena and PTP1B can be detected within breast cancer cells by PLA.
FIGURE 4:
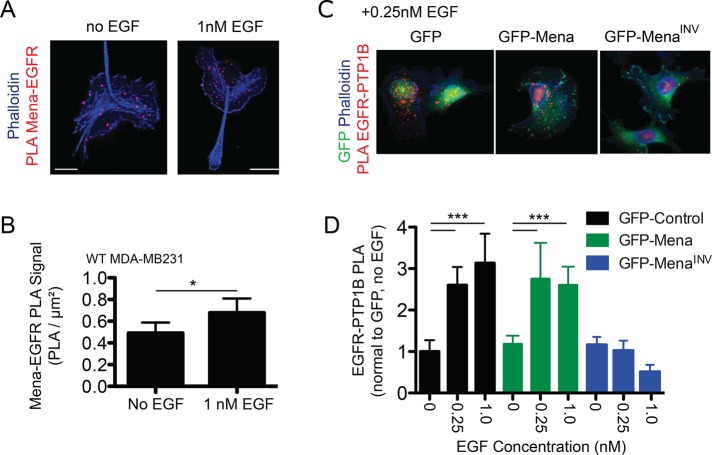
Mena interacts with PTP1B. (A) In vitro binding assay using immobilized GST-PTP1B and increasing concentrations of soluble His-Mini-Mena (containing Mena EVH1-LERER domains linked to the C-terminal coiled-coil; bottom lanes). Positive control with GST-FP4 (top lanes) and negative control with glutathione beads + GST alone (middle lanes) included to demonstrate assay specificity. Blots from representative experiment; n = 3. (B) Quantification of PLA for PTP1B and Mena in wild-type MDA-MB231 ± 1 nM EGF for 60 s. Data shown as mean ± SEM. Specificity of assay established using Mena−/− mouse embryonic fibroblasts, where background signal was negligible (data not shown). (C) Representative images for PTP1B-Mena PLA in four breast cancer cell lines: MDA-MB231, BT549, MDA-MB453, and SkBr3. (D) Mena-PTP1B PLA across four human breast cancer cell lines compared with signal measured in wild-type MDA-MB231 cells. Data shown as mean PLA/μm2 normalized to MDA-MB231 ± SEM (E) Western blot showing expression of Mena, PTP1B, and EGFR in four human breast cancer cell lines. See Supplemental Figure S4.
Inhibition of PTP1B phosphatase activity phenocopies MenaINV expression
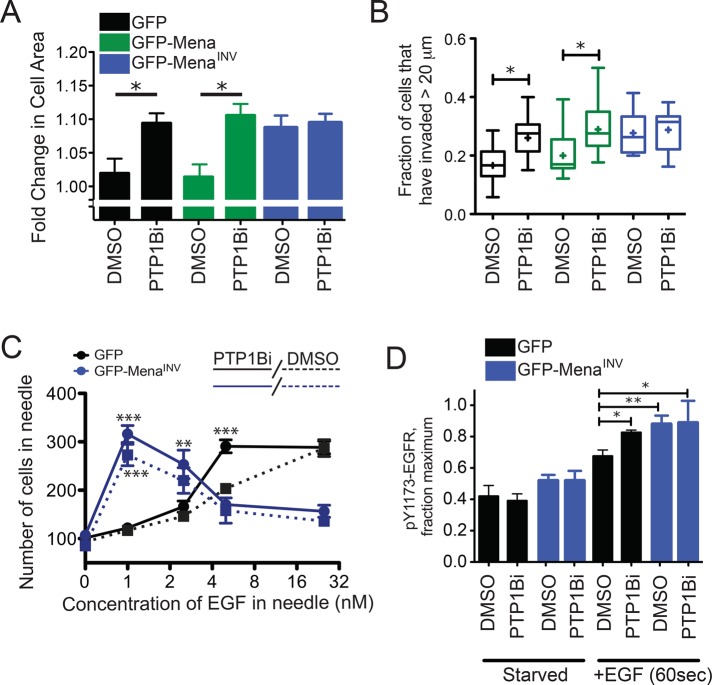
We next hypothesized that MenaINV decreases negative regulation of RTK activity by PTP1B and, thus, that specific inhibition of PTP1B will mimic some or all of the effects of MenaINV on EGF responses. Treatment with a specific PTP1B inhibitor increased membrane protrusion in response to 0.25 nM EGF by both MDA-MB231-GFP-Mena– and GFP-expressing control cells but not in MDA-MB231-GFP-MenaINV cells (Figure 5A). Therefore PTP1B inhibition mimics MenaINV-dependent increased sensitivity to EGF but cannot induce EGF responses of greater magnitude in cells expressing MenaINV. PTP1B inhibition also increased the magnitude and kinetics of membrane protrusion in parental MTLn3 cells in response to 0.5 nM EGF, confirming that the effects of PTP1B inhibition were not cell-type dependent (Supplemental Figure S5A). Further, PTP1B inhibition did not increase membrane protrusion in cells expressing MenaINV in response to 0.125 nM EGF (Supplemental Figure S5B), a concentration below the threshold required to elicit significant membrane protrusion or EGFR activation in MenaINV-expressing cells (Figure 1B), suggesting that PTP1B inhibition alone is not sufficient to drive membrane protrusion independently of EGFR activation.
FIGURE 5:
Inhibition of PTP1B mimics the effects of MenaINV expression. (A) Lamellipodial protrusion of MDA-MB231 cells 8 min poststimulation with 0.25 nM EGF after incubation with 0.1% DMSO or 10 μM PTP1B inhibitor for 60 min. Results are mean with 95% confidence intervals; 48 cells/condition. Asterisk indicates significant difference by ANOVA with Tukey multiple-comparison test (*p < 0.05). (B) 3D collagen invasion after 24 h in the presence of 0.25 nM EGF and 0.1% DMSO or 10 μM PTP1B inhibitor. Results are represented as box and whiskers at 5 and 95% percentiles; cross indicates mean value; five assays/condition. Asterisk indicates significant difference by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test (*p < 0.05). (C) EGF dose-response of invasive cells collected from MTLn3 xenograft tumors expressing GFP (black solid and dotted lines) or GFP-MenaINV (blue solid or dotted lines). Needles contained EGF and 0.01% DMSO without (dotted lines) or with10 μM PTP1B inhibitor (solid lines). Results are mean ± SEM and plotted on log 2 x-axis; more than three tumors for conditions with error bars; one tumor for conditions without error bars. Asterisks indicate significant difference by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test (**p < 0.01 or ***p < 0.001) at each concentration of EGF for conditions with more than three tumors. (D) EGFR phosphorylation at Y1173 (0.25 nM EGF, 3 min) after 60 min of preincubation with 0.1% DMSO or 10 μM PTP1B inhibitor after 4 h of serum starvation. Results are mean ± SEM; three experiments. Asterisks indicate significant difference by ANOVA with Student-Newman-Keuls multiple-comparison test (*p < 0.05, **p < 0.01). See also Supplemental Figure S5 and Supplemental Table S6.
We next asked whether decreasing PTP1B activity mimics the effects of MenaINV expression on EGF-dependent tumor cell invasion in vitro or in vivo. PTP1B inhibition increased EGF-elicited invasion of MDA-MB231-GFP and -GFP-Mena–expressing cells into collagen I gels but had no significant effect on cells expressing MenaINV (Figure 5B). Expression of MenaINV in xenografted tumor cells decreases the concentration of EGF required for efficient chemotaxis/invasion by 25-fold in vivo (Roussos et al., 2011a); we thus asked whether PTP1B inhibition similarly increases the EGF sensitivity of invading cells in tumors. Consistent with the in vitro assays, a dose-response analysis using the in vivo invasion assay (Wyckoff et al., 2000) on xenografts showed that PTP1B inhibition increased the sensitivity of control cells to EGF: maximal tumor cell invasion was elicited into needles containing 5 nM EGF, a fivefold-lower concentration than normally required for such a response (Figure 5C). Furthermore, PTP1B inhibition had no effect at any of the EGF concentrations assayed on invasion in xenograft tumors arising from MenaINV-expressing cells (Figure 5C). In sum, these data indicate that PTP1B inhibition mimics the effects of MenaINV on EGF-elicited motility, chemotaxis, and invasion in vitro and in vivo but does not increase EGF responses in cells that express MenaINV.
Because the effects of MenaINV on biophysical cell responses to EGF are mimicked by PTP1B inhibition, we asked whether MenaINV similarly dysregulated EGFR signaling. Pretreatment with PTP1B inhibitor increased EGFR Y1173 phosphorylation in control cells stimulated with 0.25 nM EGF but failed to increase Y1173 phosphorylation further in MenaINV-expressing cells (Figure 5D). In addition, wild-type MTLn3 cells treated with the PTP1B inhibitor exhibited significantly greater PI(4,5)P2 hydrolysis after EGF stimulation (Supplemental Figure S5C), similar to the increased PLCγ activity exhibited by cells expressing MenaINV (Figure 3).
To identify other network participants that are dysregulated by MenaINV expression, we used liquid chromatography–tandem mass spectrometry (LC-MS/MS)–based phosphoproteomics to quantify tyrosine phosphorylation after incubation with or without 0.25 nM EGF. We detected 54 tyrosine phosphorylation sites across 41 proteins in at least biological duplicate (Supplemental Figure S6); phosphorylation across the 54 sites was significantly higher in cells expressing MenaINV versus control cells (p = 0.015). Of the identified proteins, 12 are known PTP1B substrates; as a group, these exhibited significantly higher phosphorylation in MenaINV-expressing cells than in controls (p = 0.0078); however, these 12 were not significantly more phosphorylated than the 54 phosphorylation sites overall (p = 0.06).
A PTP1B-Mena-SHIP2-EGFR complex is dysregulated upon MenaINV expression
Because EGFR is a known PTP1B substrate (Haj et al., 2003; Mertins et al., 2008) and we identified complexes containing both Mena and PTP1B in breast cancer cells, we examined whether Mena facilitates formation of EGFR-PTP1B complexes. In wild-type MDA-MB231 cells, complexes containing endogenous Mena-EGFR were detected by PLA (Figure 6A). EGF stimulation increased abundance of the PLA signal, indicating that EGFR activation induces formation of additional Mena-EGFR–containing complexes (Figure 6B). We then used PLA to quantify EGFR-PTP1B–containing complexes in GFP-, GFP-Mena–, and GFP-MenaINV–expressing cells before and after EGF treatment. EGF stimulation increased EGFR-PTP1B PLA in cells expressing GFP or GFP-Mena but had no effect on cells expressing MenaINV (Figure 6, C and D). Transient knockdown of PTP1B expression in MDA-MB231-GFP-Mena cells reduced the EGFR-PTP1B PLA signal, indicating that assay detected PTP1B/EGFR-containing complexes specifically (Supplemental Figure S7A). Therefore MenaINV expression blocks EGF-elicited recruitment of PTP1B to EGFR, providing a potential explanation for the increased receptor phosphorylation in cells expressing MenaINV (Figure 3).
FIGURE 6:
PTP1B recruitment to EGFR is abrogated in cells expressing MenaINV. (A) Representative images for Mena-EGFR PLA ± EGF (1 nM, 60 s). Phalloidin shown in blue, and Mena-EGFR PLA shown in red. (B) Quantification of Mena-EGFR PLA in wild-type MDA-MB231 cells ± EGF (1 nM, 60 s). Asterisk indicates significant difference by two-tailed t test (*p < 0.05). (C) Representative images for EGFR-PTP1B PLA for MDA-MB231 cells expressing GFP, GFP-Mena, or GFP-MenaINV stimulated with 0.25 nM EGF for 60 s. GFP signal shown in green, phalloidin shown in blue, and EGFR-PTP1B PLA shown in red. (D) Quantification of EGFR-PTP1B PLA ± 0.25 nM EGF for 60 s. Data are mean with 95% confidence interval; >20 cells/condition. Asterisks indicate significant difference by ANOVA with Tukey multiple-comparison test (***p < 0.001).
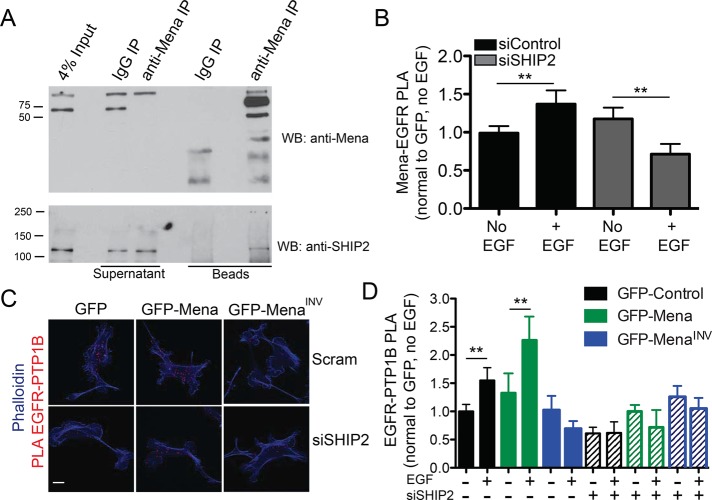
All Mena isoforms are likely maintained as stable tetramers by their conserved C-terminal tetramerization domains (Gertler and Condeelis, 2011) and thus contain EVH1 domains at the N-terminal ends of each of the four subunits. Given their modular composition, the subunits of a Mena tetramer could potentially bind and link together up to four distinct EVH1 ligands. Therefore we asked whether another Mena EVH1-binding protein recruits Mena-PTP1B complexes to EGFR upon receptor activation. We identified SHIP2, a 5′ inositol phosphatase that dephosphorylates phosphatidylinositol 3,4,5-trisphosphate to generate phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2) in our mass spectrometry screen as one of the 41 proteins phosphorylated in EGF-stimulated MDA-MB231 cells (Supplemental Figure S6). On EGF stimulation, SHIP2 is recruited rapidly into complexes with activated EGFR (Pesesse et al., 2001). We inspected the SHIP2 sequence, found four canonical EVH1-binding motifs, and verified by coimmunoprecipitation (coIP) that endogenous SHIP2 associates with Mena (Figure 7A).
FIGURE 7:
A SHIP2-Mena-PTP1B complex regulates EGFR-PTP1B interaction in MDA-MB231 cells. (A) Total Mena protein was immunoprecipitated from Rat2 fibroblast lysate and immunoblotted for SHIP2. (B) Quantification of Mena-EGFR PLA in wild-type MDA-MB231 cells starved or stimulated with 1 nM EGF for 60 s. Experiment performed 72 h posttransfection with 25 nM control (siControl) or SHIP2 (siSHIP2)–targeted SMARTPool siRNA. Results are shown as mean with 95% confidence intervals; >20 cells/condition. Asterisks indicate significant difference by ANOVA with Tukey multiple-comparison test (**p < 0.01). (C) Representative images of EGFR-PTP1B PLA in MDA-MB231 cell lines 72 h posttransfection with siControl or siSHIP2. (D) EGFR-PTP1B PLA in MDA-MB231 cell lines 72 h posttransfection with siControl (indicated by “–“ in siSHIP2 line) or siSHIP2. Cells stimulated ± 0.25 nM EGF for 60 s. Results are shown as mean with 95% confidence intervals; > 50 cells/condition. Asterisks indicate significant difference by nonparametric Kruskal–Wallis test and Dunn’s multiple-comparison test (**p < 0.05). See also Supplemental Figure S7.
The foregoing observations led us to test the hypothesis that SHIP2 can recruit Mena-PTP1B complexes to activated EGFR by binding to one or more EVH1 domains in Mena tetramers. If SHIP2 links Mena-PTP1B complexes to activated EGFR, then SHIP2 depletion should abolish EGF-induced recruitment of PTP1B into complexes with EGFR. RNA interference–mediated SHIP2 depletion from wild-type MDA-MB231 cells (Supplemental Figure S7B) eliminated the EGF-induced increase in Mena-EGFR complexes, as indicated by PLA assays (Figure 7B). SHIP2 depletion in GFP- and GFP-Mena–expressing cells also eliminated the increase in EGFR-PTP1B complexes normally induced by stimulation with 0.25 nM EGF, reducing their abundance to a level similar to that observed in GFP-MenaINV cells transfected with control small interfering RNA (siRNA; Figure 7, C and D). In addition to recruiting Mena-PTP1B complexes to activated EGFR, we wondered whether SHIP2 enzymatic activity might also contribute to the observed MenaINV-driven changes in breast cancer cell motility and invasion. We used the highly specific SHIP2 inhibitor AS1949490 (Suwa et al., 2009) to determine whether its 5′ inositol phosphatase activity was required for MenaINV-enhanced membrane protrusion. SHIP2 inhibition attenuated protrusion at later time points in MDA-MB231–expressing MenaINV (Supplemental Figure S7D) and MTLn3 cells (Supplemental Figure S7C).
In sum, these data are consistent with a model in which activated EGFR rapidly recruits a SHIP2-Mena-PTP1B complex, which leads to receptor dephosphorylation by PTP1B. In cells that express MenaINV, however, SHIP2-dependent recruitment of PTP1B to EGFR is abolished, eliminating PTP1B-mediated dephosphorylation of EGFR (Figure 8). Thus we propose that MenaINV disrupts negative feedback to the EGFR and thereby increases signaling and, consequently, cellular responses elicited by low EGF concentrations.
FIGURE 8:
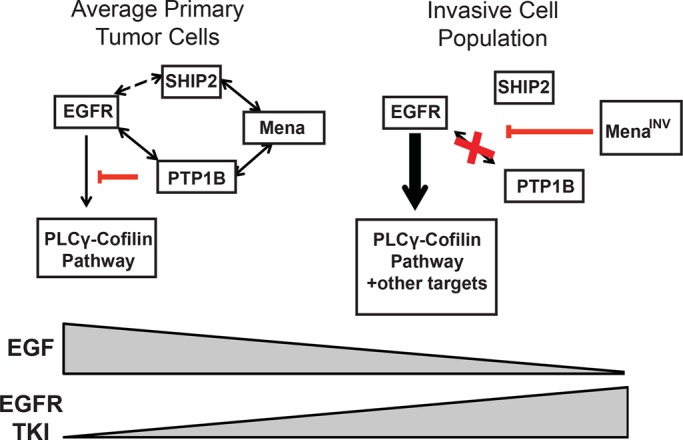
Model for Mena-dependent regulation of RTK signaling. In average primary tumor cells, activation of EGFR leads to rapid recruitment of a SHIP2-Mena-PTP1B complex, which leads to receptor dephosphorylation by PTP1B and sensitivity to TKIs. However, in the invasive tumor cell population, where levels of MenaINV are high, SHIP2-dependent recruitment of PTP1B to EGFR is abolished, eliminating PTP1B-mediated dephosphorylation of EGFR, causing sensitivity to EGF, as well as resistance to TKIs.
DISCUSSION
Mena regulates signaling by specific RTKs
Mena and the other Ena/VASP proteins regulate motility and adhesion of numerous cell types (Pula and Krause, 2008); at a mechanistic level, their most extensively characterized functions influence actin polymerization and modulate the morphology and dynamics of membrane protrusions (Bear and Gertler, 2009). We found that Mena serves an additional, unanticipated, isoform-dependent role as a regulator of at least three RTKs known to drive invasion and metastasis in breast cancer (EGFR, IGFR and Met; Alexander and Friedl, 2012). Cellular responses to EGF and other growth factors are tightly regulated through control of receptor trafficking and activation state (Avraham and Yarden, 2011), with tyrosine phosphorylation of RTKs being regulated by a dynamic interplay between kinases and phosphatases (Kleiman et al., 2011). Here we show that complexes containing Mena and PTP1B are recruited to activated EGFR in a SHIP2-dependent manner. Because PTP1B is known to regulate EGFR, IGFR, and Met signaling (Feldhammer et al., 2013), our findings are consistent with a model in which ligand-induced, SHIP2-mediated recruitment of Mena- and PTP1B-containing complexes contributes to attenuation of signaling by these RTKs. Further, MenaINV disrupts EGF-elicited recruitment of PTP1B to EGFR, providing a basis for the enhanced signaling, motility and invasion observed in response to stimulation with low EGF concentrations in cells expressing MenaINV.
Our PLA analysis indicated that complexes containing Mena and PTP1B present in unstimulated cells are recruited to the activated EGFR. We attempted to detect Mena and PTP1B in complex by coIP without success, perhaps due to low affinity or stoichiometry of the complex. It is possible, however, that Mena associates directly with PTP1B, as the Mena EVH1 domain can bind purified PTP1B directly in vitro (Figure 4A). Future studies will address which region in Mena specifically regulates the interaction between these two proteins and whether the interaction is direct or requires additional components. Most of the PTP1B in cells is anchored to the outer membrane of the endoplasmic reticulum, where it can contact internalized transmembrane receptors (Eden et al., 2010). However, in platelets (Frangioni et al., 1993) and in MTLn3 rat adenocarcinoma cancer cells (Cortesio et al., 2008), cleavage by calpain has been shown to release catalytically active PTP1B, which can then localize throughout the cytoplasm (Frangioni et al., 1993; Feldhammer et al., 2013). Therefore it is possible that calpain-dependent release of PTP1B into the cytoplasm could contribute to Mena-dependent association of PTP1B with RTKs in these cells. Despite the relatively small fraction present in the cytoplasm, PTP1B clearly functions at a variety of structures within the cytosol (Feldhammer et al., 2013), including focal adhesions, filopodia on neuronal growth cones (Fuentes and Arregui, 2009), and invadopodia (Cortesio et al., 2008), which are actin-driven protrusions that degrade the ECM during invasion. Of interest, these structures are also sites where Mena is localized (Philippar et al., 2008; Pula and Krause, 2008).
In breast cancer cells, as little as 0.25 nM EGF induces rapid (within 60 s) recruitment of Mena-PTP1B complexes to EGFR, thereby dephosphorylating the receptor and attenuating its activity. Whereas PTP1B can regulate early endosome fusion and trafficking of Met and EGFR (Sangwan et al., 2011), expression of Mena in MDA-MB231 cells was not associated with changes in EGFR surface abundance or distribution. The biochemical and biophysical effects of Mena on the EGF response occur within a time frame that is likely too brief to allow for receptor endocytosis and vesicle scission (Taylor et al., 2011; Zheng et al., 2013), although a novel mechanism for rapid endocytosis has recently been described (Boucrot et al., 2014). Mena-dependent effects on EGFR signaling may occur while the receptor is still on the plasma membrane. Thus, in breast cancer cells, Mena, which enhances the actin polymerization underlying EGF-elicited lamellipodial protrusion (Philippar et al., 2008), also directly participates in EGFR signal attenuation.
SHIP2 mediates EGF-induced recruitment of Mena and PTP1B to EGFR
We also find that ligand-triggered recruitment of Mena-PTP1B complexes to EGFR requires SHIP2, an SH2 domain–containing inositol phosphatase with known scaffolding functions (Erneux et al., 2011) that is rapidly recruited (via the adaptor protein SHC) to activated EGFR (Pesesse et al., 2001). The kinetics of RTK-dependent recruitment of SHIP2 is very fast, and it occurs within 20 s of stimulation (Zheng et al., 2013). Mena translocation to lamellipodia can also be observed within 20 s after EGF stimulation and depends on availability of F-actin barbed ends (Philippar et al., 2008), which are generated by cofilin severing upon its PLCγ-mediated release from the plasma membrane (Van Rheenen et al., 2007). The EGF-induced, EGFR-PTP1B–containing complexes form with kinetics similar to that of the Mena-dependent effects on both EGFR signaling and lamellipodial protrusion. Our data support a model in which individual Mena tetramers associate with both PTP1B and SHIP2 potentially through direct binding of their N-terminal EVH1 domains. This raises the interesting possibility that such complexes may also interact with F-actin barbed ends through the actin-binding motifs within the C-terminal EVH2 domains of Mena.
Mena binds other EVH1 ligands that influence its subcellular localization and mediates interactions with signaling and scaffolding proteins (Pula and Krause, 2008; Bear and Gertler, 2009). Mena localization to the leading edge of lamellipodia and tips of filopodia depends, in part, on EVH1-mediated binding to lamellipodin (Lpd; Krause et al., 2004). Recently Lpd was found in complexes with EGFR, and knockdown of Lpd or Mena disrupted scission of clathrin-coated pits, a late step in clathrin-mediated endocytosis (CME) of the receptor (Vehlow et al., 2013). Lpd-EGFR associated constitutively in the HeLa cells used in that study, and, since CME of EGFR initiates only after EGF stimulation, the results of Vehlow et al. (2013) imply that growth factor stimulation induces Mena recruitment to the receptor. Our data are consistent with this model; however, we observed Mena recruitment to EGFR and effects on receptor signaling within 60s after stimulation, a time that precedes vesicle scission, the first step in CME affected by Mena depletion (Vehlow et al., 2013). It is possible that the SHIP2-EGFR interaction helps to bring Mena into complex with Lpd or that Lpd in complex with EGFR increases Mena-PTP1B recruitment to the receptor at later times after EGF stimulation. Consistent with this possibility, we found that inhibition of SHIP2 5′ inositol phosphatase activity, which produces PI(3,4)P2, affects EGF-stimulated protrusion. Of interest, Lpd contains one of the few PH domains specific for PI(3,4)P2 (Krause et al., 2004), and both SHIP2 and PI(3,4)P2 regulate CME dynamics (Nakatsu et al., 2010; Posor et al., 2013). In sum, these data support a key role for Mena, via EVH1-mediated interactions, in the recruitment of important signaling proteins downstream of EGFR.
Expression of MenaINV dysregulates phosphotyrosine signaling
Expression of MenaINV reduces PTP1B recruitment to the receptor and its dephosphorylation, thereby eliciting EGFR-dependent protrusion and motility-related responses at substantially reduced ligand concentration. Within tumors, spontaneous up-regulation of MenaINV expression likely enhances both paracrine signaling with macrophages and the autocrine loops that provide EGFR ligands to tumor cells, increasing prometastatic behaviors (Wyckoff et al., 2004; Patsialou et al., 2009; Zhou et al., 2014). MenaINV expression also increases tumor cell sensitivity to HGF and IGF-1 by 20- and 5-fold, respectively, suggesting that tumor cells expressing MenaINV in vivo may be sensitized to HGF and IGF-1 secreted by tumor-associated fibroblasts or other cells in the tumor microenvironment (Yee et al., 1989; Mueller et al., 2012). Selective inhibition of PTP1B by MenaINV likely affects a set of proteins that are normally dephosphorylated by PTP1B-containing Mena complexes. MenaINV, like Mena, localizes to filopodia, lamellipodia, and invadopodia, as well as to sites of cell matrix and cell:cell adhesion; MenaINV-mediated attenuation of PTP1B function (or localization) within these structures could have a large effect on tumor cell invasion and motility. Consistent with this idea, several PTP1B targets that localize to these structures and function in tumor cell migration and invasion were among the proteins detected in our mass spectrometry screen for MenaINV-induced dysregulated tyrosine phosphorylation. We also found that treatment of breast cancer cells in vitro or in vivo with a targeted PTP1B inhibitor induced phenotypes that are strikingly similar to those caused by MenaINV expression.
The roles that PTP1B plays in cancer are complex and likely context dependent. Complete PTP1B deletion in the HER2/Neu model of murine mammary delays or prevents tumor formation (Bentires-Alj and Neel, 2007). However, mammary-specific ablation of PTP1B after tumor formation does not affect tumor maintenance or growth (Balavenkatraman et al., 2011). More work will be required to determine exactly how selective dysregulation of PTP1B by MenaINV contributes to tumor progression. The exact mechanism underlying MenaINV-mediated dysregulation of tyrosine phosphorylation remains to be determined. Given that the INV exon is located near the EVH1 domain of Mena, it is possible that inclusion of INV affects formation of complexes with PTP1B, SHIP2, or both, which in turn could affect recruitment of both proteins to RTKs. Because all Mena isoforms form stable tetramers (Gertler and Condeelis, 2011), it is possible that inclusion of INV could alter interactions with up to four distinct EVH1 ligands within a MenaINV tetramer. Future studies will address how inclusion of the sequence encoded by the INV exon regulates these interactions. Regardless of the outcome, our findings indicate that Mena and its isoforms act to modulate RTK signaling pathways in addition to their known role in regulating actin dynamics.
MATERIALS AND METHODS
Antibodies, growth factors, and inhibitors
Growth factors were purchased from Life Technologies, Carlsbad, CA (EGF), and PeproTech, Rocky HIll, NJ (HGF, NRG-1, and IGF). The compounds used were Met inhibitor SU11274 (Selleck Chemicals, Houston, TX), EGFR inhibitor erlotinib (LC Labs, Woburn, MA; PeproTech), PTP1B inhibitor (539741; Calbiochem, Billerica, MA), and SHIP2 inhibitor AS1949490 (Tocris, Minneapolis, MN). The following antibodies were used: EGFR (Cell Signaling Technology, Danvers, MA), pEGFR Y1173 (Cell Signaling Technology; Epitomics), vimentin (BD Transduction Laboratories, San Jose, CA), E-cadherin (Cell Signaling Technology), PTP1B (Millipore, Burlington, MA), SHIP2 (Cell Signaling Technology), and panMena (Lebrand et al., 2004).
Cell culture
All breast cancer cell lines were obtained from the American Type Culture Collection and cultured in DMEM with 10% heat-inactivated fetal bovine serum. Cell lines were engineered to stably express 10- to 15-fold-higher levels of Mena isoforms more than for wild-type cell lines. For siRNA-mediated knockdown experiments, 25 nM SHIP2– or PTP1B–targeted ON-TARGETplus SMARTpool siRNA (Dharmacon, GE Lifesciences, Lafayette, CO) was transfected in serum-free OptiMeM using Dharmafect4 with assays performed 72 h posttransfection. The ON-TARGETplus nontargeted control pool (25 nM) was used in those conditions labeled “scramble.”
PTP1B and Mena-EVH1 in vitro binding assay
Constructs used for recombinant protein production were GST-PTP1B (Addgene, Cambridge, MA; plasmid #8602), GST, and GST-FP4 His-Mini-Mena (His-TEV-EVH1-LERER-6xGly-CoCo), which contains mouse Mena (Gertler et al., 1996) residues 1–258 fused, via a 6xGly linker, to the C-terminal 31-residue coiled-coil from Mena. The coiled-coil, which mediates Mena tetramerization, was included to allow for the potential effects of avidity in the binding assay (Gupton et al., 2012), and intervening sequences were omitted to enable recovery of soluble, purified protein. Detailed protocols for protein expression and purification are given in the Supplemental Experimental Procedures.
GST, GST-FP4, and GST-PTP1B (3 μM) were immobilized on glutathione beads and blocked for 2 h at 4°C (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) [HEPES], pH 7.6, 150 mM NaCl, 1% NP-40, 3% bovine serum albumin [BSA]). Beads were washed once and incubated with 50, 100, 500 nM His-Mini-Mena, rotating for 2 h at 4°C in binding buffer (20 mM HEPES, pH 7.6, 150 mM NaCl, 1% NP-40). Beads were washed three times in binding buffer while rotating at 4°C for 5 min. Proteins were eluted in 4× SDS–PAGE sample buffer and assayed by Western blotting.
EGFR internalization assays
Surface proteins were biotinylated using the method of Roberts et al. (2001; see the Supplemental Experimental Procedures). Cells were lysed in 75 mM HEPES (pH 7.5), 200 mM NaCl, 10% glycerol, 1.5% Triton-X 100, and 0.75% NP-40 with a protease Mini-Complete protease inhibitor tablet (Roche, Indianapolis, IN) on ice. To measure the surface level of EGFR, cells were lysed immediately after labeling with biotin. EGFR STAR enzyme-linked immunosorbent assay (ELISA; Millipore) was used to quantify total and biotinylated EGFR by modifying the protocol to replace the anti-rabbit immunoglobulin G (IgG)–horseradish peroxidase conjugate with anti-rabbit IR800 and streptavidin-IR680 (Li-Cor, Lincoln, NE) antibodies to facilitate dual detection of total and biotinylated EGFR using the Li-Cor Odyssey scanning system. Negative controls of nonlabeled samples or surface-stripped samples (without the internalization step) were included for each experiment.
Protrusion and 3D invasion assays
Protrusion assays were performed as described (Meyer et al., 2012). Briefly, serum-starved cells plated on collagen- and Matrigel-coated dishes were imaged every 20 s for 10 min at 37°C (TE2000 microscope; Nikon) with a 20× objective. Growth factor/inhibitor solutions were added after 80 s. Cell areas were traced immediately before stimulation and 9 min after stimulation using ImageJ (National Institutes of Health, Bethesda, MD). Data shown are from individual cells pooled from at least three separate experiments. The 3D invasion assay in Figure 6B was performed as described (Giampieri et al., 2009). See the Supplemental Experimental Procedures.
PI(4,5)P2 hydrolysis assay
PI(4,5)P2 hydrolysis was assayed as described (Van Rheenen et al., 2007). See the Supplemental Experimental Procedures.
Cofilin:actin fluorescence resonance energy transfer
F-actin:cofilin binding was measured by fluorescence resonance energy transfer (FRET) efficiency using acceptor photobleaching as described (Van Rheenen et al., 2007). See the Supplemental Experimental Procedures.
Immunoprecipitation and Western blotting
Standard procedures were used for protein electrophoresis, Western blotting, and IP. For IP, cells were lysed in 20 mM HEPES, pH 7.4, 200 mM NaCl, 2 mM MgCl2, 5% glycerol, 1% NP-40, and phosphatase inhibitors. See the Supplemental Experimental Procedures. For the Mena-SHIP2 IP, cells were harvested in chilled lysis buffer (20 mM HEPES, pH 7.4, 200 mM NaCl, 2 mM MgCl2, 5% glycerol) supplemented with Mini-Complete EDTA-free protease inhibitors (Roche) and a phosphatase inhibitor cocktail (PhosSTOP; Roche) at 4°C. Precleared lysate was incubated with 40 μl of 50% slurry of Protein A Plus beads (Pierce, Grand Island, NY) and Mena antibody or IgG control for 3 h at 4°C. Beads were washed three times in lysis buffer, eluted in 2× SDS sample buffer, and resolved by SDS–PAGE.
Proximity ligation assay
Cells were prepared as for protrusion assay and stimulated with growth factor as indicated. Cells were fixed for 20 min in 4% paraformaldehyde in PHEM buffer (60 mM PIPES [piperazine-N,N’-bis(ethanesulfonic acid)], pH 7.0, 25 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.0, 10 mM EGTA [ethylene glycol tetraacetic acid], pH 8.0, 2 mM MgCl2, 0.12 M sucrose), permeabilized with 0.2% Triton X-100, blocked with 10% BSA, and incubated with primary antibodies overnight at 4°C. PLA was then performed according to the manufacturer’s protocol (Olink Biosciences, Uppsala, Sweden). The spots per cell were counted, and the number was normalized relative to cell area and/or GFP expression. Data are pooled from at least three different experiments.
In vivo invasion assay
The in vivo invasion assay was performed in three mice per condition as described (Wyckoff et al., 2000). Briefly, needles were held in place by a micromanipulator around a single mammary tumor of an anesthetized mouse. Needles contained a mixture of Matrigel, 0–25 nM EGF, buffer, and EDTA with either 0.01% dimethyl sulfoxide (DMSO), or 10 μM PTP1B inhibitor. After 4 h of cell collection, the contents of the needles were extruded. Cells were stained with 4′,6-diamidino-2-phenylindole and counted on an Olympus IX70 inverted microscope.
Phosphotyrosine analysis via LC-MS/MS
Serum-starved MDA-MB231 cells expressing GFP or GFP-MenaINV were stimulated with 0, 0.25, 0.5, or 15.8 nM EGF for 1 min. Cells were immediately lysed in 8 M urea (Sigma-Aldrich, St. Louis, MO) and quantified using the BCA assay (Pierce). As previously described, quantitative LC-MS/MS analyses of tyrosine-phosphorylated peptides using iTRAQ 8plex and an Orbitrap Elite mass spectrometer (Thermo) were performed (Johnson et al., 2012). In brief, proteins were reduced, alkylated, and digested with modified trypsin (Promega, Madison, WI). Peptides were desalted, lyophilized, and labeled with iTRAQ 8plex (AB Sciex, Framingham, MA). Tyrosine-phosphorylated peptides were enriched using IP and immobilized metal affinity chromatography. Mascot peptide identifications, phosphorylation-site assignments, and quantification were verified manually with the assistance of computer-aided manual validation software (Curran et al., 2013). Supplemental Table S1 contains the sequences of identified phosphotyrosine peptides along with relative quantification and statistical analysis. See the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Evanthia Roussos, Michele Balsamo, and Aaron Meyer for advice and John Lamar for advice and for gifts of tumor tissue. This work was supported by DOD Breast Cancer Research Program Grants W81XWH-10-1-0040 to S.K.H. and W81XWH-13-1-0031 to M.J.O., National Institutes of Health Grants U54-CA112967 to F.B.G., F.W., and D.A.L. and GM58801 to F.B.G., funds from the Ludwig Center at the Massachusetts Institute of Technology to F.B.G., Koch Institute National Cancer Institute Core Grant P30-CA14051, and CA150344 to J.C. and CA100324 for J.J. We thank the Flow Cytometry, Microscopy and Histology facilities in the Koch Institute Swanson Biotechnology Center for support and the Analytical Imaging Facility in the Biophotonics Center at the Albert Einstein College of Medicine.
Abbreviations used:
- CME
clathrin-mediated endocytosis
- ECM
extracellular matrix
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EVH
Ena/Vasp homology
- GFP
green fluorescent proteins
- GST
glutathione-S-transferase
- HGF
hepatocyte growth factor
- IGF
insulin growth factor
- IGFR
insulin growth factor receptor
- Immuno-EM
immuno-electron microscopy
- IP
immuno-precipitation
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- Lpd
lamellipodin
- PLA
proximity ligation assay
- PLCγ
phospholipase C gamma
- RTK
receptor tyrosine kinase
- TKI
tyrosine kinase inhibitor
- TMEM
tumor microenvironment of metastasis.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0442) on September 2, 2015.
REFERENCES
- Alexander S, Friedl P. Cancer invasion and resistance: interconnected processes of disease progression and therapy failure. Trends Mol Med. 2012;18:13–26. doi: 10.1016/j.molmed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Bailly M, Wyckoff J, Bouzahzah B, Hammerman R, Sylvestre V, Cammer M, Pestell R, Segall JE. Epidermal growth factor receptor distribution during chemotactic responses. Mol Biol Cell. 2000;11:3873–3883. doi: 10.1091/mbc.11.11.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balavenkatraman KK, Aceto N, Britschgi A, Mueller U, Bence KK, Neel BG, Bentires-Alj M. Epithelial protein-tyrosine phosphatase 1B contributes to the induction of mammary tumors by HER2/Neu but is not essential for tumor maintenance. Mol Cancer Res. 2011;9:1377–1384. doi: 10.1158/1541-7786.MCR-11-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LJ, Jarchau T, Oschkinat H, Walter U. EVH1 domains: structure, function and interactions. FEBS Lett. 2002;513:45–52. doi: 10.1016/s0014-5793(01)03291-4. [DOI] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Haugh JM. Directed migration of mesenchymal cells: where signaling and the cytoskeleton meet. Curr Opin Cell Biol. 2014;30C:74–82. doi: 10.1016/j.ceb.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M, Neel BG. Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer. Cancer Res. 2007;67:2420–2424. doi: 10.1158/0008-5472.CAN-06-4610. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Ferreira APA, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2014;517:460–465. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TEB, Small JV, Curth U, Dickinson RB, Faix J. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 2011;30:456–467. doi: 10.1038/emboj.2010.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesio CL, Chan KT, Perrin BJ, Burton NO, Zhang S, Zhang Z-Y, Huttenlocher A. Calpain 2 and PTP1B function in a novel pathway with Src to regulate invadopodia dynamics and breast cancer cell invasion. J Cell Biol. 2008;180:957–971. doi: 10.1083/jcb.200708048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran TG, Bryson BD, Reigelhaupt M, Johnson H, White FM. Computer aided manual validation of mass spectrometry-based proteomic data. Methods. 2013;61:219–226. doi: 10.1016/j.ymeth.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- Erneux C, Edimo WE, Deneubourg L, Pirson I. SHIP2 multiple functions: a balance between a negative control of PtdIns(3,4,5)P3 level, a positive control of PtdIns(3,4)P2 production, and intrinsic docking properties. J Cell Biochem. 2011;112:2203–2209. doi: 10.1002/jcb.23146. [DOI] [PubMed] [Google Scholar]
- Feldhammer M, Uetani N, Miranda-Saavedra D, Tremblay ML. PTP1B: a simple enzyme for a complex world. Crit Rev Biochem Mol Biol. 2013;48:430–445. doi: 10.3109/10409238.2013.819830. [DOI] [PubMed] [Google Scholar]
- Frangioni J V, Oda A, Smith M, Salzman EW, Neel BG. Calpain-catalyzed cleavage and subcellular relocation of protein phosphotyrosine phosphatase 1B (PTP-1B) in human platelets. EMBO J. 1993;12:4843–4856. doi: 10.1002/j.1460-2075.1993.tb06174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Fuentes F, Arregui CO. Microtubule and cell contact dependency of ER-bound PTP1B localization in growth cones. Mol Biol Cell. 2009;20:1878–1889. doi: 10.1091/mbc.E08-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F, Condeelis J. Metastasis: tumor cells becoming MENAcing. Trends Cell Biol. 2011;21:81–90. doi: 10.1016/j.tcb.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Riquelme D, Hughes-Alford SK, Tadros J, Rudina SS, Hynes RO, Lauffenburger D, Gertler FB. Mena binds α5 integrin directly and modulates α5β1 function. J Cell Biol. 2012;198:657–676. doi: 10.1083/jcb.201202079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusenbauer S, Vlaicu P, Ullrich A. HGF induces novel EGFR functions involved in resistance formation to tyrosine kinase inhibitors. Oncogene. 2013;32:3846–3856. doi: 10.1038/onc.2012.396. [DOI] [PubMed] [Google Scholar]
- Haj FG, Markova B, Klaman LD, Bohmer FD, Neel BG. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J Biol Chem. 2003;278:739–744. doi: 10.1074/jbc.M210194200. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H, Del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol Cell Proteomics. 2012;11:1724–1740. doi: 10.1074/mcp.M112.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman LB, Maiwald T, Conzelmann H, Lauffenburger DA, Sorger PK. Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol Cell. 2011;43:723–737. doi: 10.1016/j.molcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- Mertins P, Eberl HC, Renkawitz J, Olsen J V, Tremblay ML, Mann M, Ullrich A, Daub H. Investigation of protein-tyrosine phosphatase 1B function by quantitative proteomics. Mol Cell Proteomics. 2008;7:1763–1777. doi: 10.1074/mcp.M800196-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Hughes-Alford SK, Kay JE, Castillo A, Wells A, Gertler FB, Lauffenburger DA. 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J Cell Biol. 2012;197:721–729. doi: 10.1083/jcb.201201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KL, Madden JM, Zoratti GL, Kuperwasser C, List K, Boerner JL. Fibroblast-secreted hepatocyte growth factor mediates epidermal growth factor receptor tyrosine kinase inhibitor resistance in triple-negative breast cancers through paracrine activation of Met. Breast Cancer Res. 2012;14:R104. doi: 10.1186/bcr3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Perera RM, Lucast L, Zoncu R, Domin J, Gertler FB, Toomre D, De Camilli P. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J Cell Biol. 2010;190:307–315. doi: 10.1083/jcb.201005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer. 2011;11:177–187. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]
- Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–9506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesse X, Dewaste V, De Smedt F, Laffargue M, Giuriato S, Moreau C, Payrastre B, Erneux C. The Src homology 2 domain containing inositol 5-phosphatase SHIP2 is recruited to the epidermal growth factor (EGF) receptor and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate in EGF-stimulated COS-7 cells. J Biol Chem. 2001;276:28348–28355. doi: 10.1074/jbc.M103537200. [DOI] [PubMed] [Google Scholar]
- Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. J Cell Sci. 2012;125:5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim H-D, Giampieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell. 2008;15:813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli J, Goswami S, Jones JG, Rohan TE, Pieri E, Chen X, Adler E, Cox D, Maleki S, Bresnick A, et al. Invasive breast carcinoma cells from patients exhibit MenaINV- and macrophage-dependent transendothelial migration. Sci Signal. 2014;7:ra112. doi: 10.1126/scisignal.2005329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- Pula G, Krause M. Role of Ena/VASP proteins in homeostasis and disease. Handb Exp Pharmacol. 2008;2008:39–65. doi: 10.1007/978-3-540-72843-6_3. [DOI] [PubMed] [Google Scholar]
- Roberts M, Barry S, Woods A, van der Sluijs P, Norman J. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol. 2001;11:1392–1402. doi: 10.1016/s0960-9822(01)00442-0. [DOI] [PubMed] [Google Scholar]
- Robinson BD, Sica GL, Liu Y-F, Rohan TE, Gertler FB, Condeelis JS, Jones JG. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res. 2009;15:2433–2441. doi: 10.1158/1078-0432.CCR-08-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohan TE, Xue X, Lin H-M, D’Alfonso TM, Ginter PS, Oktay MH, Robinson BD, Ginsberg M, Gertler FB, Glass AG, et al. Tumor microenvironment of metastasis and risk of distant metastasis of breast cancer. J Natl Cancer Inst. 2014;106:1–11. doi: 10.1093/jnci/dju136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, Pozzuto M, Stobezki R, Goswami S, Segall JE, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci. 2011a;124:2120–2131. doi: 10.1242/jcs.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011b;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos ET, Goswami S, Balsamo M, Wang Y, Stobezki R, Adler E, Robinson BD, Jones JG, Gertler FB, Condeelis JS, et al. Mena invasive (Mena(INV)) and Mena11a isoforms play distinct roles in breast cancer cell cohesion and association with TMEM. Clin Exp Metastasis. 2011c;28:515–527. doi: 10.1007/s10585-011-9388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos ET, Wang Y, Wyckoff JB, Sellers RS, Wang W, Li J, Pollard JW, Gertler FB, Condeelis JS. Mena deficiency delays tumor progression and decreases metastasis in polyoma middle-T transgenic mouse mammary tumors. Breast Cancer Res. 2010;12:R101. doi: 10.1186/bcr2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan V, Abella J, Lai A, Bertos N, Stuible M, Tremblay ML, Park M. Protein-tyrosine phosphatase 1B modulates early endosome fusion and trafficking of Met and epidermal growth factor receptors. J Biol Chem. 2011;286:45000–45013. doi: 10.1074/jbc.M111.270934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K, Murakami I, Katayama T, Tomizawa K, Osada H, Sekido Y, Maehara Y, Yatabe Y, Mitsudomi T. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res. 2010;16:5489–5498. doi: 10.1158/1078-0432.CCR-10-1371. [DOI] [PubMed] [Google Scholar]
- Suwa A, Yamamoto T, Sawada A, Minoura K, Hosogai N, Tahara A, Kurama T, Shimokawa T, Aramori I. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br J Pharmacol. 2009;158:879–887. doi: 10.1111/j.1476-5381.2009.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, Yip S-CC, Backer JM, Eddy RJ, Condeelis JS. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehlow A, Soong D, Vizcay-Barrena G, Bodo C, Law A-L, Perera U, Krause M. Endophilin, Lamellipodin, and Mena cooperate to regulate F-actin-dependent EGF-receptor endocytosis. EMBO J. 2013;32:2722–2734. doi: 10.1038/emboj.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60:5401–5404. [PubMed] [Google Scholar]
- Yee D, Paik S, Lebovic GS, Marcus RR, Favoni RE, Cullen KJ, Lippman ME, Rosen N. Analysis of insulin-like growth factor I gene expression in malignancy: evidence for a paracrine role in human breast cancer. Mol Endocrinol. 1989;3:509–517. doi: 10.1210/mend-3-3-509. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhang C, Croucher DR, Soliman MA, St-Denis N, Pasculescu A, Taylor L, Tate SA, Hardy WR, Colwill K, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N, Kenny PA, Wiley HS, Condeelis JS, Segall JE. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene. 2014;33:3784–3793. doi: 10.1038/onc.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.