Sorting nexin 9 (SNX9) is shown to differentiate Smad3 from Smad2 nuclear delivery by mediating the association of phosphorylated Smad3 with importin 8 and the nuclear membrane. While the absence of SNX9 had negligible effects on transforming growth factor β receptor activity or Smad2 signaling, Smad3-dependent targets and phenotypes were inhibited.
Abstract
Transforming growth factor β (TGFβ) is a pleiotropic protein secreted from essentially all cell types and primary tissues. While TGFβ’s actions reflect the activity of a number of signaling networks, the primary mediator of TGFβ responses are the Smad proteins. Following receptor activation, these cytoplasmic proteins form hetero-oligomeric complexes that translocate to the nucleus and affect gene transcription. Here, through biological, biochemical, and immunofluorescence approaches, sorting nexin 9 (SNX9) is identified as being required for Smad3-dependent responses. SNX9 interacts with phosphorylated (p) Smad3 independent of Smad2 or Smad4 and promotes more rapid nuclear delivery than that observed independent of ligand. Although SNX9 does not bind nucleoporins Nup153 or Nup214 or some β importins (Imp7 or Impβ), it mediates the association of pSmad3 with Imp8 and the nuclear membrane. This facilitates nuclear translocation of pSmad3 but not SNX9.
INTRODUCTION
Transforming growth factor β (TGFβ) regulates a variety of cellular processes, including matrix deposition, mitosis, development, differentiation, and apoptosis (Roberts and Wakefield, 2003; Elliott and Blobe, 2005). The primary intracellular mediators of TGFβ action are the Smad proteins, although non-Smad pathways have been reported, often in a cell type–specific context (Rahimi and Leof, 2007; Ross and Hill, 2008). Three general categories of Smads have been identified: receptor-regulated Smads (R-Smads; Smad2 and Smad3 for TGFβ or Activin and Smad1, Smad5, and Smad8 for bone morphogenetic proteins); common-mediator Smad (Co-Smad; Smad4); and inhibitory Smads (I-Smads; Smad6 and Smad7). The R- and Co-Smad proteins shuttle continuously between the nucleus and cytoplasm in unstimulated cells and in the presence of TGFβ (Inman et al., 2002; Xu et al., 2002; Schmierer and Hill, 2005).
The import and nuclear translocation of cargo generally requires association with specific transport receptors (karyopherins) and interactions with various nuclear pore complex proteins (nucleoporins) (Chook and Suel, 2011; Marfori et al., 2011). Although a great deal of information concerning Smad nuclear import has been generated, and there is evidence for various Karyopherins, nucleoporins, and the dynein light chain km23-2 in Smad3 trafficking (Hill, 2009; Jin et al., 2009), it is still not settled how R-Smads are translocated to the nucleus or whether distinct mechanisms are used by Smad2 and Smad3.
While the manner by which the Smad proteins traffic from the cytoplasm to the nucleus is unresolved, the sorting nexins (SNXs) are one family (>30 in human) of structurally related trafficking proteins with proposed roles in receptor degradation, sorting, internalization, and recycling (Carlton et al., 2005; Badour et al., 2007; Verges, 2007). All SNXs are defined by the presence of a phox homology (PX) domain that binds phosphoinositides and aids in targeting SNXs to particular membranes (Worby and Dixon, 2002). SNX9, however, also contains an amino terminal SH3 domain required for dynamin, cdc42-associated kinase (ACK2), WASp, and Itch binding; a Bin/Amphiphysin/Rvs (BAR) domain that senses membrane curvature and is required for dimerization; and a low-complexity region that binds AP-2α and clathrin (Supplemental Figure S1A; Worby and Dixon, 2002; Carlton et al., 2005; Badour et al., 2007; Lundmark and Carlsson, 2009; Baumann et al., 2010). Because SNX9 is known to modulate the trafficking responses of several transmembrane receptors (Worby and Dixon, 2002; Verges, 2007), and Smad phosphorylation has been shown to be coupled to TGFβ endocytic activity in various systems (Hayes et al., 2002; Penheiter et al., 2002; Di Guglielmo et al., 2003), studies were initiated to examine the role(s) of SNX9 in TGFβ receptor action. Although we found negligible effects on receptor activity, we show that SNX9 has an obligate role in the cellular response to TGFβ and differentially regulates Smad3- from Smad2-dependent phenotypes. The results expose a new role for SNX9 downstream of its canonical plasma membrane action and define a mechanism by which specificity in profibrotic TGFβ signaling can be controlled through SNX9 promoting Smad3 nuclear import following ligand-induced phosphorylation.
RESULTS
SNX9 specifically regulates Smad3-dependent TGFβ signaling
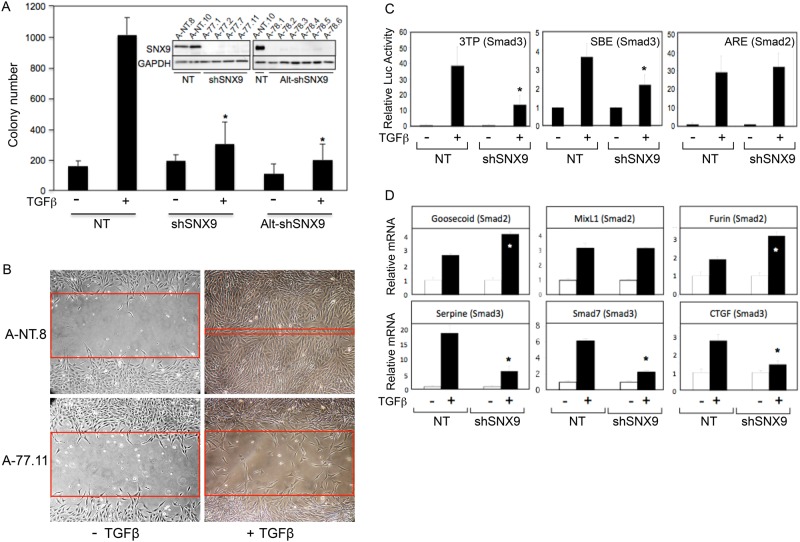
TGFβ receptor (TGFβR) signaling has been shown to be intimately connected to the cellular trafficking machinery in various systems (Hayes et al., 2002; Penheiter et al., 2002; Di Guglielmo et al., 2003). As SNX9 is known to function at similar plasma membrane and endosomal locales (Verges, 2007; Cullen, 2008; Lundmark and Carlsson, 2009), studies were initiated to determine whether SNX9 had a role in the mesenchymal cell response to TGFβ. AKR-2B clones expressing two distinct SNX9 short hairpin RNAs (shRNAs) or dominant-negative (DN)-SNX9 constructs were isolated, and the ability of TGFβ to stimulate anchorage-independent growth (AIG) was determined (Supplemental Figure S1A provides a schematic of SNX9 and the dominant negatives). Although basal growth was unaffected, ligand-induced AIG was reduced to control levels by SNX9 knockdown (KD) or DN expression (Figure 1A and Supplemental Figure S1B). TGFβ also has a major role in cell migration and wound repair (Rahimi and Leof, 2007; Barrientos et al., 2008). Analogous to that observed for AIG, SNX9 KD significantly impaired the in vitro migratory action of TGFβ (Figure 1B).
FIGURE 1:
SNX9 regulates TGFβ-stimulated soft-agar colony formation, wound closure, and Smad3-dependent transcriptional activity. (A) AKR-2B cells stably integrated with nontargeting (NT) or SNX9 shRNA were seeded in soft agar in the absence (−) or presence (+) of 10 ng/ml of TGFβ. Data depict the number of colonies from two pooled NT (A-NT.8 and A-NT.10), four pooled SNX9 shRNA1 (shSNX9; A-77.1, A-77.2, A-77.7, and A-77.11), and four pooled SNX9 shRNA2 (Alt-shSNX9; A-78.1, A-78.2, A-78.5, and A-78.6) clones. Inset shows SNX9 knockdown. (B) Scratch assays were performed on NT (A-NT.8) and SNX9 KD (A-77.11) clones as described in Materials and Methods and are representative of three separate experiments. Red bands indicate the leading edge after 24 h in the presence or absence of TGFβ. (C) NT or SNX9 KD AKR-2B cultures were transfected with the indicated luciferase reporters and either left untreated (−) or stimulated (+) with TGFβ for 24 h. Data are from three independent experiments (A-NT.8 and pooled data from A-77.7 and A-77.11 clones) and represent the mean fold induction ± SD relative to untreated. (D) A-NT.8 or A-77.7 cells were arrested and treated (+) for 6 h with TGFβ. RNA was isolated and qPCR performed for the indicated Smad2- or Smad3-dependent targets. Data reflect the normalized fold induction above unstimulated NT controls ± SD (n = 3). * denotes significant difference between stimulated NT and shSNX9 cultures.
As Figure 1, A and B, and Supplemental Figure S1B documented that two TGFβ phenotypes studied in fibroproliferative models were dependent on SNX9, we next investigated whether Smad transcriptional responses displayed a similar SNX9 requirement. Initial studies showed a 50–70% reduction from transfected Smad3-dependent reporters (3TP or SBE), yet no effect on Smad2-regulated Activin response element (ARE) signaling (Figure 1C and Supplemental Figure S1C). Because this was quite surprising, qPCR evaluation of three endogenous Smad2- or Smad3-responsive genes was undertaken. While none of the Smad2-responsive genes were inhibited, reduction in each of the Smad3-responsive genes was observed (Figure 1D). In fact, for two of the three Smad2-responsive genes, there was a statistically significant increase with reduced SNX9, likely reflecting positive and negative actions of Smad2 and Smad3, respectively (Labbe et al., 1998).
SNX9 is required for Smad3 nuclear import
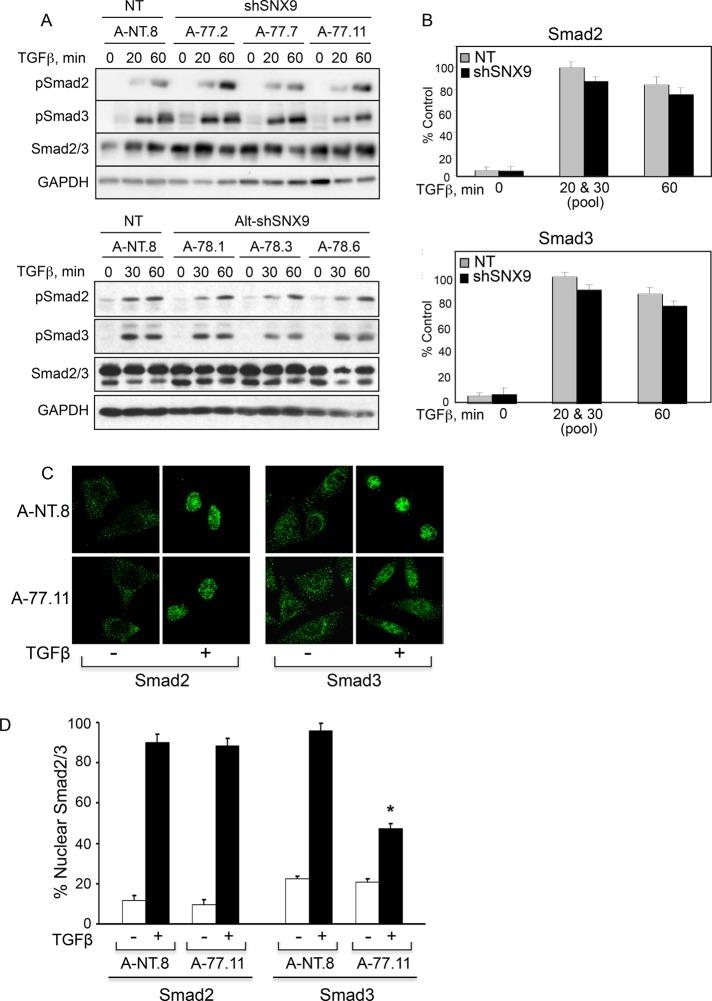
SNX9 is essential for the endocytosis of various cargoes (Lundmark and Carlsson, 2009). As TGFβ-stimulated Smad2/3 phosphorylation occurs downstream of dynamin action (Hayes et al., 2002; Penheiter et al., 2002; Di Guglielmo et al., 2003), it seemed reasonable that SNX9 might modulate TGFβ signaling through specific inhibition of Smad3 phosphorylation. Contrary to our expectations, no difference in either the kinetics or extent of R-Smad phosphorylation was observed in SNX9 KD or DN clones relative to control (Figures 2, A and B, and Supplemental Figure S1D). Because R-Smad phosphorylation enhances their nuclear accumulation (Feng and Derynck, 2005; Schmierer and Hill, 2007; Hill, 2009), we next investigated whether SNX9 was required for Smad3 nuclear import. While SNX9 loss significantly reduced nuclear Smad3 following addition of TGFβ, nuclear Smad2 was unaffected (Figure 2C). Quantitation of this response demonstrated an approximate 50–60% decrease in the nuclear accumulation of Smad3 with no appreciable effect on Smad2 or basal nuclear R-Smads (Figure 2D). This finding was further supported using DN-SNX9 constructs. Consistent with the loss in nuclear Smad3 observed in the KD cells, DN-SNX9 specifically prevented ligand-stimulated Smad3 nuclear delivery (Supplemental Figure S1E).
FIGURE 2:
SNX9 functions downstream of R-Smad phosphorylation. (A) AKR-2B clones expressing nontargeting (NT) or SNX9 shRNA were left untreated (0) or stimulated with TGFβ. Western blot analysis was performed for the indicated phosphorylated (pSmad2, pSmad3) or total (Smad2/3, GAPDH) proteins. (B) Quantitation of pSmad2 (n = 9) or pSmad3 (n = 11) at each time compared with NT and normalized to GAPDH. The quantitation of the 20- and 30-min time points shown in A were pooled and are presented as 20 & 30 (pool). (C) NT (A-NT.8) or SNX9 KD (A-77.11) cells were stimulated in the absence (−) or presence (+) of TGFβ for 45 min, and Smad2 or Smad3 immunofluorescence was determined. (D) Quantitation of nuclear R-Smad staining ± SD from 20 cells in each of two experiments from C. The y-axis represents the nuclear pixel signal obtained with 100%, defined as the signal from 7 A-NT.8 cells that showed no cytoplasmic R-Smad staining following TGFβ stimulation. * denotes significant difference between stimulated NT and shSNX9 cultures.
SNX9 specifically impacts phosphorylated Smad3 nuclear entry
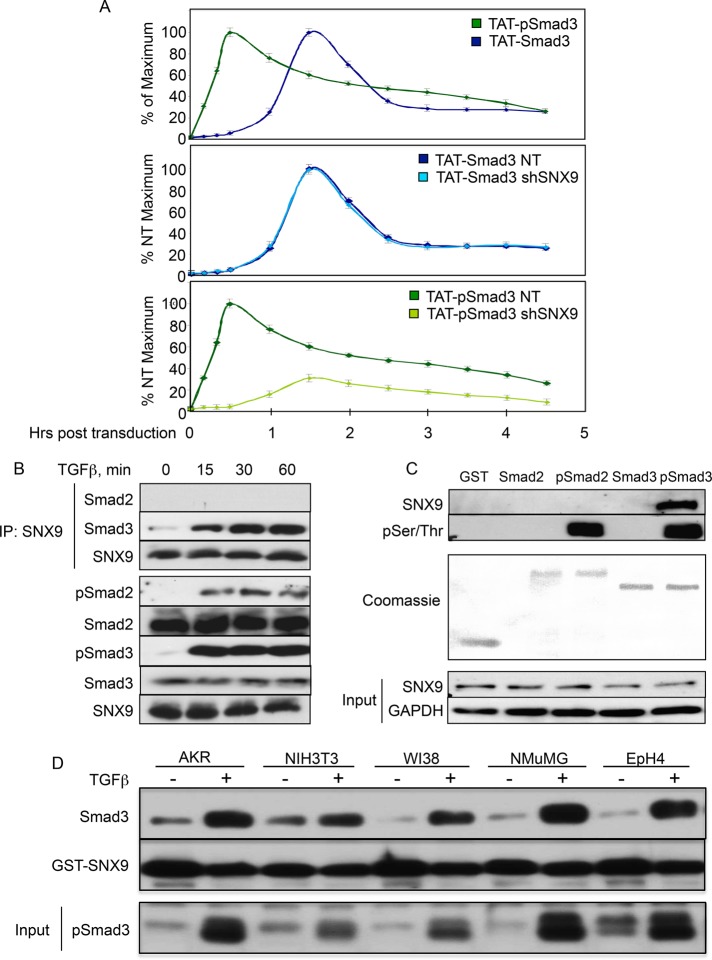
While it has been suggested that R-Smad nuclear accumulation is due primarily to retention of phosphorylated over nonphosphorylated R-Smads (Schmierer and Hill, 2005, 2007), other studies have also observed increased rates of R-Smad nuclear trafficking in stimulated versus unstimulated cells (Kurisaki et al., 2001; Schmierer et al., 2008). To address these issues in vivo without the complication of unknown ratios of unphosphorylated to phosphorylated Smad3 produced after ligand stimulation, we generated a Smad3 construct fused to the cell-transducing TAT domain from HIV (Becker-Hapak et al., 2001), labeled the purified protein with 125I, and phosphorylated half with the immunopurified/activated type I TGFβ receptor (TβRI) before it was added to mesenchymal cells that then underwent nuclear purification. After normalizing for initial cellular uptake (Supplemental Figure S2), we observed clear differences in both the kinetics of nuclear import as well as retention between TAT-pSmad3 and TAT-Smad3 (Figure 3A, top panel). For instance, while half-maximal nuclear translocation of TAT-pSmad3 occurred within ∼20 min, unphosphorylated TAT-Smad3 was ∼3.5 times slower. Furthermore, consistent with a role for Smad phosphorylation in also enhancing nuclear retention, the rate of TAT-pSmad3 and TAT-Smad3 nuclear loss was 28.6 counts/min and 235.7 counts/min, respectively, which was unaffected by SNX9 KD (i.e., corresponding rates of 26.6 counts/min and 211.8 counts/min). Consistent with the analyses shown in Figure 2, C and D, and Supplemental Figure S1E, while SNX9 loss had no appreciable effect on unphosphorylated TAT-Smad3 (i.e., basal shuttling), it significantly inhibited TAT-pSmad3 nuclear accumulation (Figure 3A, middle and bottom panels, respectively). Thus, while the TAT peptides allow one to differentiate the rate of basal Smad3 shuttling from pSmad3 nuclear import, they also confirm the importance of phosphorylation on Smad3 nuclear retention and the need for SNX9 in promoting pSmad3 nuclear entry.
FIGURE 3:
SNX9 specifically binds and is required for enhanced pSmad3 nuclear entry. (A) [125I]-labeled TAT-Smad3 and TAT-pSmad3 proteins were generated as described in Materials and Methods. AKR-2B cells expressing nontargeting (NT) shRNA (A-NT.8) or shRNA against SNX9 (shSNX9; A-77.7) were transduced with the indicated TAT peptides, and 125I counts in total cell and nuclear lysates were obtained from 10 to 270 min. Data reflect the mean ± SD from three experiments for the 30- to 240-min time points and from two experiments for the 10-, 20-, and 270-min time points. (B) AKR-2B cells were stimulated with TGFβ, and the indicated proteins coprecipitating with SNX9 were detected by Western blotting (top three panels). Bottom five panels: indicated proteins in total lysate. (C) AKR-2B lysates were incubated with GST or GST-Smad fusion protein untreated or in vitro phosphorylated by TβRI. Bound SNX9 (top panel) and the phosphorylated R-Smad (second panel) used in the pull down were detected using SNX9 and phospho-Ser/Thr antibodies, respectively. (D) Cell lines were stimulated in the absence (−) or presence (+) of TGFβ for 60 min. Following lysis and GST-SNX9 pull down, Western blotting was performed. Bottom panel depicts pSmad3 expression in total cell lysate.
SNX9 preferentially binds phosphorylated Smad3
As the preceding data define a new role for SNX9 in mediating Smad3 nuclear transport and biological action, we addressed the following mechanistic questions. First, does SNX9 show differential R-Smad binding, is this regulated by ligand, and is there any identifiable role for Smad4? Second, is SNX9 required for importin (Imp) and/or nucleoporin (Nup) binding to Smad3? Third, how does SNX9 function to promote pSmad3 nuclear import?
For investigation of the first of these issues, AKR-2B cells were stimulated with TGFβ, and the ability of SNX9 to coimmunoprecipitate Smad2 or Smad3 was determined. While a slight basal association with Smad3 was observed, addition of ligand enhanced the interaction (Figure 3B). In contrast, no binding of SNX9 and Smad2 could be detected basally or following TGFβ treatment. This interaction of SNX9 with pSmad3 was further confirmed using glutathione S-transferase (GST)-Smad2 or GST-Smad3 fusion proteins (Figure 3C). Analogous to coimmunoprecipitation (co-IP), SNX9/R-Smad binding was only observed with GST-pSmad3. Furthermore, the SNX9/pSmad3 association shown in AKR-2B cells (Figure 3, B and C) was additionally documented in two other nontransformed mesenchymal (NIH3T3 and WI38) and epithelial (NMuMG and EpH4) cell lines (Figure 3D).
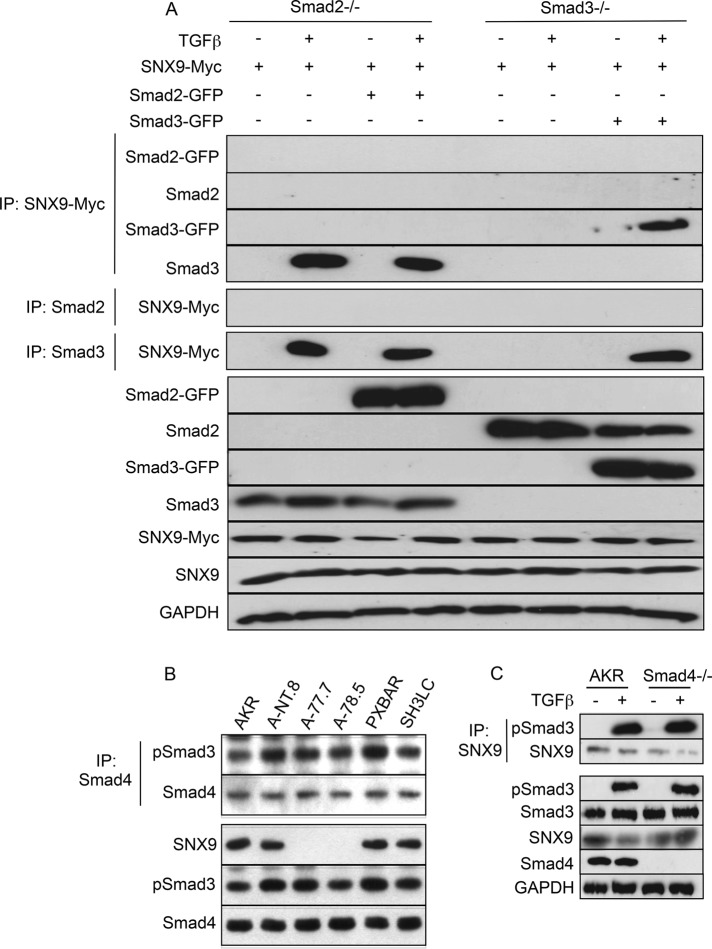
Figure 3, B and C, shows that a SNX9/Smad2 association cannot be observed either in vivo or in vitro. To address the possibility that Smad2 has a role in promoting and/or enhancing SNX9/Smad3 binding, we performed the studies provided in Figure 4A. In the absence of Smad2, there was ligand-dependent SNX9/Smad3 binding similar to that seen in AKR-2B cells. Because the canonical model of R-Smad nuclear translocation has Smad4 present to generate the most energetically favorable heterotrimer (Wu et al., 2001; Chacko et al., 2004), we determined the requirement for SNX9 or Smad4 in the formation of either the pSmad3/Smad4 or pSmad3/SNX9 complex (Figure 4, B and C); no identifiable role for either SNX9 or Smad4 was observed.
FIGURE 4:
pSmad3/SNX9 binding is independent of Smad2 or Smad4. (A) Smad2−/− and Smad3−/− null MEFs were transfected (+) with SNX9-Myc, Smad2-GFP, and/or Smad3-GFP and either left untreated (−) or stimulated (+) with TGFβ for 30 min. Co-IPs were performed using antisera to the Myc epitope tag (panels 1–4), Smad2 (panel 5), or Smad3 (panel 6); and samples were Western blotted for GFP (panels 1 and 3), Smad2 (panel 2), Smad3 (panel 4), or Myc (panels 5 and 6). Bottom seven panels: Western analysis was performed as above, except the Smad and SNX9 fusion proteins were detected using antisera to GFP or Myc, respectively. (B) Parental AKR-2B cells (AKR) and clones expressing nontargeting or SNX9 shRNA and two SNX9 DN lines were stimulated for 30 min with TGFβ and lysates immunoprecipitated with antisera to Smad4. Following Western transfer, the membrane was probed with antibodies to pSmad3 and Smad4. The bottom three panels depict expression of the indicated proteins in total cell lysate. (C) AKR-2B fibroblasts (AKR) and Smad4 KO (−/−) MEFs were left untreated (−) or stimulated (+) with TGFβ for 30 min before SNX9 immunoprecipitation. Following Western blotting, the membrane was probed for pSmad3 or SNX9. The remaining five panels depict expression of the indicated proteins in total cell lysate.
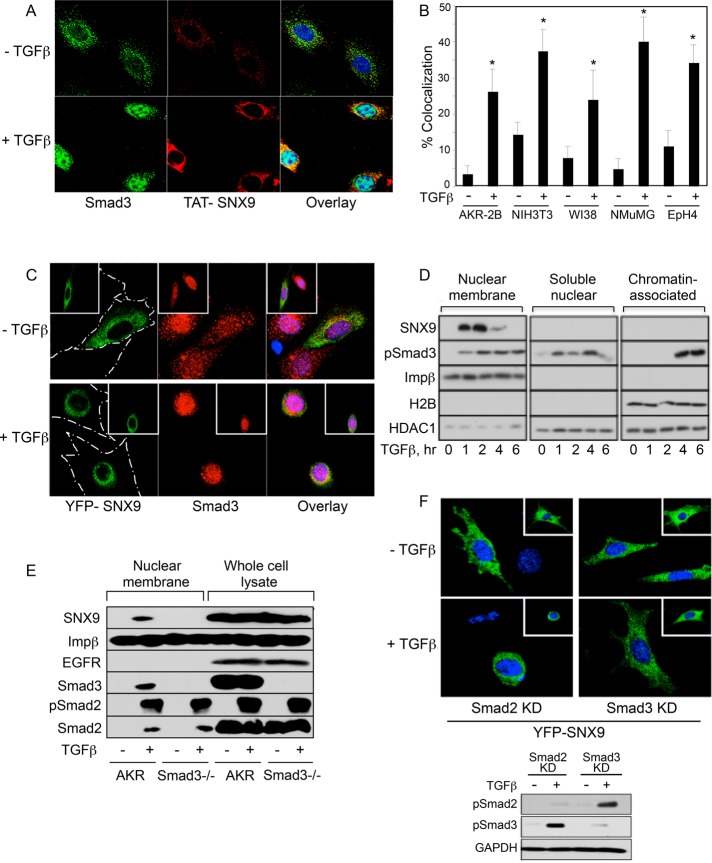
The co-IP and GST pull-down data depicted in Figures 3, B–D, and 4, A and C, show a pSmad3/SNX9 interaction in multiple murine and human cell lines. Because this indicated a novel role for a member of the SNX family, it was further confirmed using immunofluorescence microscopy. As shown in Supplemental Figure S3 and Figure 5, A–C, SNX9 and Smad3 colocalize within a similar perinuclear compartment following treatment with TGFβ. The increased colocalization ranged from 2.6-fold (NIH3T3) to 8.5-fold (NMuMG), with a significant increase observed in all lines (Figure 5B).
FIGURE 5:
TGFβ induces perinuclear SNX9/pSmad3 colocalization and nuclear membrane binding. (A) AKR-2B cells were transduced with HA-TAT-SNX9 protein (0.8 μM) for 90 min. After transduction and treatment with TGFβ for 60 min, TAT-SNX9 and endogenous Smad3 were detected by immunofluorescence analysis. Nuclei were stained with DAPI. (B) AKR-2B, NIH3T3, WI38, NMuMG, and EpH4 cells were treated as above, except stimulation with TGFβ was for 45 min for all lines other than AKR-2B. Colocalization of TAT-SNX9 and Smad3 was determined from 20 cells in each of two experiments ± SD. * denotes significant difference from unstimulated cultures of the same cell type. (C) Following YFP-SNX9 transfection, AKR-2B cells were treated in the absence (−) or presence (+) of TGFβ for 45 min and stained for Smad3 as described in Materials and Methods. Cell outline is shown in the YFP-SNX9 panels, and the inset shows an additional microscopic field. The YFP signal was pseudocolored green to enable observation of any colocalization when overlaid with Smad3. (D) AKR-2B cells were left untreated (0) or stimulated with TGFβ and processed for nuclear membrane, soluble nuclear, or chromatin-associated proteins. (E) Analogous study as in D was performed on quiescent (−) and 30-min TGFβ-treated (+) AKR-2B (AKR) cells and Smad3 null MEFs (Smad3−/−). (F) Smad2 or Smad3 knockdown (KD) AKR-2B cells were transfected with YFP-SNX9 and treated as in C before fixation and confocal microscopy. Inset reflects an additional microscopic field, and the bottom figure documents appropriate loss of pSmad signaling in the knockdowns.
SNX9 requirements for nuclear membrane association
As SNX9 has a role in pSmad3 nuclear import (Supplemental Figure S1E and Figures 2, C and D, and 3A), we investigated 1) whether SNX9 bound and/or traversed the nuclear membrane; and 2) the cis and trans requirements for the SNX9/pSmad3 complex to bind and/or undergo nuclear translocation. While TGFβ stimulated the association of SNX9 and pSmad3 with the nuclear membrane, only pSmad3 entered the nucleus and bound chromatin (Figure 5D). Because SNX9 shows minimal nuclear membrane binding in the absence of TGFβ treatment (Figure 5D), yet is necessary for pSmad3 nuclear trafficking (Supplemental Figure S1E and Figures 2, C and D, and 3A), suggests, first, that Smad3 would be required for SNX9 nuclear membrane binding; and second, in the absence of Smad3, negligible SNX9 would associate with the nuclear membrane following addition of TGFβ. As shown in Figure 5E, although SNX9 associated with the nuclear membrane following TGFβ stimulation, binding also required expression of Smad3. These biochemical findings were further confirmed by transfecting yellow fluorescent protein (YFP)-SNX9 into Smad2 or Smad3 KD cells (Figure 5F; Andrianifahanana et al., 2010). While TGFβ induced a perinuclear distribution for YFP-SNX9 dependent on Smad3, this occurred independent of Smad2 expression, and no evidence for YFP-SNX9 undergoing nuclear import was observed (Figure 5, C and F).
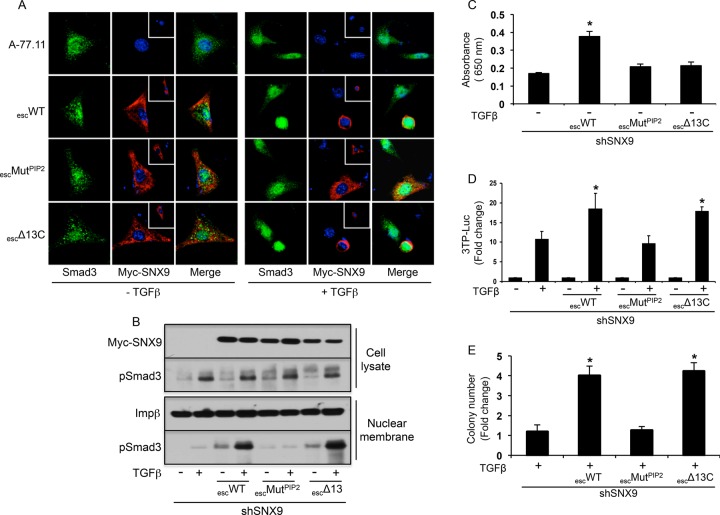
As trafficking functions of the SNXs have been shown to be dependent on both phosphoinositide binding (Yarar et al., 2008) and homodimerization (Childress et al., 2006), we investigated whether analogous SNX9 motifs were required for Smad3 nuclear recruitment and Smad3-regulated responses. SNX9 KD cells were transfected with shRNA-resistant wild type (WT)-SNX9 or escape constructs harboring either point mutations in the PX and BAR domain motifs required for high-avidity PIP2 binding (Yarar et al., 2008) or a truncation of the carboxyl-terminal 13 amino acids required for homodimerization (Childress et al., 2006). While the WT SNX9 escape construct (escWT) and homodimerization mutant (escΔ13C) both colocalized with Smad3 in a perinuclear locale following TGFβ treatment, the PIP2 mutant (escMUTPIP2) showed only diffuse cytoplasmic staining (Figure 6A). These findings provided the first indication that PIP2 binding and homodimerization could be uncoupled from SNX9 action. The interactions with pSmad3 and the nuclear membrane were further established biochemically. As shown in Figure 6B, although pSmad3 nuclear membrane binding is dependent on SNX9, it is independent of SNX9 homodimerization but requires SNX9 binding motifs for PIP2.
FIGURE 6:
Nuclear membrane association of pSmad3/SNX9 is dependent on phosphoinositide binding motifs in SNX9 and independent of SNX9 homodimerization. (A) SNX9 KD A-77.11 cells were transfected with the indicated Myc-tagged SNX9 escape constructs, treated in the absence (−) or presence (+) of TGFβ for 45 min, and stained for Smad3 or Myc-SNX9. Nuclei were stained with DAPI; the inset shows an additional transfected cell. (B) Following A-77.11 cell (shSNX9) transfection and TGFβ treatment as in A, total cell lysate and nuclear membrane preparations were Western blotted for the indicated proteins. (C) Dynamin GTPase assay was performed as described in Materials and Methods on SNX9 KD A-77.7 cells alone or transfected with the escape constructs as in A. Data reflect mean ± SD GTPase activity from three experiments. (D) SNX9 KD A-77.7 cells were transiently transfected with the Smad3-regulated (3TP) luciferase reporter alone or in conjunction with the indicated escape constructs. Cells were either left untreated (−) or stimulated (+) with TGFβ for 24 h, and normalized luciferase activity was determined. Data represent the mean fold induction ± SD relative to untreated from four experiments. (E) SNX9 KD A-77.7 cells were transiently transfected as in A, and fold soft-agar colony induction relative to no TGFβ from four experiments was determined as in Figure 1A. * denotes statistical difference from TGFβ-treated shSNX9 cells.
Figure 6, A and B, indicates that the action of SNX9 in pSmad3 signaling is distinct from its conventional role(s) in regulating intracellular trafficking. This was confirmed by assessing the ability of the SNX9 WT and mutant escape constructs to reconstitute canonical SNX9 action and/or Smad3-dependent signaling (Figure 6, C–E). Although re-expression of WT SNX9 enhanced basal dynamin GTPase activity, neither the PX/BAR nor the dimerization mutant possessed this capability (Figure 6C). This is consistent with previous publications (Childress et al., 2006; Yarar et al., 2008) and reflects established SNX9 function. In contrast, while escWT- and escΔ13C-SNX9 reconstituted Smad3-dependent 3TP-luciferase activity and soft-agar colony formation, this was not observed with the PX/BAR mutant (Figure 6, D and E).
SNX9 is required for Smad3/Imp8 complex formation
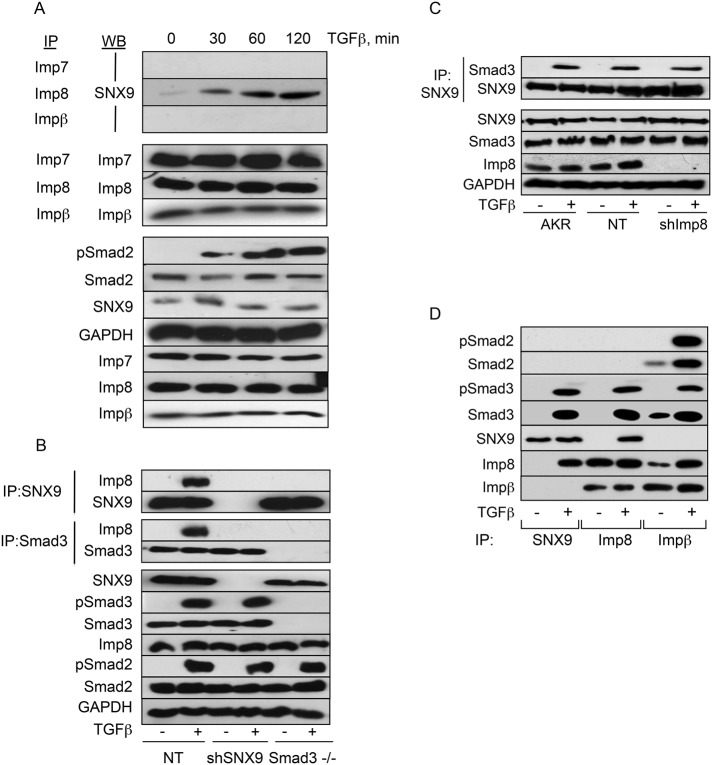
Prior studies have reported roles for the importins (Imp7, Imp8, and Impβ) and/or nucleoporins (Nup153 and Nup214) in pSmad3 nuclear translocation (Xiao et al., 2000; Kurisaki et al., 2001; Xu et al., 2002, 2007; Yao et al., 2008). Because we identified a similar function for SNX9, we evaluated whether these findings might be related. As shown in Figure 7A, ligand-dependent binding of Imp8 and SNX9 was observed with similar kinetics to that observed for the SNX9/Smad3 association (Figure 3B). In contrast, no significant binding of Imp7 or Impβ with SNX9 could be detected (Figure 7A). Owing to difficulties immunoprecipitating endogenous nucleoporins, we transiently transfected hemagglutinin (HA)-tagged Nup153 and Nup214 and examined the ability of these to coimmunoprecipitate with SNX9. No binding between SNX9 and either Nup153 or Nup214 was observed (Supplemental Figure S4, A and B).
FIGURE 7:
SNX9 is required for pSmad3/Imp8 binding, but SNX9/pSmad3 association is independent of Imp8. (A) AKR-2B cells were stimulated with TGFβ and immunoprecipitated (IP) with the indicated importin (Imp) antibodies. Following Western transfer (WB), the membrane was probed for associated SNX9 (first three panels) or the immunoprecipitated protein (panels 4–6). In the bottom seven panels, total cell lysate was immunoblotted for the indicated targets. (B) AKR-2B cells stably expressing nontargeting (NT; A-NT.8) or SNX9 shRNA (shSNX9; A-77.7) and Smad3 null MEFs (Smad3−/−) were left untreated (−) or stimulated with TGFβ for 30 min. Cultures were lysed and immunoprecipitated for SNX9 or Smad3 before Western analysis for bound Imp8, SNX9, or Smad3. Remaining panels show expression of the indicated phosphorylated (p) or total protein in total cell lysate. (C) AKR-2B cells (AKR) or clones stably expressing nontargeting (NT) or Imp8 (shImp8) shRNA were treated as in B. Following SNX9 IP, coprecipitating Smad3 or SNX9 was determined. Remaining panels depict expression of the indicated proteins in total cell lysate. (D) AKR-2B cells were treated as in B. Lysates were immunoprecipitated with SNX9, Imp8, or Impβ antibodies and probed for the indicated targets.
The TGFβ-induced association of SNX9 with Imp8 and pSmad3 indicates that Smad3 nucleocytoplasmic shuttling is even more complex than previously thought. To further address that issue, we investigated the Smad3, SNX9, and/or Imp8 requirement to generate SNX9/Imp8, Smad3/Imp8, or SNX9/Smad3 complexes (Figure 7, B and C). While the absence of Smad3 prevented the association of SNX9 with Imp8 (Figure 7B, compare lanes 2 and 6 in panel 1), and loss of SNX9 significantly diminished the formation of Smad3/Imp8 complexes (Figure 7B, compare lanes 2 and 4 in panel 3), knockdown of Imp8 had minimal impact on SNX9/Smad3 association (Figure 7C, compare lanes 2 or 4 with 6 in panel 1).
Imp8 links pSmad3 to Impβ for nuclear entry.
The requirement for Impβ family members (i.e., Impβ and Imp8) in R-Smad nuclear import is unsettled. For instance, 1) there are reports both indicating (Xiao et al., 2000; Kurisaki et al., 2001) as well as rejecting (Xu et al., 2003, 2007) a need for Impβ; and 2) it was surprising that we found no effect of Imp8 KD on SNX9/pSmad3 binding (Figure 7C), since Imp8 could be coprecipitated with both proteins following TGFβ treatment (Figure 7, A and B). To address these issues, we determined the interrelation of Smad2 and Smad3 with SNX9, Imp8, and/or Impβ (Figure 7D). While SNX9, Smad3, and Imp8 showed the TGFβ-dependent interactions depicted in Figures 3–5 and 7, and previously documented (Strom and Weis, 2001), Imp8/Impβ heteromers were readily observed and Impβ bound both Smad2 and Smad3 with ligand increasing the association. The functional significance of this interaction was addressed in Figure 8, A and B. Although Impβ KD abrogated TGFβ-stimulated as well as basal nuclear import of Smad2 and Smad3, SNX9 or Imp8 KD specifically prevented ligand-induced Smad3 nuclear accumulation, further documenting that basal and stimulated Smad import are distinctly regulated (Figures 3A, 7, and 8, A and B; Xu et al., 2007; Yao et al., 2008).
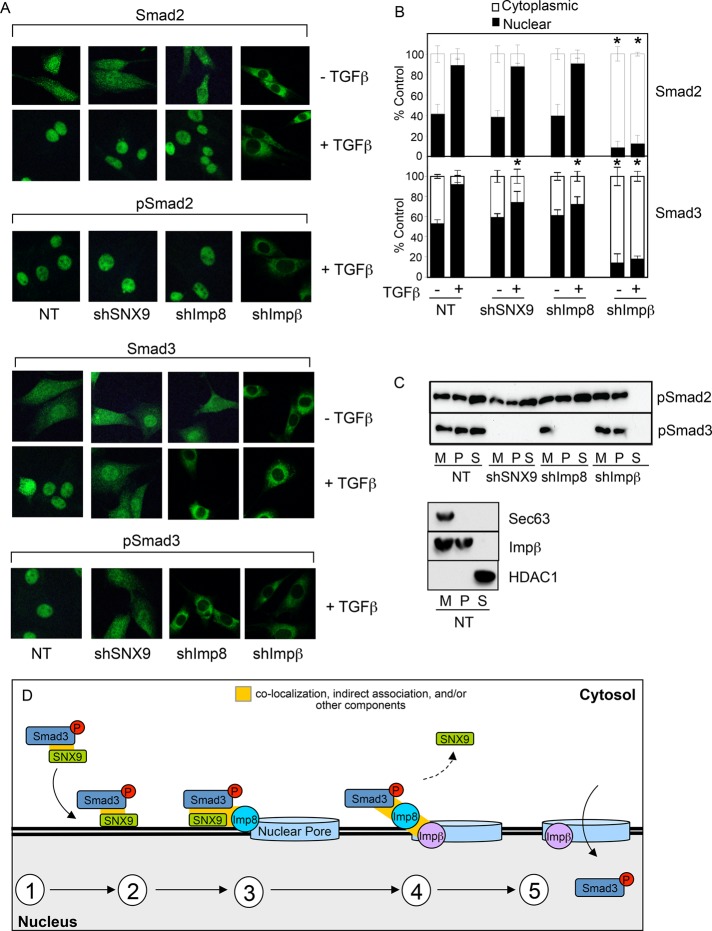
FIGURE 8:
SNX9 and Imp8 differentially regulate pSmad3 association with the nuclear membrane and pore. (A) AKR-2B cells stably expressing nontargeting shRNA (NT; A-NT.8) or shRNA to SNX9 (shSNX9; A-77.7), Imp8 (shImp8; A-89.4), or Impβ (shImpβ; nonclonal population) were either left untreated or stimulated for 30 min with TGFβ and immunostained as indicated. (B) Quantification of nuclear and cytoplasmic Smad2 and Smad3 from 2 experiments ± SD. * denotes significant difference in nuclear R-Smad from similarly treated NT cells. (C) Top, AKR-2B cells described in A were stimulated with TGFβ for 30 min. Nuclear membrane (M), crude nuclear pore (P), and nuclear-soluble (S) fractions were prepared, and pSmad2 and pSmad3 were assessed. Bottom panel, determination of fraction purity from NT control cells. (D) First, Smad3 phosphorylation results in the binding of pSmad3 with SNX9 independent of Smad4. As the binding interactions, stoichiometry, and/or overall composition of the complex are currently unknown, this is indicated by the absence of any direct association depicted between the components. Second, SNX9 mediates pSmad3/SNX9 binding with the nuclear membrane. Third, nuclear membrane–associated Imp8 targets pSmad3 and SNX9 to the nuclear pore. Fourth, SNX9 dissociates from the nuclear membrane, while pSmad3 undergoes Impβ-nuclear translocation (fifth).
The preceding data (Supplemental Figure S1E and Figures 2D, 3A, and 8B) demonstrate a requirement for SNX9, Imp8, and/or Impβ in pSmad3 nuclear entry. It does not address whether each blocks pSmad3 trafficking at defined sites. For investigating this further, KD cells were treated with TGFβ, and the association of pSmad2 or pSmad3 with the nuclear membrane, crude nuclear pore, or soluble nuclear fraction was determined. As expected, pSmad2 was unaffected by SNX9 or Imp8 KD (i.e., found in all fractions) and was only excluded from the soluble nuclear fraction in the absence of Impβ (Figure 8C). This is consistent with pSmad2 activity/trafficking being independent of SNX9 or Imp8 (Figures 1–3, 7D, 8, A and B, and Supplemental Figure S1) yet requiring Impβ for translocation through the nuclear pore (Figures 8, A–C). While a similar block was observed for pSmad3 in Impβ KD cells, loss of SNX9 or Imp8 had distinct phenotypes. Although SNX9 KD prevented pSmad3 association with the nuclear membrane (Supplemental Figure S5 and Figures 6B and 8C), the absence of Imp8 (which had no effect on pSmad3 nuclear membrane binding) abrogated pSmad3 entry into the nuclear pore and subsequent translocation (Supplemental Figure S5 and Figure 8C). A model to help put these findings in context is proposed in Figure 8D.
DISCUSSION
As the majority of TGFβ-regulated R-Smad transcriptional activity and biological action is associated with Smad3 (Feng and Derynck, 2005), it would seem likely that mechanisms exist that distinguish Smad3 from Smad2 nuclear trafficking. The importance of that question is best exemplified for kidney fibrosis and skin cancer, for which it has been documented that, while Smad3 signaling is profibrotic/procarcinogenic, Smad2 is antifibrotic/anticarcinogenic (Hoot et al., 2008; Meng et al., 2010). Thus, rather than designing studies to simply “inhibit TGFβ signaling,” in the current study, we provide evidence that targeting the SNX/pSmad3 interaction could provide a means to directly impact Smad3-dependent pathologies.
R-Smad proteins are known to continuously shuttle between the cytoplasm and nucleus (Inman et al., 2002; Xu et al., 2002; Schmierer and Hill, 2005), however, upon TGFβ stimulation the ratio of cytoplasmic to nuclear R-Smads decreases dramatically. Mechanisms of increased nuclear trafficking (Kurisaki et al., 2001; Chen et al., 2005; Schmierer and Hill, 2007), increased nuclear retention (Inman et al., 2002; Nicolas et al., 2004; Schmierer and Hill, 2005), or a combination thereof could all account for this, and evidence exists supporting each (Schmierer et al., 2008). To further investigate whether Smad phosphorylation impacts the rate of nuclear import, we used a new approach based on the cell-penetrating activity of the HIV TAT peptide (Becker-Hapak et al., 2001; Sawant and Torchilin, 2010; Arif et al., 2014). We reasoned that in the context of a system in which basal (i.e., constitutive) nuclear shuttling of nonphosphorylated Smad3 occurs, immunofluorescence (or Western blotting) is not designed to address whether phosphorylation (i.e., inducible) affects the kinetics of nuclear import, as it is difficult to determine time 0 (i.e., “start”) for a pool of nonphosphorylated Smad3 in a constitutive system. Furthermore, even though adding ligand would give a time 0 for phosphorylated Smad3, it routinely takes ∼20–30 min for sufficient pSmad3 to be detected. In contrast, TAT peptides allow one to quantitatively assess similarly synchronized and normalized pools of phosphorylated and nonphosphorylated Smad3. As shown in Figure 3A, when normalized for initial uptake, phosphorylated TAT-Smad3 (i.e., analogous to ligand stimulated) entered the nucleus ∼3.5 times faster than nonphosphorylated TAT-Smad3 (i.e., analogous to basal shuttling). Moreover, in agreement with a number of studies (Hill, 2009), 1) both phosphorylated and nonphosphorylated TAT fusions underwent nuclear translocation; and 2) increased nuclear retention of phosphorylated TAT-Smad3 was observed. Thus, while TAT-Smad3 peptides provide a new methodology to further address some of these unresolved issues, they independently confirm canonical findings regulating R-Smad action. Our determination that SNX9 functions to both regulate and provide specificity for this process should hopefully help clarify this issue.
The SNXs represent a family of proteins with diverse functions (Verges, 2007; Cullen, 2008; Lundmark and Carlsson, 2009). Because earlier work documented an association of SNXs 2, 4, and 6 with type I and type II TGFβ receptors (Parks et al., 2001), and SNX9 is known to function at both the plasma membrane and the early endosome (Lundmark and Carlsson, 2009), studies were initiated to determine whether these findings were related. Contrary to our expectations, while loss of SNX9 had only minimal effect on either R-Smad phosphorylation (Supplemental Figure S1D and Figure 2, A and B) or TGFβ receptor internalization (unpublished observations), Smad3-dependent signaling (Supplemental Figure S1C and Figure 1, C and D) as well as nuclear membrane binding (Supplemental Figure S5, A and B, and Figures 6B and 8C) was significantly diminished. Of particular note, Smad3 signaling, SNX9 attaining a perinuclear locale, and pSmad3 nuclear association occurred independent of SNX9 homodimerization but required the ability to bind phosphoinositides (Figure 6). In that SNX9 homodimerization (in conjunction with phosphoinositide binding) has been reported to be essential for correct plasma membrane/early endosome targeting, dynamin binding, and cargo endocytosis (Soulet et al., 2005; Childress et al., 2006; Yarar et al., 2008; Lundmark and Carlsson, 2009), these results separated SNX9’s canonical functions from its role in pSmad3 signaling.
SNX9 was found to preferentially bind and/or colocalize with phosphorylated Smad3 in vivo by co-IP or transfection of epitope-tagged SNX9 and in vitro using GST pull down in a number of cell types (Figures 3, B–D, 4, A and C, 5, A–C and F, 6A, and 7, C and D, and Supplemental Figure S3). When direct association, however, was assessed using in vitro–generated SNX9 and phosphorylated GST-Smad3, no interaction could be detected (unpublished observations). While this could simply reflect the significant technical issues associated with such an in vitro approach, it might also indicate the requirement for a higher-order complex. Although these two possibilities are not mutually exclusive, and we are examining that issue, in this initial study, we investigated two relevant possibilities. First, would SNX9/pSmad3 binding only occur in the context of a heteromeric R-Smad interaction (i.e., also require Smad2 and/or Smad4); and second, might this reflect a need for a previously identified importin or nucleoporin implicated in R-Smad nuclear import?
To address the first question, we performed studies in Smad2, Smad3, and Smad4 null mouse embryo fibroblasts (MEFs; Figure 4). Although an obligate requirement for Smad3 was observed, neither the presence nor absence of Smad2 or Smad4 had any detectable impact on the coprecipitation of SNX9 and Smad3 following TGFβ treatment. While this supports the hypothesis that SNX9 can regulate homomeric pSmad3 complexes, heteromeric complexes containing pSmad3 and Smad4 are just as likely to be regulated by SNX9, as the heterotrimer (consisting of two pSmad3s and a single Smad4 molecule) represents the most energetically favorable structure (Chacko et al., 2004).
Investigating the second possibility, however, is somewhat more problematic, as evidence supports a role(s) for Imp7, Imp8, Impβ, Nup153, and/or Nup214 in R-Smad nuclear entry (Xu et al., 2002; Moustakas and Heldin, 2008; Yao et al., 2008; Hill, 2009). As such, we determined whether any of the aforementioned importins and nucleoporins could be coprecipitated with SNX9 following addition of TGFβ. Such an interaction was observed only with Imp8 (Supplemental Figure S4, A and B, Figure 7, A, B, and D), supporting previous work suggesting that Imp8 is required for nuclear import of Smad3 in stimulated cells (Xu et al., 2007). Of particular note was that, in the absence of SNX9, we were not only unable to co-IP Imp8 with pSmad3 (Figure 7B), but pSmad3 was not associated with the nuclear membrane (Supplemental Figure S5 and Figures 6B and 8C). This is in contrast to studies using Imp8 KD cells, in which no detectable impact on SNX9/pSmad3 binding (Figure 7C) or association with the nuclear membrane was observed (Supplemental Figure S5 and Figure 8C). These findings suggest that SNX9 functions to promote the interaction of pSmad3 with Imp8 by recruiting it to the nuclear membrane. However, although Imp8 can associate with SNX9 and Impβ (Figure 7, A, B, and D), SNX9 is never found with Impβ (Figure 7, A and D), indicating that the formation of pSmad3/SNX9 and pSmad3/Impβ complexes may be mutually exclusive. Collectively, these data suggest nuclear delivery of pSmad3 occurs sequentially whereby, first, SNX9 is necessary for pSmad3 nuclear membrane binding and presentation to Imp8; second, Imp8 promotes pSmad3 complexing with the nuclear pore machinery; and third, Impβ is required for pSmad3 translocation through the nuclear pore (Figure 8D).
In summary, we provide evidence that SNX9 is involved in Smad3 (but not Smad2) signaling and acts as an adaptor for the nuclear translocation of pSmad3. As the majority of TGFβ-regulated transcriptional activity and biological action is associated with Smad3, the ability to specifically impact Smad3-dependent phenotypes would provide a previously unavailable degree of specificity to modulate and/or treat various TGFβ-dependent disorders.
MATERIALS AND METHODS
Cell lines and constructs
The following cell lines were used: murine AKR-2B (mesenchymal), NIH3T3 (mesenchymal), NMuMG (mammary epithelial), and EpH4 (mammary epithelial), and human WI38 lung fibroblasts. Cultures were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and stimulated with 5 ng/ml TGFβ unless stated otherwise.
For assessing the effect of DN-SNX9 expression on TGFβ responses, A105 cells (an AKR-2B subclone expressing endogenous and chimeric TGFβ receptors consisting of the ligand-binding domain of the GM-CSF alpha or beta receptors fused to the transmembrane and cytoplasmic domain of TβRI and TβRII, respectively, generated to investigate aspects of TGFβ receptor trafficking; Anders et al., 1997, 1998) were stably cotransfected with pBabe-Puro and the indicated SNX9 plasmids. Clones were selected using 1.2 μg/ml puromycin. For SNX9 knockdown, shRNA was generated using the lentiviral system from Sigma’s MISSION library, purchased from the Mayo Clinic Jacksonville RNA Interference Technology Resource. Briefly, plasmid constructs were transfected into 293FT cells and packaged using ViraPower reagents (Invitrogen, Carlsbad, CA) as described by the manufacturer. Following a 72-h incubation, viruses were harvested and added to AKR-2B plated at 2.5 × 105 cells/six-well plate. The medium was changed the following day, and 48 h after infection, clones were selected in 1.2 μg/ml puromycin. A nontargeting (NT) shRNA was used as control. AKR-2B knockdown of Imp8 and Impβ was performed in a similar manner. As two distinct shRNAs were used (#77 and #78) and multiple clones selected, the designation A.NT.8 and A.77.11, for example, refers to AKR-2B (A) clone 8 or 11 expressing NT or the #77 SNX9 shRNA, respectively. When the pooled result from multiple clones is assessed (Figures 1 and 2), the #78 shRNA is referred to as Alt (alternative)-shRNA to differentiate the shRNAs.
shRNA sequences included NT: CGAAAGTAGGTACATCCCTTA; SNX9 77 clones: CCTGACTTGGATTTGATAGAA; SNX9 78 clones: CCTACTGACTACGTGGAAATT; Imp8 89 clones: GCACATTGTTAGAGAGACAAT; Impβ pool 5: GCGCTGTTAGACATGAGCTAA.
SNX9 rescue constructs were generated using QuickChange II XL (Agilent Technologies, Santa Clara, CA) by mutating nucleotides 1164, 1167, and 1170 from T, C, G to G, T, A, respectively, while phosphoinositide binding and homodimerization mutants were prepared essentially as described (Childress et al., 2006; Yarar et al., 2008). Transfections used Transit 2020 (Mirus Bio, Madison, WI) and 2.5 μg DNA. Primers are provided in Supplemental Table S1.
Growth/migration studies
Soft-agar assays were performed as previously described (Rahimi et al., 2009). Briefly, 4 × 104 cells were seeded in a six-well plate in the presence or absence of 10 ng/ml TGFβ (R&D Systems, Minneapolis, MN). Following 10-d growth at 37°C, the number of colonies >100 μm in diameter were counted using an Optronix Gelcount (Oxford Optronics, Milton, Abingdon, UK). For scratch assays, cultures in 10% FBS/DMEM were seeded into six-well plates (5 × 105 cells/well) and incubated overnight at 37°C. Following monolayer disruption using a sterile 200 μl pipette tip, the medium was changed to 0.5% FBS/DMEM, and cultures were incubated in the presence or absence of TGFβ for 24 h. Images (×100) were taken at 0 and 24 h.
Luciferase reporter assays
Cultures were transfected with 2 μg of the indicated luciferase constructs together with 0.5 μg CMV-β-galactosidase using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). For the ARE transfections, 2 μg of FAST1 was also cotransfected. The cells were serum starved in 0.1% FBS/DMEM overnight and either left untreated or stimulated with TGFβ for 24 h. Normalized luciferase activity was determined as described (Anders and Leof, 1996; Rahimi and Leof, 2007; Ross and Hill, 2008).
Immunofluorescence microscopy
Cells (2 × 104) were plated onto coverslips in 10% FBS/DMEM and incubated overnight at 37°C. Cultures were placed in 0.5% FBS/DMEM alone or containing TGFβ for 45 min, rinsed twice with cold phosphate-buffered saline (PBS), and fixed for 30 min in 4% formaldehyde at room temperature. Subsequent to 0.2% Triton X-100 permeabilization (3 min at room temperature) and washing (0.2% bovine serum albumin [BSA] in PBS, wash buffer), cultures were blocked in wash buffer containing 10% donkey serum for 1 h at room temperature before the addition of primary antibody to Smad2 or Smad3 in 0.2% BSA/PBS, 10% normal donkey serum for 1 h at room temperature. Antibody information is listed in Supplemental Table S2. The coverslips were rinsed three times in wash buffer before room temperature incubation (10 min) in 50 mM NH4Cl to quench background fluorescence. Following an additional wash buffer rinse, samples were placed in either anti-AF488 (green) or AF-594 (red) secondary antibody (Invitrogen) to detect R-Smads or TAT-SNX9, respectively, for 1 h at room temperature. Coverslips were washed three times (10 min each) with wash buffer and once in PBS before addition of mounting media containing 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Fluorescence images were collected on an LSM510 confocal microscope using MetaMorph software for image analysis and quantitation (Molecular Devices, Sunnyvale, CA).
YFP-SNX9 was detected as described for Smad2/3 with the following minor modifications. Subsequent to plating 5 × 105 cells/well (six-well plate) and 12 h of incubation at 37°C, cells were transfected with YFP-SNX9 (1.5 μg/well) and cultured for an additional 12 h before trypsination and seeding (2 × 104 cells) onto glass coverslips. Following 12 h of growth in 10% FBS/DMEM, the medium was removed and replaced with 0.5% FBS/DMEM ± TGFβ (5 ng/ml) for 45 min. Fixation, permeabilization, and visualization was as above.
Quantitative reverse transcription (RT)-PCR analysis
Following TGFβ stimulation, total RNA was isolated using Trizol reagent (Invitrogen), and 2 μg was reverse transcribed (SuperScript III Reverse Transcriptase system; Invitrogen, CA). Samples were diluted 1:5 with water and one-fiftieth was used as template for qPCR with platinum SYBR green qPCR superMix-UDG (Invitrogen). Primers are presented in Supplemental Table S1; sample induction was normalized to GAPDH.
Western blotting, co-IP, and GST pull down
Cells were lysed for 30 min on ice in RIPA buffer (50 mM Tris [pH 7.4], 1% Triton X-100, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA [pH 8] and 10 mM NaF) containing 50 μg/ml phenylmethylsulfonyl fluoride (PMSF), 100 μM sodium vanadate, and 1 μg/ml leupeptin. Insoluble materials were pelleted by centrifugation at 16,060 × g for 10 min, and 20–80 μg of protein was separated by SDS–PAGE. Antibodies are provided in Supplemental Table S2. Protein markers for fractionation studies included Impβ (nuclear membrane marker), histone 2B (chromatin-associated marker), histone deacetylase 1 (soluble nuclear marker), and epidermal growth factor receptor (plasma membrane marker).
For co-IP, cells were lysed in kinase lysis buffer (50 mM Tris [pH 7.4], 0.1% Triton X-100, 250 mM NaCl, 5 mM EDTA [pH 8] and 50 mM NaF) containing 50 μg/ml PMSF, 100 μM sodium vanadate, and 1 μg/ml leupeptin on ice for 30 min. Protein (500–800 μg) was precleared for 1 h with either protein A or G agarose beads (Millipore, Charlottesville, VA) and incubated overnight (O/N) at 4°C with the indicated antibody. For prevention of masking of desired proteins by the immunoglobulin light chain, immunoprecipitating antibodies were cross-linked (before incubation) with BS3 (Thermo Fisher Scientific, Rockford, IL) per the manufacturer’s instructions, using a 15-fold molar excess of cross-linker (final cross-linker concentration of 0.25 mM). Immune complexes were collected by addition of protein A or G agarose beads (50 μl) and incubation for 2 h at 4°C. Following three washes in kinase lysis buffer, coimmunoprecipitated proteins were eluted in 2× Laemmli buffer and analyzed by Western blotting. For the detection of pSmad3 following immunoprecipitation, blots were probed with pSmad3-specific antibodies (Cell Signaling Technology, Danvers, MA) and incubated in protein A–horseradish peroxidase conjugate (GE Healthcare, Pittsburgh, PA) (1:500 dilution), before processing for chemiluminescence detection.
GST fusion proteins were purified using the BugBuster GST Bind Purification Kit following the manufacturer’s instructions (Novagen, EMD4 Biosciences, Darmstadt, Germany). For assessment of SNX9 binding to GST constructs, Cos7 cells were transfected with Flag-TβRII (3 μg) and HA-TβRI (1 μg) using Lipofectamine 2000 (Invitrogen). Following O/N incubation, cultures were treated for 30 min with TGFβ before addition of kinase lysis buffer and HA immunoprecipitation (O/N at 4°C). TβRI was purified per the manufacturer’s recommendation using Catch and Release version 2.0 (Upstate Biotechnology, Lake Placid, NY) and incubated in 50 μl kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, 1 mM dithiothreitol [DTT]) containing 5 μM ATP, 5 μCi of [γ-32P]ATP per μl, and 2 μg substrate (GST-Smad2 or -Smad3). The kinase assay was allowed to proceed for 10 min at 37°C before incubation with AKR-2B lysate for 1 h at room temperature. Following the addition of glutathione-agarose beads (1 h at 4°C, rocking) and three washes with lysis buffer, bound SNX9 was assessed by immunoblotting. For assessment of GST-SNX9 binding of endogenous pSmad3, 500 μg of cell lysate was incubated with 10 μg of GST-SNX9 (O/N at 4°C). Pull down and Western blotting was as described above.
Isolation of nuclear and chromatin-bound fractions
Nuclear membranes were prepared from NE-PER Nuclear and Cytoplasmic Extraction–purified nuclei (Thermo Scientific, Rockford, IL), followed by membrane extraction using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific) with the addition of 50 μg/ml PMSF, 100 μM sodium vanadate, 0.1 TIU/ml aprotinin, and 1 μg/ml leupeptin to the lysis buffers. Following removal of the cytoplasmic extract, the nuclear pellet was washed three times in PBS containing 50 μg/ml PMSF, 100 μM sodium vanadate, 0.1 TIU/ml aprotinin, and 1 μg/ml leupeptin before nuclear lysis and Western blot analysis. An alternative nuclear membrane fraction and a crude nuclear pore complex were generated as described by Aaronson and Blobel (1975).
Nuclear soluble and chromatin-bound fractions were prepared using the Subcellular Protein Fractionation Kit (Thermo Scientific). Cell samples normalized by number were incubated on ice for 10 min with gentle mixing in supplied cytoplasmic extraction buffer (CEB). Following centrifugation at 500 × g for 5 min, the samples were suspended in ice-cold membrane extraction buffer (MEB; supplied) before centrifugation (1000 × g, 2 min) and incubation (10 min) of the pellet with gentle mixing in supplied CEB. Nuclei were pelleted at 3000 × g (5 min), and a soluble nuclear fraction was obtained by 30-min incubation in nuclear extraction buffer (NEB; supplied). The chromatin-associated fraction was acquired by suspending the remaining pellet in NEB containing CaCl2 and micrococcal nuclease (5 min, 37°C) and collecting the supernatant following a 16,000 × g spin for 5 min.
Dynamin GTPase assay
SNX9 KD cells were seeded at 2 × 105 cells per six-well culture dish and incubated at 37°C for 12 h before transient transfection with 2.5 μg of the indicated SNX9 escape construct. Following a 24-h incubation, cultures were lysed in RIPA buffer for 30 min on ice, and SNX9 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was added to 500 μg cell protein before O/N incubation at 4°C. Immune complexes were precipitated by addition of agarose G beads for 1 h at 4°C and washed three times with cold PBS, and bound proteins were eluted with 50 mM glycine (pH 2.7). Dynamin GTPase activity was performed as described by Leonard et al. (2005). Briefly, 4 μl of eluted protein was incubated in 192 μl of GTPase assay buffer (20 mM HEPES-KOH [pH 7.5], 150 mM KCl, 2 mM MgCl2, 1 mM DTT) to which 4 μl GTP stock solution (100 mM GTP, 20 mM HEPES [pH 7.4]) was added. Following 10 min at room temperature, 100 μl was transferred to a 96-well microtiter plate containing 5 μl 0.5 M EDTA (pH 8.0) to stop the reaction. An equal volume (100 μl) of Malachite Green stock solution (1 mM Malachite Green, 10 mM ammonium molybdate in 1 N HCl) was added to each well, and GTPase activity (i.e., Malachite Green detection of free phosphate) was measured by absorbance at 650 nm using a microplate reader.
Generation of TAT fusion proteins
TAT-HA-SNX9 and TAT-HA-Smad3 proteins were prepared in BL21(DE3)pLysS Escherichia coli (OD600 of 0.4) following addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mM. After a 4-h TAT protein induction at 37°C, cells were harvested, washed, and suspended in 15 ml 50 mM sodium phosphate buffer (pH 7.4) containing 300 mM NaCl and 20 mM imidazole. The cell suspension was sonicated, and the lysate was cleared by 12,000 × g centrifugation at 4°C for 20 min. The supernatant was then poured into a TALON Metal Affinity Resin column (Clontech, Mountain View, CA), washed with 50 mM sodium phosphate buffer containing 40 mM imidazole, and eluted with 150 mM imidazole.
TAT-Smad3 nuclear entry/retention assays
Purified TAT-Smad3 was labeled with 125I using the Bolton Hunter reaction. After the samples were split, 50 μg was incubated in kinase buffer containing 5 μM ATP, 20 μCi [γ-32P]ATP per microliter and activated TβR1-HA (purified by Catch and Release, as described above in the Western blotting, Co-IP, and GST pull down section, from TβRI- and TβRII-transfected Cos7 cells) to generate 32P-TAT-Smad3 phosphorylated in the COOH terminal SSXS domain (S423/S425). AKR-2B cells were grown to confluence in 24-well plates and transduced with 0.8 μM TAT-Smad3 or TAT-pSmad3 for the indicated times. Plates were washed twice with binding buffer (0.2M HEPES, 2.5% BSA in DMEM [pH7.4]) containing 75% horse serum and twice with PBS before normalization for cell number. A third of the normalized sample was used for determination of total intracellular TAT protein by cell lysis (0.2 M NaOH, 40 mg/ml salmon sperm DNA), while the remaining sample underwent nuclear fractionation and protein extraction as per NE-PER nuclear extraction recommendations (Pierce, Rockford, IL). Nuclear counts were normalized to total cell counts (i.e., to account for the time delay in TAT-protein transduction; Supplemental Figure S2A), and maximal counts in NT cells were defined as 100%. Raw and normalized 125I and 32P TAT-Smad3 cell transduction counts are provided in Supplemental Figure S2.
Statistical analysis
All p values were obtained through Student’s t test (two-tailed) comparisons.
Supplementary Material
Acknowledgments
We thank Sven Carlsson, Ralf Janknecht, Katie Ullman, and André Hoelz for the indicated SNX9, Smad, Nup153-HA, and Nup214-HA constructs, respectively. This work was supported by Public Health Service grants GM-55816 and GM-54200 from the National Institute of General Medical Sciences, the Caerus Foundation, the Scleroderma Foundation, and the Mayo Foundation (to E.B.L.).
Glossary
Abbreviations used:
- AIG
anchorage-independent growth
- ARE
Activin response element
- BAR
Bin/Amphiphysin/Rvs
- BSA
bovine serum albumin
- CEB
cytoplasmic extraction buffer
- co-IP
coimmunoprecipitation
- Co-Smad
common-mediator Smad
- DAPI
4′,6-diamidino-2-phenylindole
- DN
dominant negative
- DTT
dithiothreitol
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- HA
hemagglutinin
- Imp
importin
- IP
immunoprecipitation
- I-Smads
inhibitory Smads
- KD
knockdown
- MEB
membrane extraction buffer
- NEB
nuclear extraction buffer
- NT
nontargeting
- Nup
nucleoporin
- O/N
overnight
- PBS
phosphate-buffered saline
- PMSF
phenylmethylsulfonyl fluoride
- PX
phox homology
- R-Smads
receptor-regulated Smads
- shRNA
short hairpin RNA
- SBE
Smad-binding element
- SNX
sorting nexin
- TβRI
type I transforming growth factor β receptor
- TβRII
type II transforming growth factor β receptor
- TGFβ
transforming growth factor β
- WT
wild type
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0545) on September 2, 2015.
REFERENCES
- Aaronson RP, Blobel G. Isolation of nuclear pore complexes in association with a lamina. Proc Natl Acad Sci USA. 1975;72:1007–1011. doi: 10.1073/pnas.72.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders RA, Arline SL, Doré JJE, Leof EB. Distinct endocytic responses of heteromeric and homomeric transforming growth factor β receptors. Mol Biol Cell. 1997;8:2133–2143. doi: 10.1091/mbc.8.11.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders RA, Doré JEJ, Arline SA, Garamszegi N, Leof EB. Differential requirements for type I and type II TGFβ receptor kinase activity in ligand-mediated receptor endocytosis. J Biol Chem. 1998;273:23118–23125. doi: 10.1074/jbc.273.36.23118. [DOI] [PubMed] [Google Scholar]
- Anders RA, Leof EB. Chimeric granulocyte/macrophage colony-stimulating factor/transforming growth factor-β (TGF-β) receptors define a model system for investigating the role of homomeric and heteromeric receptors in TGF-β signaling. J Biol Chem. 1996;271:21758–21766. doi: 10.1074/jbc.271.36.21758. [DOI] [PubMed] [Google Scholar]
- Andrianifahanana M, Wilkes MC, Repellin CE, Edens M, Kottom TJ, Rahimi RA, Leof EB. ERBB receptor activation is required for profibrotic responses to transforming growth factor beta. Cancer Res. 2010;70:7421–7430. doi: 10.1158/0008-5472.CAN-10-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif E, Rathore YS, Kumari B, Ashish F, Wong HN, Holzman LB, Nihalani D. Slit diaphragm protein Neph1 and its signaling: a novel therapeutic target for protection of podocytes against glomerular injury. J Biol Chem. 2014;289:9502–9518. doi: 10.1074/jbc.M113.505743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badour K, McGavin MK, Zhang J, Freeman S, Vieira C, Filipp D, Julius M, Mills GB, Siminovitch KA. Interaction of the Wiskott-Aldrich syndrome protein with sorting nexin 9 is required for CD28 endocytosis and cosignaling in T cells. Proc Natl Acad Sci USA. 2007;104:1593–1598. doi: 10.1073/pnas.0610543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- Baumann C, Lindholm CK, Rimoldi D, Levy F. The E3 ubiquitin ligase Itch regulates sorting nexin 9 through an unconventional substrate recognition domain. FEBS J. 2010;277:2803–2814. doi: 10.1111/j.1742-4658.2010.07698.x. [DOI] [PubMed] [Google Scholar]
- Becker-Hapak M, McAllister SS, Dowdy SF. TAT-mediated protein transduction into mammalian cells. Methods. 2001;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- Carlton J, Bujny M, Rutherford A, Cullen P. Sorting nexins—unifying trends and new perspectives. Traffic. 2005;6:75–82. doi: 10.1111/j.1600-0854.2005.00260.x. [DOI] [PubMed] [Google Scholar]
- Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K. Structural basis of heteromeric Smad protein assembly in TGF-beta signaling. Mol Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Chen HB, Rud JG, Lin K, Xu L. Nuclear targeting of transforming growth factor-beta-activated Smad complexes. J Biol Chem. 2005;280:21329–21336. doi: 10.1074/jbc.M500362200. [DOI] [PubMed] [Google Scholar]
- Childress C, Lin Q, Yang W. Dimerization is required for SH3PX1 tyrosine phosphorylation in response to epidermal growth factor signalling and interaction with ACK2. Biochem J. 2006;394:693–698. doi: 10.1042/BJ20050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9:574–582. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo GM, Leroy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-β receptor signaling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Elliott RL, Blobe GC. Role of transforming growth factor beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Hayes S, Chawla A, Corvera S. TGFβ receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol. 2002;158:1239–1249. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 2009;19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- Hoot KE, Lighthall J, Han G, Lu SL, Li A, Ju W, Kulesz-Martin M, Bottinger E, Wang XJ. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118:2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Jin Q, Gao G, Mulder KM. Requirement of a dynein light chain in TGFβ/Smad3 signaling. J Cell Physiol. 2009;221:707–715. doi: 10.1002/jcp.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki A, Kose S, Yoneda Y, Heldin CH, Moustakas A. Transforming growth factor-beta induces nuclear import of Smad3 in an importin-beta1 and Ran-dependent manner. Mol Biol Cell. 2001;12:1079–1091. doi: 10.1091/mbc.12.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Leonard M, Song BD, Ramachandran R, Schmid SL. Robust colorimetric assays for dynamin’s basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR. SNX9—a prelude to vesicle release. J Cell Sci. 2009;122:5–11. doi: 10.1242/jcs.037135. [DOI] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Boden M, Kobe B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta. 2011;1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Meng XM, Huang XR, Chung AC, Qin W, Shao X, Igarashi P, Ju W, Bottinger EP, Lan HY. Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21:1477–1487. doi: 10.1681/ASN.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Dynamic control of TGF-beta signaling and its links to the cytoskeleton. FEBS Lett. 2008;582:2051–2065. doi: 10.1016/j.febslet.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Nicolas FJ, De Bosscher K, Schmierer B, Hill CS. Analysis of Smad nucleocytoplasmic shuttling in living cells. J Cell Sci. 2004;117:4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- Parks WT, Frank DB, Huff C, Renfrew Haft C, Martin J, Meng X, de Caestecker MP, McNally JG, Reddi A, Taylor SI, et al. Sorting nexin 6, a novel SNX, interacts with the transforming growth factor-beta family of receptor serine-threonine kinases. J Biol Chem. 2001;276:19332–19339. doi: 10.1074/jbc.M100606200. [DOI] [PubMed] [Google Scholar]
- Penheiter SG, Mitchell H, Garamszegi N, Edens M, Doré JJE, Jr, Leof EB. Internalization-dependent and -independent requirements for transforming growth factor β receptor signaling via the Smad pathway. Mol Cell Biol. 2002;22:4750–4759. doi: 10.1128/MCB.22.13.4750-4759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi R, Andrianifahanana M, Wilkes MC, Edens ME, Kottom TJ, Blenis J, Leof EB. Distinct roles for mammalian target of rapamycin complexes in the fibroblast response to transforming growth factor-β. Cancer Res. 2009;69:84–93. doi: 10.1158/0008-5472.CAN-08-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Wakefield LM. The two faces of transforming growth factor β in carcinogenesis. Proc Natl Acad Sci USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Sawant R, Torchilin V. Intracellular transduction using cell-penetrating peptides. Mol Biosyst. 2010;6:628–640. doi: 10.1039/b916297f. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor beta-dependent nuclear accumulation of Smads. Mol Cell Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Tournier AL, Bates PA, Hill CS. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci USA. 2008;105:6608–6613. doi: 10.1073/pnas.0710134105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:2058–2067. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges M. Retromer and sorting nexins in development. Front Biosci. 2007;12:3825–3851. doi: 10.2741/2355. [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, et al. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell. 2001;8:1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of Smad 3. J Biol Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- Xu L, Alarcon C, Col S, Massague J. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem. 2003;278:42569–42577. doi: 10.1074/jbc.M307601200. [DOI] [PubMed] [Google Scholar]
- Xu L, Kang Y, Col S, Massague J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- Xu L, Yao X, Chen X, Lu P, Zhang B, Ip YT. Msk is required for nuclear import of TGF-b/BMP-activated Smads. J Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Chen X, Cottonham C, Xu L. Preferential utilization of Imp7/8 in nuclear import of Smads. J Biol Chem. 2008;283:22867–22874. doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.