Abstract
Aortic aneurysm is a cardiovascular condition with a serious risk of mortality and the dismal prognosis of any type of major cardiovascular disease. The present study found that tissues from aortic aneurysm patients and hypoxic aortic smooth muscle cells showed aberrant high expression of the cyclic RNA hsa-circ-000595, as demonstrated by polymerase chain reaction array screening. Knockdown of hsa-circ-000595 resulted in a decreased apoptotic rate of human aortic smooth muscle cells. Furthermore, it was determined that miR-19a is a target of hsa-circ-000595. The results of the present study laid an epigenetic foundation for exploring the underlying mechanisms of the development of aortic aneurysm.
Keywords: hsa-circ-000595, aortic aneurysm, microRNA-19a, hypoxia
Introduction
Aortic aneurysm is a leading cause of cardiovascular mortality and has the dismal prognosis of any major type of cardiovascular condition (1–3). Aortic aneurysm, determined by a pathological aortic expansion by >50% of the normal vascular diameter, is divided into true aortic aneurysm and aortic pseudo-aneurysm (4). Although considerable progress has been made in improving cardiovascular survival rates over the past few decades, the five-year survival rate has remained <35% (5). The poor prognosis of cardiovascular conditions is in part due to the emergence of drug resistance (6). Consequently, there is an urgent requirement to explore the underlying molecular mechanisms of aortic aneurysm.
As is commonly known, mRNA is a type of single-stranded RNA, which carries genetic information that guides protein expression. Gene transcription occurs via the expression of mRNA molecules in each cell (7). However, abnormal RNA splicing (trans-splicing) may occur, leading to the covalent attachment of a single RNA molecule to its other end and resulting in the formation of cyclic RNA (circRNA); this is usually a rare biochemical reaction. Initially, circRNAs were identified in plants, in which they are capable of encoding sub-viral factors (8). In unicellular organisms, circRNAs occur mostly due to the self-splicing intron of ribosomal RNA, but may also be produced by evolutionarily conserved protein-coding genes (9,10). In animal cells, circRNAs are formed in the splicing process by joining the 5′ and 3′ end of the downstream exon (10,11). By far the most well-known circRNA is that formed by anti-sense transcription of the mRNA of the sex determining region Y gene, which was shown to be highly expressed in the testes (12). Analysis of gene expression in archaea and mammals showed that circRNAs are more abundant than expected; however, the biological functions of circRNAs in animals and plants have remained elusive. CircRNAs have been frequently found by RNA seqencing, and it has been attempted to identify all circRNAs present in animal cell systems. It has been revealed that certain circRNAs act as molecular 'sponges' by combining and encapsulating microRNAs (miRNAs), thereby performing their regulatory effect on gene expression (13,14). In addition, it was speculated that circRNAs have numerous other functions, pointing out that the functions of RNA go beyond that of a messenger between DNA and the encoded proteins (15). CircRNAs have not been discovered at any earlier stages as traditional methods to isolate RNA from cells discarded of these cyclic molecules. It has been wrongly assumed that only linear RNAs are the products of gene transcription; however, circRNA are the predominant transcript form of hundreds of genes (16). CircRNAs have also been widely ignored by classical RNA sequencing studies due to their lack of a tail end (17). CircRNAs are more abundant than expected and may be more important than previously thought; however, their expression and function in numerous conditions, including aortic aneurysm, have remained elusive.
The circRNA labeled hsa-circ-000595 was first identified by Memczak et al (18). It is located on chromosome 14, and the gene coding starts from position 93753822 and ends on position 93762503 of the chromosome. In addition, it has been demonstrated that hsa-circ-000595 has a connection with miR-19a and miR-19b (18). miR-19a is a member of the miR-17-92 cluster, which is located on chromosome 13q31.3 and is involved in the pathogenesis of numerous types of tumor. It has previously been reproted that miR-19a is upregulated in various types of tumors (19). The present study investigated the role of circRNAs in aortic aneurysm using an epigenetic PCR array. As hypoxia is implicated in aortic aneurysm (20), the effect of CoCl2-induced hypoxia on the expression of hsa-circ-000595, a circRNA that was found to be differentially expressed in aortic aneurism compared with normal aortic tissues, was examined. Furthermore, RNA interference was employed to knockdown circRNA expression, and its effect on hypoxia-associated apoptosis of a human aortic smooth muscle cell line was assessed. The present study preliminarily explored the underlying mechanisms of aortic aneurysm from the perspective of epigenetics.
Materials and methods
Cell line
Human aortic smooth muscle cells (V-SMC-6110) were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in low-glucose Dulbecco's modified Eagle's Medium (DMEM; Invitrogen Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen Life Technologies), 100 units/ml penicillin sodium and 100 µg/ml streptomycin sulfate (Invitrogen Life Technologies) at 37°C in a humidified chamber supplemented with 5% CO2.
Patient samples
Aortic tissues from patients with aortic aneurysm were obtained from Zhejiang Second Hospital (Huangzhou, China) between January 2005 and December 2013. The patients consisted of 28 males and 7 females aged between 35 and 84 years old with a mean age of 54 years. The Bioethics Committee of the Zhejiang Second Hospital approved all experiments, which were in accordance with the National Institutes of Health Guide (Bethesda, MD, USA).
Small interfering (si)RNA transfection
Prior to transfection, V-SMC were seeded at a density of 3.0×105, and 2 ml serum-free DMEM was added into six-well plates. siRNA (Funeng Biological Technology Co., Ltd., Shanghai, China) (forward, 5′-GGCCUAAGAUAAGAAAUAUUU-3′, and reverse, 5′-AUAUUUCUUAUCUUAGGCCUU-3′) was added to a final concentration of 50 nM. siRNA targeted to circRNA hsa-circ-000595 was transfected into V-SMC cells with Lipofectamine 2000 (Invitrogen Life Technologies) following the manufacturer's instructions. Knockdown was then confirmed by assessing the expression of circRNA and miRNA by reverse transcription quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR
Total RNA was extracted from cultured cells using TRIzol (Invitrogen Life Technologies) and determined its concentration was determined. Total RNA (0.5 µg) was used as a template to prepare cDNA (Reverse Transcription System, Promega Corporation, Madison, WI, USA; cat. no. A3500). The mRNA expression of target genes (hsa-circ-000595 and miR-19a) was quantified using SYBR Premix EX Taq (Takara Bio, Inc., Shanghai, China) on the ABI 7500 squence detection system (Advanced Biosystem, Thermo Fisher Scientific, Waltham, MA, USA). PCR was performed with the following thermocycling conditions: An initial 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec. The thermocycler used in the present study was the StepOnePlus™ Real-Time PCR system (Applied Biosystems Life Technologies, Foster City, CA, USA) The primers were obtained from Funengbio Co. (Shanghai, China), and the sequences were as follows: hsa-circ-000595, forward 5′-ACGCGGCCTAAATATGAGCA-3′, reverse 5′-GAAGCTTCCAGACTGAGCCAC-3′; m i R-19a, forward 5′-ACGTGTGCAAATCTATGCAAAAC-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′; β-actin, forward 5′-CCTCGCCTTTGCCGATCC-3′ and reverse 5′-GGATCTTCATGAGGTAGTCAGTC-3′. Housekeeping gene β-actin was used as an internal reference to normalize the results. All experiments were performed in triplicate. Finally, the 2−ΔΔCt method was performed to calculate the relative expression (21).
Western blot analysis
Western blotting was performed according to standard protocols. Briefly, the total protein from tissue or cell was extracted using radioimmunoprecipitation lysis buffer containing 1 mM phenylmethanesulfonylfluo-ride and the protein concentration was determined using the Bradford method (Beyotime Institute of Biotechnology, Nantong, China) according to the manufacturer's instructions. 20 µg total protein sample was separated by 10% SDS-PAGE (Fdbio Science, Hangzhou, China) and transferred into a nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). The membrane was blocked in phosphate-buffered saline containing Tween 20 (PBST) with 5% non-fat milk for 1 h at 4°C. The membrane was then incubated with the following primary antibodies for 12 h at 4°C: Anti-caspase 8 (mouse anti-human monoclonal antibody; 1:1,000; cat. no. sc-56070), anti-caspase 3 (mouse anti-human monoclonal antibody; 1:800; cat. no. sc-65496) and anti-B-cell lymphoma (Bcl)-2 (rabbit anti-human polyclonal antibody; 1:1,000; cat. no. sc-492) (all Santa Cruz Biotechnology, Inc., Dallas, TX, USA), followed by three washes with PBST and incubation for 30 min at 4°C with secondary antibody labeled with horseradish peroxidase. Finally, the membrane was washed three times with PBST and the protein bands were visualized with Super Signal. Antibody binding was detected using an enhanced chemiluminescence kit (EMD Millipore). The blots were then incubated in a commercial stripping solution (Pierce Biotechnology, Inc. Rockford, IL, USA) for 10 min. The membranes were re-probed with a rabbit anti-human anti-β-actin polyclonal antibody (1:1,000; cat. no. A2668; Sigma-Aldrich, St. Louis, MO, USA) as a loading control.
Hematoxylin and eosin (HE) staining
The aortic tissue samples from patients with aortic aneurysm were fixed in 4% paraformaldehyde (Aladdin, Shanghai, China), dehydrated with graded ethanol, cleared in dimethylbenzene (Aladdin) and embedded in paraffin (Aladdin). Paraffin sections (5 µm) were prepared using a Leica histotome (Leica Microsystems, Wetzlar, Germany) and deparaffinized with immersion in dimethylbenzene prior to rehydration. HE staining was subsequently performed according to standard procedures (22).
Apoptosis assays
The V-SMC cells were pre-treated with CoCl2, or without CoCl2 if they were to serve as a control. The apoptosis levels of the V-SMC cells treated with scramble or hsa-circ-000595 siRNA were subsequently analyzed using an Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit (cat. no. C1063; Beyotime Institute of Biotechnology) according to the manufacturer's instructions. The cells were seeded in 6-well plates at a density of 1×105 cells/well in DMEM medium for 24 h. The cells were then digested with 0.25% trypsin (Invitrogen Life Technologies) and resuspended in 300 µl binding buffer (Beyotime Institute of Biotechnology) containing 5 µl Annexin V-FITC and 5 µl propidium iodide solution, and incubated at room temperature in the dark for 20 min. The stained cells were analyzed by flow cytometry (FACScan; BD Biosciences, Franklin Lakes, NJ, USA).
TUNEL assay
The V-SMC cells were pre-treated with CoCl2 or without CoCl2 if they were to serve as a control. The cells were then treated with scramble siRNA or hsa-circ-000595 siRNA for 24 h. The adherent cells were subsequently detached using 0.1% trypsin/0.04% EDTA (Invitrogen Life Technologies) and the cell apoptosis levels were assessed using a DeadEnd™ Fluorometric TUNEL assay kit (cat. no. C1088; Beyotime Institute of Biotechnology). Data were acquired using an Infinite 200 PRO Multimode reader (Tecan Group, Ltd., Maennedorf, Switzerland) and were analyzed using i-control 1.9 software (Tecan Group, Ltd.).
Bioinformatics analysis
The bioinformatics data presented in the present study may be obtained from GEO (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE24194.
Statistical analysis
Values are expressed as the mean ± standard deviation. Differences between groups were analyzed by 1- or 2-way analysis of variance. Differences in the rates of tumor inhibition were validated by the χ2 test. P<0.05 was considered to indicate a significant difference between groups. The analytical software is SPSS 13.0 (SPSS Inc, Chicago, IL, USA) was used for statistical analyses.
Results
Expression of circRNA in aortic tissues from patients with aortic aneurysm
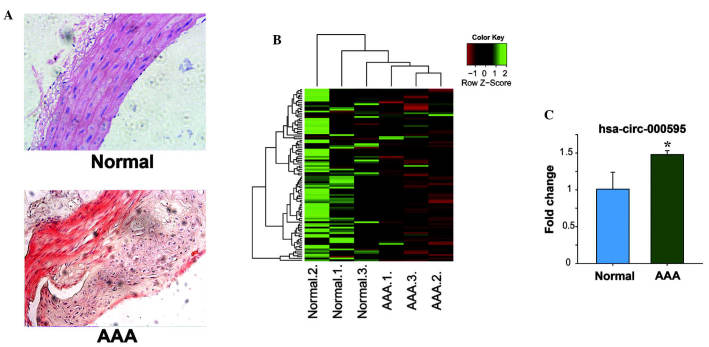
Aortic tissues from patients with aortic aneurysm were assessed by histochemical analysis and RT-qPCR. H&E staining of aortic aneurysm and normal tissues was performed (Fig. 1A). In the PCR array assays, 94 circRNAs were assessed and the results revealed that the expression of 12 circRNAs was increased by at least 1.5-fold of that in normal tissues (Fig. 1B). In aortic tissues from patients with aortic aneurysm, hsa-circ-000595 was most significantly increased by 1.5-fold compared with that in normal tissues (P<0.05) (Fig. 1C).
Figure 1.
CircRNA hsa-circ-000595 is increased in aortic tissue from patients with aortic aneurysm. (A) CircRNA levels in aortic tissue from aortic aneurysm patients (magnification, ×40). (B) Heatmap of circRNA levels in aortic tissue from aortic aneurysm patients were determined by PCR array. (C) The levels of circRNA in aortic tissue from aortic aneurysm patients were determined by reverse transcription quantitative PCR. The data are expressed as the mean ± standard deviation. *P<0.05 versus normal tissue. circRNA, cyclic RNA; hsa, Homo sapiens; PCR, polymerase chain reaction; AAA, aorta from patient with aortic aneurysm.
Hypoxia upregulates the expression of hsa-circ-000595 in v-SMCs
As hypoxia may be one of the causes of aortic aneurysm, the present study determined the expression of hsa-circ-000595 during hypoxia, which was simulated by incubation with CoCl2. The expression levels of hsa-circ-000595 in v-SMCs treated with/without CoCl2 were assessed by RT-qPCR. As shown in Fig. 2A, V-SMC expressed circRNAs normally under normoxic conditions, while treatment with CoCl2 (300 µM) led to a modest upregulation of the expression of hsa-circ-000595 in V-SMCs in a time-dependent manner (Fig. 2A; P<0.05). Of note, high doses of CoCl2 significantly increased the expression of hsa-circ-000595 (Fig. 2B; P<0.05). These results indicated that hsa-circ-000595 expression was increased under hypoxic conditions, further suggesting its implication in aortic aneurysm.
Figure 2.
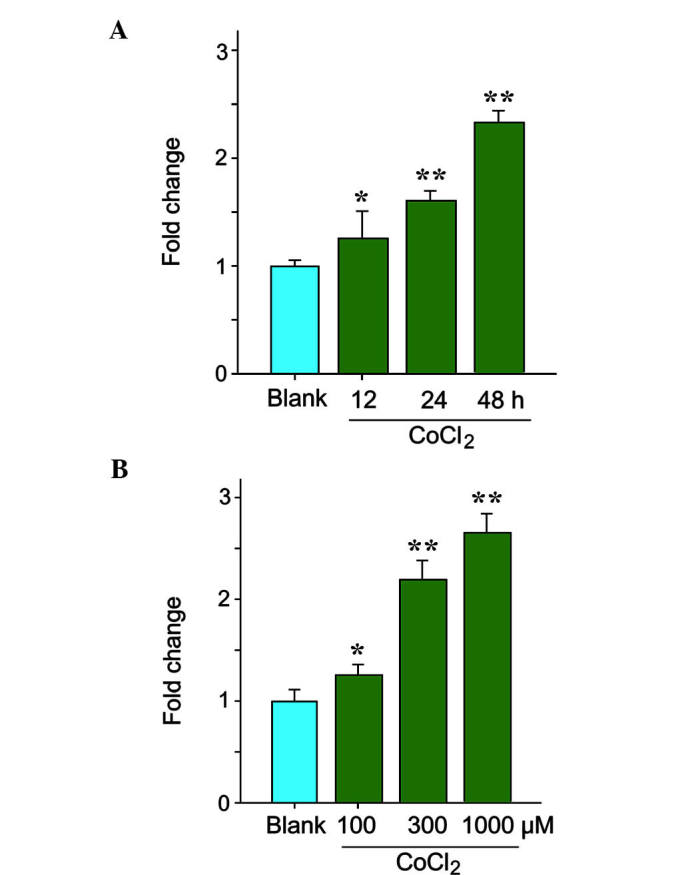
CircRNA hsa-circ-000595 was increased in hypoxic V-SMCs. (A) CircRNA hsa-circ-000595 in cobalt dichloride-induced hypoxic V-SMCs was increased in a time-dependent manner. (B) CircRNA hsa-circ-000595 was increased in hypoxic V-SMCs in a cobalt dichloride dose-dependent manner. The data are expressed as the mean ± standard deviation. *P<0.05 and **P<0.01 versus normal tissues. V-SMC, human aortic smooth muscle cell; circRNA, cyclic RNA; hsa, Homo sapiens.
Knockdown of hsa-circ-000595 attenuates hypoxia-induced apoptosis of V-SMCs
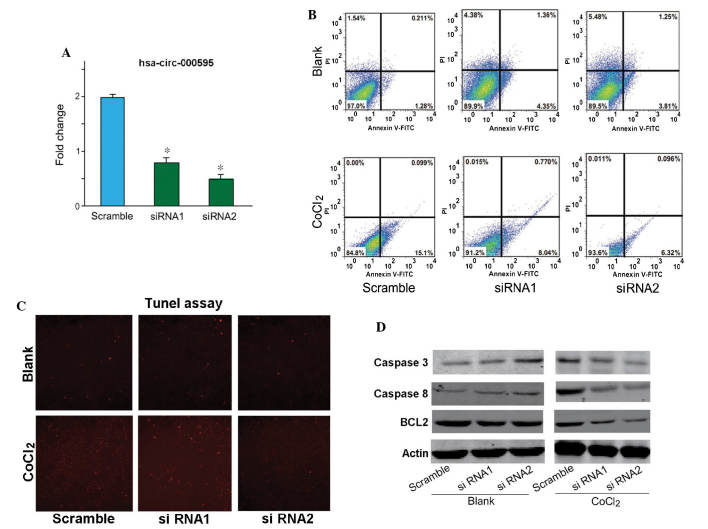
To further explore how hsa-circ-000595 influences the viability of V-SMCs, hsa-circ-000595 knockdown was performed in V-SMCs. RT-qPCR confirmed that the expression of hsa-circ-000595 was obviously downregulated in V-SMCs transfected with two different siRNAs directed towards hsa-circ-000595 for 48 h compared with the expression in cells transfected with scrambled control vector (P<0.05) (Fig. 3A). In order to clarify the role of hsa-circ-000595 in apoptosis of V-SMCs, the apoptotic rate was assessed by flow cytometry and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays. As shown in Fig. 3B, trans-fection with either of the two siRNAs of hsa-circ-000595 did not affect the apoptotic rate of V-SMC cells in the absence of CoCl2; however, in the presence of CoCl2, the apoptotic rate was decreased in hsa-circ-000595-knockdown cells compared with that in cells transfected with scrambled control. These results were validated by TUNEL assays (Fig. 3C). Under hypoxic conditions only, hsa-circ-000595 knockdown can also reduced the protein levels of caspase 3, caspase 8 and Bcl-2, which are apoptosis-associated proteins (Fig. 3D). Therefore, knockdown of hsa-circ-000595 attenuated hypoxia-induced apoptosis of aortic cells, which may potentially reduce the occurrence of aortic aneurysm.
Figure 3.
Effect of has-circ-000595 knockdown on hypoxia-induced damage of V-SMCs. (A) Knockdown efficiency of two siRNAs targeted to has-circ-000595 in V-SMCs. The data are expressed as the mean ± standard deviation. *P<0.05 vs. scrabled control. (B) Flow cytometric detection of the apoptotic rate of V-SMCs transfected with siRNAs. The two siRNAs targeted to hsa-circ-000595 did not affect the apoptotic rate of V-SMCs in the absence of CoCl2; however, under CoCl2-induced hypoxia, the hsa-circ-000595 siRNAs decreased the apoptotic rate of V-SMCs compared with that of CoCl2-treated controls. (C) Tunel analysis of the apoptotic rate of V-SMCs transfected with siRNAs in the absence or presence of CoCl2. The amount of red fluorescence is indicative of the apoptotic rate (magnification, ×20). (D) Expression levels of caspases 3, caspase 8 and Bcl-2 in scramble-, siRNA1- and siRNA2-transfected cells with or without CoCl2 were detected by western blot analysis, and GAPDH was used as a loading control. siRNA, small interfering RNA; V-SMC, human aortic smooth muscle cell; circRNA, cyclic RNA; hsa, Homo sapiens; Bcl-2, B-cell lymphoma 2; Tunel, terminal deoxynucleotidyl transferase dUTP nick end labeling; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Hsa-circ-000595 regulates miR-19a expression
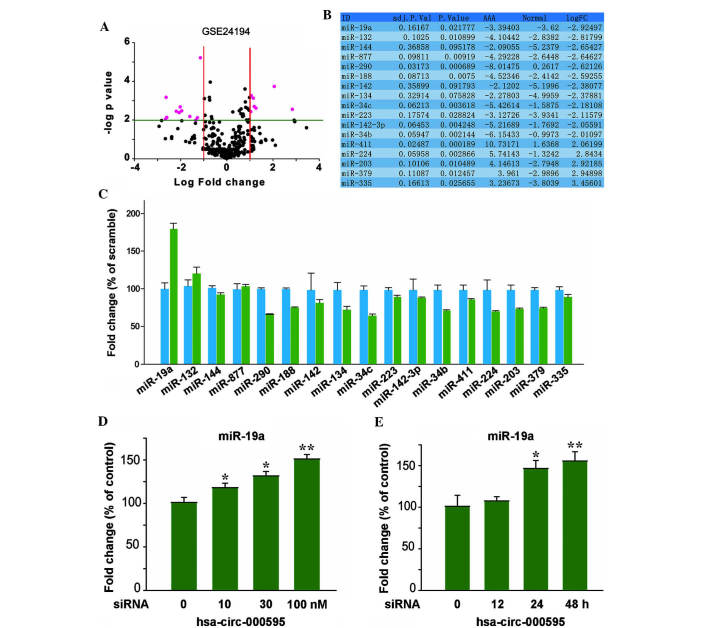
Growing evidence indicated that hsa-circ-000595 is strongly associated with apoptosis (23). CircRNAs were reported to be silencers of miRNAs. By using bioinformatics tools, the present study demonstrated that expression of 17 miRNAs was affected by hsa-circ-000595 (Fig. 4A–C), and that there was a statistically significant difference between the expression levels of miR-19a and Hsa-circ-000595. These results were obtained from the GSE24194 database. RT-PCR was then performed to determine the expression levels of miR-19a in V-SMCs treated with CoCl2. As shown in Fig. 4D and E, prior to CoCl2 treatment of V-SMCs, the cells were transfected with hsa-circ-000595 siRNA, and subsequently the expression of miR-19a was significantly increased in hsa-circ-000595-knockdown-treated V-SMCs in a dose- and time-dependent manner, as compared with that in native V-SMCs. These results suggest that knockdown of hsa-circ-000595 is able to increase the expression of miR-19a.
Figure 4.
CircRNA regulates miRNA in V-SMCs. (A-C) Aberrant expression of miRNAs in AAA (red) was assessed by bioinformatics analysis. Differences in the expression of miRNA19a between AAA (green) and normal controls (blue) were most significant. (D) Dose-dependent effects of has-circ-000595 on the expression of miR-19a in V-SMCs. (E) Time-dependent effects of hsa-circ-000595 on the expression of miR-19a in V-SMCs. The data are expressed as the mean ± standard deviation. *P<0.05 and **P<0.01 versus normal tissues. AAA, aortic tissues from patients with aortic aneurism; FC, fold change; siRNA, small interfering RNA; V-SMC, human aortic smooth muscle cell; miRNA, microRNA; circRNA, cyclic RNA; hsa, Homo sapiens.
Discussion
Evidence has shown that a combination of hsa-circ-000595 knockdown with miR-19a was more effective in preventing V-CMC cell apoptosis than either agent (24). In the present study, the expression levels of hsa-circ-000595 were higher in tissue from patients with aortic aneurysm, as compared with in normal tissue. In addition, there was a negative correlation between hsa-circ-000595 and miR-19a; the expression of miR-19a was enhanced after silencing of hsa-circ-000595 expression. However, the therapeutical value of this finding has not been exploited for any clinical applications due to the low bioavailability of hsa-circ-000595 (25,26). In order to overcome its low bioavailability, numerous analogues of hsa-circ-000595 have been created over the past two decades, but the outcomes have been disappointing (27). The present study performed a preliminary analysis in order to elucidate the synergistic mechanism of hsa-circ-000595 and miR-19a from an epigenetics perspective aimed to lay a theoretical foundation for improving the sensitivity to miRNA treatment in cardiovascular conditions.
Previous studies indicated that sensitivity to miRNA was increased by upregulation of miR200 and miR21, which blocked hsa-circ-000595 (28). The present study showed that hsa-circ-000595-knockdown upregulated miR-19a expression in hypoxic V-SMC cells. Previous studies reported that cyclo-oxygenase (COX)-2 and nuclar factor (NF)-κB, are strongly associated with cardiovascular diseases, are the target genes of hsa-circ-000595 (18,29).
The present study generated a hypoxic cell model using a cardiovascular V-SMC line. In these V-SMCs, hypoxia-inducible factor (HIF)1α is strongly linked with hypoxia-induced apoptosis and is aberrantly highly expressed at the mRNA and protein level compared with that in normal cells. According to previous studies, this is in part due to the inactivation/activation of NF-κB, COX-2 and their downstream target molecules (15). However, previous studies found that miR-19a was markedly downregulated in V-SMCs treated with a combination of circRNAs and hsa-circ-000595 compared with circRNAs alone or not (30,31). This suggested that circRNAs may increase the sensitivity to miRNA treatment not only through modulation of miR21 and miR200 expression, but also through modulation of miR-19a expression by hsa-circ-000595. In addition, hsa-circ-000595-knockdown was associated with high miR-19a expression levels, as indicated by the results of V-SMC cells treated with a combination of circRNAs and hsa-circ-000595 compared with circRNAs alone or not. Therefore, it is indicated that the levels of miR-19a may be altered in V-SMCs treated with a combination of miR-19a and hsa-circ-000595. All of these results suggested that aortic aneurysm can be induced through upregulation of miR-19a expression by hsa-circ-000595 (32–34).
In conclusion, the present study showed that hsa-circ-000595 was highly expressed aortic tissues of patients with aortic aneurysm, and that hsa-circ-000595 can inhibit the expression of miR-19a. These results laid a theoretical foundation for exploring the mechanisms of the involvement of circRNAs in aortic aneurysm and their roles in the regulation of miRNAs, which may be harnessed in order to tune the sensitivity of aortic aneurysm to miRNA treatments.
References
- 1.Quickley HA, Vitale S. Models of open-angle glaucoma prevalence and incidence in the United States. Invest Ophthalmol Vis Sci. 1997;38:83–91. [PubMed] [Google Scholar]
- 2.Flammer J, Orgül S, Costa VP, Orzalesi N, Krieglstein GK, Serra LM, Renard JP, Stefánsson E. The impact of ocular blood flow in glaucoma. Prog Retin Eye Res. 2002;21:359–393. doi: 10.1016/S1350-9462(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 3.Kniestedt C, Kanngiesser HE. Dynamic contour tonometry. Ophthalmologe. 2006;103:713–721. doi: 10.1007/s00347-006-1387-7. In German. [DOI] [PubMed] [Google Scholar]
- 4.Romppainen T, Kniestedt C, Bachmann LM, Stürmer J. Ocular pulse amplitude: A new biometrical parameter for diagnose of glaucoma. Ophthalmologe. 2007;104:230–235. doi: 10.1007/s00347-006-1467-8. In German. [DOI] [PubMed] [Google Scholar]
- 5.He G, Qing H, Tong Y, Cai F, Ishiura S, Song W. Degradation of nicastrin involves both proteasome and lysosome. J Neurochem. 2007;101:982–992. doi: 10.1111/j.1471-4159.2007.04449.x. [DOI] [PubMed] [Google Scholar]
- 6.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNAmolecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabowski PJ, Zaug AJ, Cech TR. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell. 1981;23:467–476. doi: 10.1016/0092-8674(81)90142-2. [DOI] [PubMed] [Google Scholar]
- 8.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 11.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 12.Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: Abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 13.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Li C, Zhang R, Gao X, Qu X, Zhao M, Qiao C, Xu J, Li J. miR-17-92 cluster microRNAs confers tumorigenicity in mutiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q, Ji YS, Schmedtje JF., Jr Sp1 increases expression of cyclo-oxygenase-2 in hypoxic vascular endothelium Implications for the mechanisms of aortic aneurysm and heart failure. J Biol Chem. 2000;275:24583–24589. doi: 10.1074/jbc.M003894200. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Sun L, Wang X, Chen S, Kong Y, Liu N, Chen Y, Jia Q, Zhang L, Zhang L. Activin B promotes BMSC-mediated cutaneous wound healing by regulating cell migration via the JNK-ERK signaling pathway. Cell Transplant. 2014;23:1061–1073. doi: 10.3727/096368913X666999. [DOI] [PubMed] [Google Scholar]
- 23.Brown JR, Sanseau P. A computational view of microRNAs and their targets. Drug Discov Today. 2005;10:595–601. doi: 10.1016/S1359-6446(05)03399-4. [DOI] [PubMed] [Google Scholar]
- 24.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 26.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding- independent function of gene and pseu-dogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiertz E, Hill A, Tortorella D, Ploegh H. Cytomegaloviruses use multiple mechanisms to elude the host immune response. Immunol Lett. 1997;57:213–216. doi: 10.1016/S0165-2478(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet. 2008;9:503–507. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- 33.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y, McCaffrey AP. Rational design leads to more potent RNA interference against hepatitis B virus: Factors effecting silencing efficiency. Mol Ther. 2009;17:538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]