Abstract
Prostate cancer (PCa) is the second leading cause of cancer-related mortality among American males. Studies suggest that cigarette smoking is associated with the progression of PCa; however, the molecular mechanisms underlying this process have not been extensively investigated. PCa progression is characterized by increased cell migration and alterations in extracellular matrix (ECM)- and cell adhesion molecule (CAM)-related gene expression. In the present study, the influence of cigarette smoke medium (SM) on cell migration and on the expression of ECM- and CAM-related genes in PC3 prostate adenocarcinoma cells was investigated. According to a wound-healing assay, SM treatment promoted PC3 cell migration. RNA expression levels from SM-treated and control cells were analyzed using a polymerase chain reaction (PCR) array. Of 84 genes analyzed, 27.38% (23/84) exhibited a ≥2-fold change in threshold cycle in PC3 cells following 0.5% SM treatment. Functional gene grouping analysis demonstrated that SM treatment modulated the RNA transcription of approximately 18.4% of CAMs and 33.93% of ECM-related genes. Quantitative PCR analysis showed that SM treatment led to a significant decrease in transcription levels of the following genes: Collagen 5 α-1(V), connective tissue growth factor, integrin β-2, kallmann syndrome 1, laminin α 3, matrix metallopeptidase 7 (MMP7), MMP13, secreted protein acidic cysteine-rich, thrombospondin-2 and versican; and that SM significantly increased the transcription levels of MMP2 and MMP12. Furthermore, MMP2 knockdown significantly reduced the migration of SM-treated PC3 cells. The present study provides novel insights into the association of cigarette smoking with PCa progression, via the alteration of ECM/CAM interactions.
Keywords: cigarette smoke, cell migration, cell adhesion molecules, extracellular matrix, prostate cancer
Introduction
Prostate cancer (PCa) is the most common tumor in males in the USA, with an estimated 240,890 new cases and 33,720 mortali-ties related to PCa in 2011 (1). However, few risk factors have been identified beyond age and family history (2,3). Despite a proven link between cigarette smoking and a number of types of tumors (4), the association between cigarette smoking and PCa remains unresolved. Observational case-control studies investigating the association between cigarette smoking and PCa have produced inconclusive results (5). Epidemiological studies suggest that the results of investigations into the correlation between cigarette smoking and the incidence of PCa may differ from the results of those measuring PCa-related mortality (6). An observational cohort study confirmed that while smoking does not appear to be associated with PCa incidence, there is increased mortality due to PCa among heavy smokers (7). Furthermore, cigarette smokers have a higher risk of developing advanced stage and high-grade PCa, as compared with non-smokers (8). Hence, cigarette smoking is a potential contributing factor to PCa progression.
Tumor progression is a multistep process that includes cell migration, invasion and metastasis, to which alterations in the expression of extracellular matrix (ECM) and cell adhesion molecule (CAM)-related genes contribute (9). During PCa progression, tumor cell migration and invasion occur following the degradation of the ECM, which is composed of collagen, proteoglycans, fibronectin, laminin and other glycoproteins (9–13). Tumor metastasis involves the detachment of cancer cells from the primary tumor, disruption of the basement membrane and invasion of the surrounding stroma by tumor cells. Cancer cells enter the vascular and lymphatic systems and distal sites, including the liver, lungs and brain, where they extravasate, proliferate and undergo angiogenesis (9,14–16). ECM proteins such as matrix metalloproteinases (MMPs) are associated with tumor growth, cancer progression and metastasis (17–23).
Tobacco smoke contains at least 3,500 chemical compounds, many of which are toxic, carcinogenic and/or mutagenic. It consists of a particulate solid phase (tar) and a gaseous phase, containing volatile organic compounds (VOCs), free radicals and other volatile and semi-volatile compounds (24,25). Exposing prostate cancer cells to cigarette smoke promotes cell proliferation and the secretion of vascular endothelial growth factor (VEGF), a potent angiogenic factor (26). Angiogenesis is the process of new blood vessel formation, which is fundamental to the progression, invasion, and metastasis of numerous cancers, including PCa (27).
Interaction between the ECM and CAMs helps determine tissue shape, structure and cellular function. ECM and CAMs are involved in a range of disorders and diseases. ECM- and CAM-related gene expression on cell surfaces is involved in pathophysiological processes, such as wound healing, inflammation, and disease (9,28–34). Levels of ECM and CAM-related gene expression vary. For example, studies have shown that an increase in the expression of ECM-related genes, including MMP2 and MMP9, promotes cell invasion and the progression of PCa (35–37). The transition from healthy prostatic intraepithelial neoplasia to invasive PCa is associated with a decrease in the expression of MMP7 and the cell matrix adhesion protein, integrin β 4 (ITGB4) (38). The loss of cell adhesion molecules, such laminin α 3 (LAMA3), is associated with high Gleason scores, high preoperative prostate-specific antigen (PSA) levels and more advanced stages of PCa (39).
Cigarette smoke enhances prostate cancer cell proliferation and VEGF secretion (26). The present study examined the direct effects of cigarette smoke on cell migration, and the expression of ECM- and CAM-related genes in PC3 cells.
Materials and methods
Preparation of SM
3R4F cigarettes (Reference Cigarette Program, University of Kentucky Research Institute; Lexington, KY, USA) contained 11.0 mg/cigarette (mg/cig) total particulate matter (TPM), 9.4 mg/cig tar, 0.73 mg/cig nicotine, and 12 mg/cig carbon monoxide (CO). SM was generated by collecting whole smoke from 20 reference cigarettes into 100 ml of cell culture media. Briefly, cigarette smoke from 20 cigarettes was bubbled into cell culture medium. The medium was then filtered, and was designated as SM. Fresh SM was used in the specified final concentrations.
Cell culture
PC3 cells [American Type Culture Collection (ATCC), Manassas, VA, USA] were plated onto a 12-well tissue culture plate and cultured in ATCC-formatted F12K medium (cat. no. CRL-1435 and cat. no. 30-2004; ATCC, Rockville, MD, USA; respectively) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA, USA). Cells were then incubated at 37°C, at 5% CO2.
siRNA transfection
Small interfering RNA for MMP2 (MMP2 siRNA; cat. no. sc-29398; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) designed to knock down human MMP2 expression was resuspended to a final 10 µΜ concentration in a buffer containing 10µM Tris-HCl, 20 mM NaCl, and 1 mM EDTA at pH 8.0. For a control, scrambled siRNA (cat. no. sc-37007, Santa Cruz Biotechnology, Inc.) was prepared and used in the same condition. siRNA transfection was performed according to the manufacturer's instructions. In brief, PC3 cells were washed with siRNA transfection medium (Santa Cruz Biotechnology, Inc.) and incubated at 37°C in 200 µl transfection medium containing 0.8 µM of MMP2 siRNA or scrambled siRNA (siCTL). Following 24 h of transfection, MMP2 expression was assessed using western blot analysis and a wound healing assay was performed.
Wound healing assay
A scratch-wound assay was performed as previously described by Rodriguez et al (40) in order to assess cell migration in the presence of SM. Following incubation, when cells had reached ~100% confluence, they were washed with serum-free F12K medium, and replenished with ATCC-formatted medium containing 0.5% FBS. The cells were cultured for 24 h. Subsequently, a sterile 20 ml pipette tip was used to scratch the monolayer of cells in two perpendicular straight lines through the center of the wells. Wells were gently washed with serum-free culture, medium replenished with the medium containing 0.5% FBS and treated with 0 (control), 0.2, 0.5, 1 or 2% SM in cell culture medium. Cells were cultured for 24 h, after which, cells that had migrated into the gaps were counted using a microscope (Diaphot 300; Nikon Corporation, Tokyo, Japan).
RNA isolation
Isolation of total RNA was performed using TRIzol® Reagent (cat. no. 15596-026; Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Cells were seeded on 6-well plates and treated with SM or F12K medium supplemented with 0.5% FBS. Subsequently, chloroform (0.2 ml; Sigma-Aldrich, St. Louis, MO, USA) was added to the wells. Samples were incubated at room temperature for 3 min, and centrifuged at 12,000 x g at 4°C for 15 min. Subsequently, isopropanol (0.5 ml; Thermo Fisher Scientific, Waltham, MA, USA) was added to the supernatant. Following incubation at room temperature for 10 min, samples were centrifuged at 12,000 x g at 4°C for 10 min. The pellets were washed with 75% ethanol, dissolved in RNAse-free water (Thermo Fisher Scientific) and incubated at 60°C for 10 min.
Gene expression profiling
Cells were treated with 0.5% SM for 24 h. Subsequently, total RNA was extracted using TRIzol and an RNeasy mini kit (cat. no. 74104; Qiagen, Valencia, CA, USA). RNA integrity was assessed using the bioanalyzer 'Agilent 2200 Tape Station' (Agilent Technologies, Oxford, UK). The expression of 84 CAM- and ECM-related genes were profiled using an RT2 Profiler Polymerase Chain Reaction (PCR) Array for human extracellular matrix and adhesion molecules, according to the manufacturer's instructions (cat. no. PAHS-013A; SABiosciences, Qiagen). The gene expression of 25 µg RNA per plate was measured. RNA was converted into cDNA using a reverse transcription cocktail (cat. no. 330401, Qiagen) at 42°C for 15 min. cDNA was then mixed with 2 x SABioscience RT PCR Master Mix (cat. no. 330520, Qiagen) and subjected to PCR amplification using ABI 7300 and ABI 7500 platforms (AB Applied Biosystems, Foster City, CA, USA). Quantitative (q)PCR primers and DNA oligos were purchased from Real Time Primers, LLC (Elkins Park, PA, USA) and Integrated DNA Technologies (Coralville, IA, USA), respectively. Threshold cycle (Ct) was used to calculate changes in gene expression. Calculation of Ct values and statistical analyses were performed using web-based applications from SA Bioscience (Qiagen). Ct values were normalized against those of actin and GAPDH. Ct values were converted to linear values using the equation [2^ (−Ct)]. P-values were calculated using Student's t-test and a 95% confidence interval (CI). Changes in gene expression were expressed as fold change (FC) and fold regulation (FR). The PCR analysis was conducted using web-based applications for RT2 Profiler PCR Array Data Analysis version 3.5 (http://www.sabiosciences.com/RTPCR.php; Qiagen).
qPCR analysis
PCR reactions were performed using Power SYBR Green PCR Master Mix (cat. no. 4309155; AB Applied Biosystems, Warrington, UK) containing RT enzyme mix (cat. no. 4389988, AB Applied Biosystems). RNA samples were subjected to qPCR amplification using the ABI 7300 cycler (AB Applied Biosystems). Using quantitative primers, the thermal cycler conditions were as follows: 48°C for 30 min and 95°C for 10 min, followed by 42–52 cycles at 95°C for 15 sec and 60°C for 1 min.
Statistical analysis
Ct value calculations and statistical analyses were performed using a web-based application (SA Bioscience) as described above. Statistical significance was determined using Student's t-test. Data represent the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
SM-mediated migration of PC3 cells
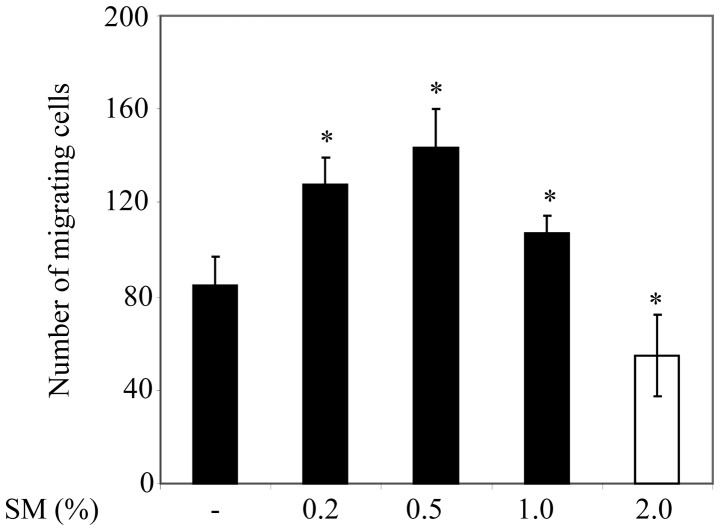
In order to examine the influence of cigarette smoke on PC3 cell migration, the cells were cultured on 12-well plates. Cells were treated with 0 (negative control), 0.2, 0.5, 1 or 2% SM, and cells that had migrated into the gap were subsequently counted. PC3 cell migration was significantly greater following 0.2, 0.5, and 1% SM treatment compared with the migration of 0% SM-treated cells or control counterparts (Fig. 1). It is notable that PC3 cell migration was significantly lower following 1% SM treatment compared with 0.2 and 0.5% SM treatment. Furthermore, PC3 cell migration was significantly lower in 2% SM-treated cells compared with that of the control cells. Since 0.5% SM induced the greatest increase in cell migration compared with control cells, 0.5% SM was used for the remaining experiments in the present study.
Figure 1.
SM-mediated migration of PC3 cells assessed using a wound-healing assay. Data are presented as the number of migrating cells per SM concentration and represent the mean ± standard deviation of the number of migrating cells. *P<0.01 of control cells vs. SM-treated cells. SM, cigarette smoke medium.
SM-modulated changes in the expression of CAM- and ECM-related genes
ECM- and CAM-related genes are involved in multiple processes that contribute to tumor progression (9). In order to investigate the effect of SM on the expression of ECM- and of CAM-related genes, PC3 cells were incubated in a starvation medium (0.5% FBS) for 24 h and then treated with 0.5% SM or 0% SM (negative control) for a further 24 h. Of the 84 genes, the expression of 23 (27.38%) exhibited an FC value of ≥2 (Table I). These genes included: C-type lectin domain family 3; collagen type I α 1 (COL1A1); COL5A1; COL11A1; COL14A1; COL16A1; contactin 1; connective tissue growth factor (CTGF); catenin, α 1; catenin Δ 2; integrin β 2 (ITGB2); kallmann syndrome 1 sequence (KAL1); laminin α 3 (LAMA3); matrix metallopeptidase 2 (MMP2); MMP3; MMP7; MMP12; MMP13; selectin E; secreted protein acidic, cysteine-rich (SPARC); transforming growth factor, β-induced; Thrombospondin 3 (THBS3); and Versican (VCAN).
Table I.
SM-modulated changes in gene expression of CAM- and ECM-related molecules.
| Gene | FC | FR | Unigene | Accession |
|---|---|---|---|---|
| CLEC3B | 0.43 | −2.33 | Hs.476092 | NM_003278 |
| CNTN1 | 0.33 | −2.99 | Hs.143434 | NM_001843 |
| COL11A1 | 2.28 | 2.28 | Hs.523446 | NM_080629 |
| COL14A1 | 0.43 | −2.34 | Hs.409662 | NM_021110 |
| COL16A1 | 0.47 | −2.14 | Hs.368921 | NM_001856 |
| COL1A1 | 0.49 | −2.03 | Hs.172928 | NM_000088 |
| COL5A1 | 0.49 | −2.04 | Hs.210283 | NM_000093 |
| CTGF | 0.47 | −2.11 | Hs.591346 | NM_001901 |
| CTNNA1 | 0.10 | −10.65 | Hs.534797 | NM_001903 |
| CTNND2 | 2.16 | 2.16 | Hs.314543 | NM_001332 |
| ITGB2 | 0.43 | −2.34 | Hs.375957 | NM_000211 |
| KAL1 | 0.48 | −2.08 | Hs.521869 | NM_000216 |
| LAMA3 | 0.41 | −2.46 | Hs.436367 | NM_000227 |
| MMP2 | 2.70 | 2.70 | Hs.513617 | NM_004530 |
| MMP3 | 6.37 | 6.37 | Hs.375129 | NM_002422 |
| MMP7 | 0.41 | 2.44 | Hs.2256 | NM_002423 |
| MMP12 | 2.74 | 2.74 | Hs.1695 | NM_002426 |
| MMP13 | 0.47 | −2.14 | Hs.2936 | NM_002427 |
| SELE | 3.49 | 3.49 | Hs.89546 | NM_000450 |
| SPARC | 0.24 | −4.16 | Hs.111779 | NM_003118 |
| TGFBI | 0.34 | −2.92 | Hs.369397 | NM_000358 |
| THBS3 | 0.38 | −2.60 | Hs.169875 | NM_007112 |
| VCAN | 0.35 | −2.82 | Hs.643801 | NM_004385 |
Gene expression was assessed using an RT2 Profiler PCR array. SM, cigarette smoke medium; FC, fold change; FR, fold regulation; RT, reverse transcription; CAMs, cell adhesion molecules; ECM, extracellular matrix; PCR, polymerase chain reaction; CLEC3B, c-type lectin domain family 3; CNTN1, contactin 1; COL1A1, collagen type I α 1; CTGF, connective tissue growth factor; CTNNA1, catenin α 1; CTNND2, catenin Δ 2; ITGB2, integrin β 2; KAL1, kallmann syndrome 1 sequence; LAMA3, laminin α 3; MMP, matrix metallopeptidase; SELE, selectin E; SPARC, secreted protein acidic, cysteine-rich; TGFB1, transforming growth factor, β-induced; THBS3, Thrombospondin 3; and VCAN, Versican.
Analysis of SM-modulated changes in the expression of CAM and ECM-related genes
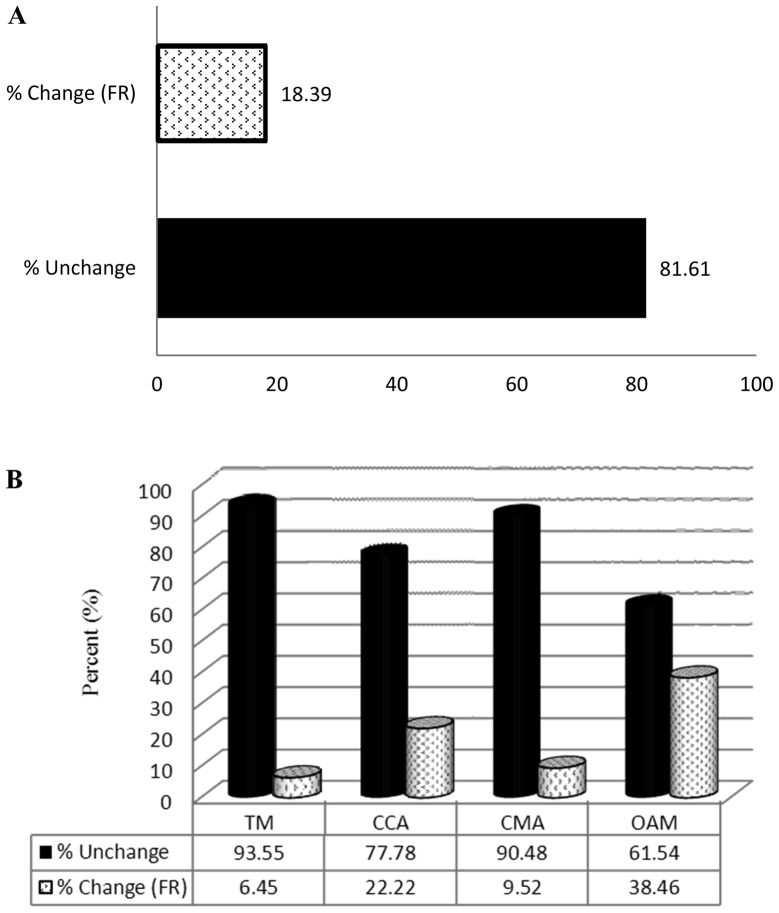
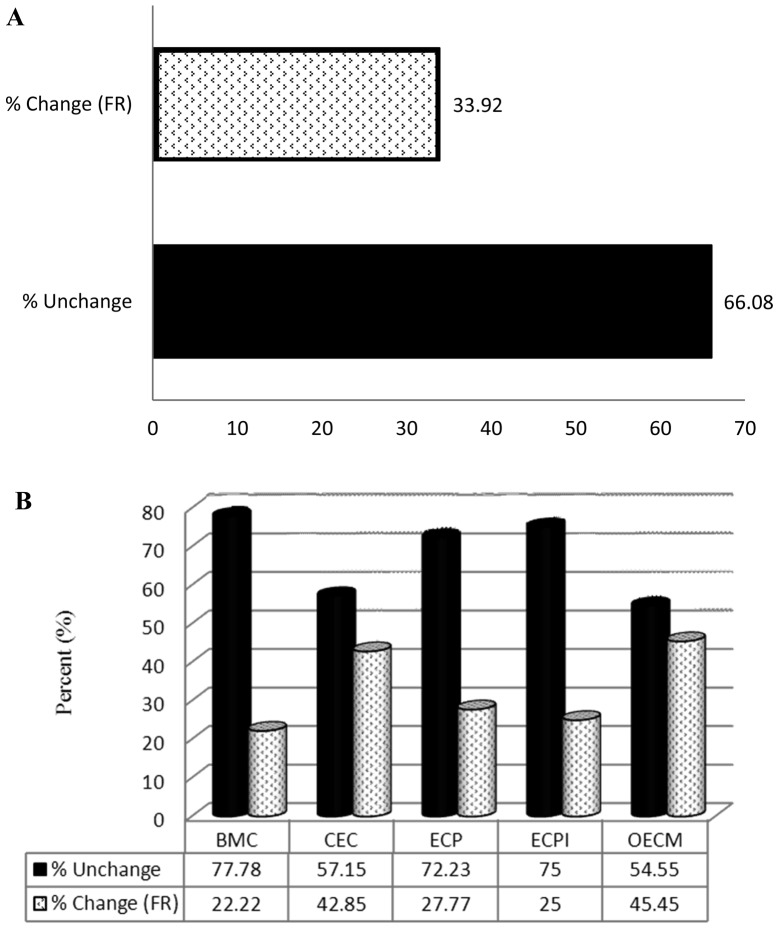
Genes with an FR of ≥2 (Table II) were sorted into subgroups. CAMs were divided into the following subgroups: Transmembrane molecules (TM); cell-to-cell adhesion molecules (CCA); cell matrix adhesion molecules (CMA); and other adhesion molecules (OAM). ECM-related molecules were divided into the following subgroups: Basement membrane constituents (BMC); collagens and extracellular matrix structural constituents (CEC); extracellular matrix proteases (ECP); extracellular matrix protease inhibitors (ECPI); and other ECM (OECM). SM altered the expression of 18.39% (16/87) of the CAM-related genes (Fig. 2A). The FR of 6.45% (2/31) of TM, 22.22% (2/9) of CCA, 9.52% (2/21) of CMA and 38.46% (10/26) of OMA genes was altered by ≥2 in the SM group compared with the control group (Fig. 2B). SM treatment altered the expression of 33.92% (19/56) of ECM-related genes (Fig. 3A). The FR of 22.22% (2/9) of BMC, 42.85% (6/14) of CEC, 27.77% (5/18) of ECP, 25% (1/4) of ECPI and 45.45% (5/11) of OECM genes was altered in the SM-treated group compared with the control group (Fig. 3B).
Table II.
SM-modulated changes in gene expression of CAMs and ECM-related molecules using qPCR analysis.
| Gene | FC | 95% CI | P-value | FR |
|---|---|---|---|---|
| COL5A1 | 0.25 | (0.08, 0.42) | 0.048875 | −2.17 |
| CTGF | 0.31 | (0.21, 0.41) | 0.000417 | −3.24 |
| ITGB2 | 0.36 | (0.30, 0.42) | 0.000338 | −2.74 |
| KAL1 | 0.26 | (0.07, 0.45) | 0.049642 | −3.91 |
| LAMA3 | 0.24 | (0.15, 0.33) | 0.001575 | −4.19 |
| MMP2 | 1.28 | (1.08, 1.48) | 0.042917 | 1.28 |
| MMP7 | 0.44 | (0.27, 0.61) | 0.027884 | −2.26 |
| MMP12 | 3.47 | (1.74, 5.20) | 0.009610 | 3.47 |
| MMP13 | 0.48 | (0.37, 0.59) | 0.007994 | −2.26 |
| SPARC | 0.15 | (0.11, 0.19) | 0.001258 | −6.59 |
| THBS3 | 0.14 | (0.09, 0.19) | 0.004143 | −7.35 |
| VCAN | 0.78 | (0.69, 0.87) | 0.015940 | −1.28 |
SM, cigarette smoke medium; FC, fold change; FR, fold regulation; CI, confidence interval; CAMs, cell adhesion molecules; ECM, extracellular matrix; qPCR, quantitative transcription-polymerase chain reaction; COL5A1, collagen type V α 1; CTGF, connective tissue growth factor; ITGB2, integrin, β 2; KAL1, kallmann syndrome 1 sequence; LAMA3, laminin α 3; MMP, matrix metallopeptidase; SPARC, secreted protein acidic, cysteine-rich; THBS3, Thrombospondin 3; and VCAN, Versican.
Figure 2.
Effect of SM on the expression of CAM-related genes assessed using an RT2 Profiler polymerase chain reaction Array. (A) Percentage of CAM-related genes in which the FR was ≥2.0 following SM treatment, and the percentage of those in which the expression was not affected by SM treatment. (B) Percentage of genes from each CAM-related subgroup in which the FR was ≥2.0 following SM treatment and the percentage of those in which expression was not affected. CAM, cell adhesion molecules; TM, transmembrane molecules CCA, cell-to-cell adhesion; SMA, cell matrix adhesion molecules; OAM, other adhesion molecules; SM, cigarette smoke medium; FR, fold regulation.
Figure 3.
Effect of SM on the expression of ECM-related genes assessed using an RT2 Profiler PCR Array. (A) Percentage of ECM-related genes in which the FR was ≥2.0 following SM treatment, and the percentage of those in which expression was not affected by SM. (B) Percentage of genes from each ECM-related subgroup in which the FR was ≥2.0 following SM treatment, and the percentage in which expression was not affected. BMC, basement membrane constituent; ECM, extracellular matrix; CEC, collagens and ECM structural constituents; ECP, extracellular proteases; ECPI, ECP inhibitors; OEM, other ECM molecules; SM, cigarette smoke medium; FR, fold regulation.
Validation of SM-modulated changes in gene expression
SM-treated and control PC3 cells were harvested. RNA was then extracted, purified and subjected to qPCR analysis using the primer sets provided in Table III. Ct values were normalized to those of GAPDH. Statistically significant differences in gene expression in SM-treated compared with control samples were determined using Student's t-test, and calculated with a 95% confidence interval. SM significantly increased MMP2 and MMP12 expression, and significantly decreased COL5A1, CTGF, ITGB2, KAL1, LAMA3, MMP7, MMP13, SPARC, THBS3 and VCAN expression in PC3 cells compared with that in control cells. The data suggest there may be a correlation between SM-induced migration (Fig. 1) and changes in the expression of ECM- and CAM-related genes. These changes in gene expression may be associated with interactions between CAM- and ECM-related molecules, which may lead to SM-enhanced migration of PC3 cells.
Table III.
List of quantitative polymerase chain reaction primers.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| COL5A1 | 5′-TGTATTTCCCCTGACCTTCA-3′ | 5′-ACCTTTAATCCATCGGGAAG-3′ |
| CTGF | 5′-TTTGGCCCAGACCCAACTAT-3′ | 5′-GTGCAGCCAGAAAGCTAAA-3′ |
| ITGB2 | 5′-ACAAGCTGGCTGAAAACAAC-3′ | 5′-GCAGAAGGAGTCGTAGGTGA-3′ |
| KAL1 | 5′-CAGCAAACACTTCCGTTCTT-3′ | 5′-GCTTCTTCTTTGTTGGGACA-3′ |
| LAMA3 | 5′-CTGCCAGTGCATCTGAATCT-3′ | 5′-TTTCTGACCATGCTCTTTGC-3′ |
| MMP2 | 5′-TTGACGGTAAGGACGGACTC-3′ | 5′-ACTTGCAGTACTCCCCATCG-3′ |
| MMP7 | 5′-AGCCAAACTCAAGGAGATGC-3′ | 5′-GCCAATCATGATGTCAGCAG-3′ |
| MMP12 | 5′-ACACATTTCGCCTCTCTGCT-3′ | 5′-CCTTCAGCCAGAAGAACCTG-3′ |
| MMP13 | 5′-TGACCCTTCCTTATCCCTTG-3′ | 5′-ATACGGTTGGGAAGTTCTGG-3′ |
| SPARC | 5′-GCACGGACTGTCAGTTCTCT-3′ | 5′-AAGAACAACCGATTCACCAA-3′ |
| THBS3 | 5′-ACACAGTTCTCCTGCGACTC-3′ | 5′-ATGGACCCACCCAGAATAAT-3′ |
| VCAN | 5′-TTGCTGTGGAAGGAACTGAG-3′ | 5′-CATAGGTGGCAGAAGCAGAA-3′ |
COL5A1, collagen type V α 1; CTGF, connective tissue growth factor; ITGB2, integrin, β 2; KAL1, kallmann syndrome 1 sequence; LAMA3, laminin α 3; MMP, matrix metallopeptidase; SPARC, secreted protein acidic, cysteine-rich; THBS3, Thrombospondin 3; and VCAN, Versican.
MMP2-modulated migration of SM-treated PC3 cells
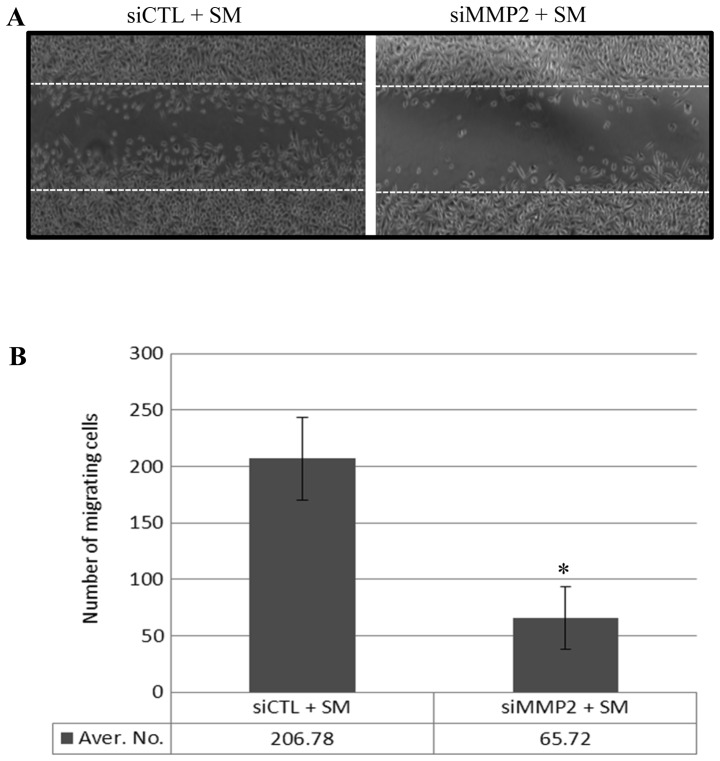
SM treatment led to an increase in the number of migrating PC3 cells (Fig. 1) and enhanced the expression of a number of MMPs, including MMP2 (Tables I and II). In order to confirm that the increase in MMP2 expression contributed to enhanced PC3 cell migration, PC3 cells were transfected either with a scrambled siRNA (siCTL) or with MMP2 siRNA (siMMP2), which inhibited MMP2 expression. Following 24 h of transfection, a reduction in MMP2 expression was measured using western blot analysis (data not shown). Following 24 h incubation in 0.5% FBS medium, the monolayer was 'wounded' and treated with 0.5% SM for 12 h. Migration of the siMMP2-transfected PC3 cells (with reduced MMP2 expression) was 68% lower than that of siCTL transfected PC3 cells (Fig. 4A and B). These data suggest that SM-enhanced MMP2 expression may contribute to the migration of PC3 cells.
Figure 4.
SM-induced MMP2 expression enhanced PC3 cell migration. (A) Effect of SM on the migration of siCTL and siMMP2 cells using a wound-healing assay. (B) Quantification of migrating siCTL and siMMP2 cells following SM treatment. Data are presented as the number of migrating cells, and represent the mean ± standard deviation of the number of migrating cells. *P<0.05 for siCTL cells vs. siMMP2 cells (with reduced MMP2 expression). CTL, control; si, small interfering; MMP2, matrix metallopeptidase; SM, cigarette smoke medium.
Discussion
A recent observational cohort study demonstrated that smoking is associated with the progression rather than the incidence of PCa (7). However, to the best of our knowledge the underlying molecular mechanism of PCa progression is yet to be investigated. A previous study suggested that cigarette smoke enhanced the growth of PCa cells and increased secretion of VEGF, a potent angiogenic factor (26). In the present study, the effect of cigarette smoke on PC3 cell migration was investigated, and the expression of genes involved in tumor progression was measured. The results suggested that cigarette smoke may increase PC3 cell migration by altering the expression of a number of CAM- and ECM-related genes.
Physiological interactions between CAMs and the ECM define the shape, structure, and function of healthy tissues, which are modulated by cellular activities, such as cell growth, division, differentiation and migration (41,42). However, ECM-CAM interactions are also involved in a range of pathophysiological disorders, including cancer (9,28–34). Changes in the expression of ECM- and CAM-related genes may alter the interaction between these molecules, which may affect PCa progression. Cigarette smoke has been shown to enhance PC3 cell growth and to increase VEGF secretion (26). In the present study SM treatment led to an increase in PC3 cell migration (Fig. 1). Therefore, exposure to cigarette smoke may accelerate PCa progression. These results are consistent with previous findings, which suggest that smokers exhibit a higher risk of developing advanced stage and high-grade PCa, and report an increased mortality among heavy smokers (7).
The results of the present investigation suggested that PCa cell growth and migration was greater following 0.5% SM treatment compared with that of PCa cells treated with 0.2, 1 and 2% SM (26; Fig. 1), which suggests that 0.5% is an optimal concentration with which to enhance PCa progression. This optimal SM concentration is estimated to contain levels of TPMs, nicotine, and CO, which are equivalent to those found in 1/100 smoke produced by one reference cigarette, according to the Reference Cigarette Program. Cigarette smoke also contains VOCs and reactive free radicals (24,26), which can dysregulate healthy physiological processes, and promote the proliferation, migration, invasion and metastasis of cancer cells (25). Chronic exposure to cigarette smoke, even at low doses, may pose a risk for advanced stage and high-grade PCa in smokers and non-smokers.
Changes in the expression of ECM- and CAM-related genes may alter the interactions of these moleules, influencing PCa progression (43,44). An RT2 Profiler PCR array analysis of PC3 cells suggested that 0.5% SM led to a ≥2-FC change in the expression of 23 out of 84 ECM- and CAM-related genes in PC3 cells (Table I). This is consistent with the results of the PCR analysis conducted in the present study.
The amplitude and type of expression change in ECM- and CAM-related genes varies between disease groups and disorders (38). The transition from healthy epithelial cells to PC3 cells is associated with a decrease in ITGB4 and MMP7 expression, and the transition from prostatic intraepithelial neoplasia to PCa is associated with a downregulation in COL6A1 and ITGB2 expression (38). Loss of the cell adhesion molecule, LAMA3, is associated with high Gleason scores, elevated preoperative PSA levels and advanced stages of PCa (39). Reduced CTGF expression promotes cell growth, migration and invasion of nasopharyngeal carcinoma (45). Elevated COL11A1 expression is detected malignant lesions of the stomach (46) and in sporadic colorectal tumors (47). COL11A1 also promotes tumor progression and is associated with a poor survival outcome in patients with ovarian cancer (48). Among ECM-related genes, an increase in MMP2 expression promotes cell invasion and PCa progression (35–37), and an upregulation of MMP12 is associated with the degradation and invasion of type I collagen in PCa cells (49). In accordance with previous reports (35–37,46–49), the results of the present study suggested that SM treatment enhances the expression of COL11A, MMP2 and MMP12 in PC3 cells (Tables I and II). These finding suggested that SM-mediated alteration of these molecules may be associated with the progression of PCa.
To the best of our knowledge, the present study was the first to examine the direct effects of cigarette smoke on PCa progression. The results of the present study are consistent with those of with epidemiological studies linking cigarette smoking to PCa progression. CAM- and ECM-related genes may serve as biomarkers for understanding PCa progression in smokers. In addition, the present study suggested that MMP inhibitors may be useful therapeutic treatments to prevent PCa progression.
Acknowledgments
This study was supported by grants from the Flight Attendant Medical Research Institute (S.S) and National Institutes of Health [grant no. 2R56HL09211 (S.T.)].
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Chan JM, Jou RM, Carroll PR. The relative impact and future burden of prostate cancer in the United States. J Urol. 2004;172:S13–S16. doi: 10.1097/01.ju.0000142068.66876.53. [DOI] [PubMed] [Google Scholar]
- 3.Huncharek M, Haddock KS, Reid R, Kupelnick B. Smoking as a risk factor for prostate cancer: a meta-analysis of 24 prospective cohort studies. Am J Public Health. 2010;100:693–701. doi: 10.2105/AJPH.2008.150508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart SL, Cardinez CJ, Richardson LC, et al. Surveillance for cancers associated with tobacco use - United States, 1999–2004. MMWR Surveill Summ. 2008;57:1–33. [PubMed] [Google Scholar]
- 5.Colditz G. Consensus conference: smoking and prostate cancer. Cancer Causes Control. 1996;7:560–562. doi: 10.1007/BF00051890. [DOI] [PubMed] [Google Scholar]
- 6.Hickey K, Do KA, Green A. Smoking and prostate cancer. Epidemiol Rev. 2001;23:115–125. doi: 10.1093/oxfordjournals.epirev.a000776. [DOI] [PubMed] [Google Scholar]
- 7.Rohrmann S, Linseisen J, Allen N, et al. Smoking and the risk of prostate cancer in the European Prospective Investigation into Cancer and Nutrition. Br J Cancer. 2013;108:708–714. doi: 10.1038/bjc.2012.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zu K, Giovannucci E. Smoking and aggressive prostate cancer: a review of the epidemiologic evidence. Cancer Causes Control. 2009;20:1799–1810. doi: 10.1007/s10552-009-9387-y. [DOI] [PubMed] [Google Scholar]
- 9.Roomi MW, Kalinovsky T, Rath M, et al. Down-regulation of urokinase plasminogen activator and matrix metalloproteinases and up-regulation of their inhibitors by a novel nutrient mixture in human prostate cancer cell lines PC-3 and DU-145. Oncol Rep. 2011;26:1407–1413. doi: 10.3892/or.2011.1434. [DOI] [PubMed] [Google Scholar]
- 10.Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J. 1990;4:1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]
- 11.Barsky SH, Siegal GP, Jannotta F, et al. Loss of basement membrane components by invasive tumors but not by their benign counterparts. Lab Invest. 1983;49:140–147. [PubMed] [Google Scholar]
- 12.Liotta LA, Tryggvason K, Garbisa S, et al. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature. 1980;284:67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]
- 13.Liotta LA, Rao CN, Barsky SH. Tumor invasion and the extracellular matrix. Lab Invest. 1983;49:636–649. [PubMed] [Google Scholar]
- 14.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 15.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 16.Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 17.Nelson AR, Fingleton B, Rothenberg ML, et al. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol. 2000;18:1135–1149. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- 18.Bérubé M, Deschambeault A, Boucher M, et al. MMP-2 expression in uveal melanoma: differential activation status dictated by the cellular environment. Mol Vis. 2005;11:1101–1111. [PubMed] [Google Scholar]
- 19.Garzetti G, Ciavattini A, Lucarini G, et al. Tissue and serum metalloproteinase (MMP-2) expression in advanced ovarian serous cystoadenocarcinomas: clinical and prognostic implications. Anticancer Res. 1995;15:2799–2804. [PubMed] [Google Scholar]
- 20.Gohji K, Fujimoto N, Hara I, Fujii A, Gotoh A, et al. Serum matrix metalloproteinase-2 and its density in men with prostate cancer as a new predictor of disease extension. Int J Cancer. 1998;79:96–101. doi: 10.1002/(SICI)1097-0215(19980220)79:1<96::AID-IJC18>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 21.Lokeshwar BL. MMP inhibition in prostate cancer. Ann NY Acad Sci. 1999;878:271–289. doi: 10.1111/j.1749-6632.1999.tb07690.x. [DOI] [PubMed] [Google Scholar]
- 22.Shariat SF, Karam JA, Roehrborn CG. Blood biomarkers for prostate cancer detection and prognosis. Future Oncol. 2007;3:449–461. doi: 10.2217/14796694.3.4.449. [DOI] [PubMed] [Google Scholar]
- 23.Achbarou A, Kaiser S, Tremblay G, et al. Urokinase overproduction results in increased skeletal metastasis by prostate cancer cells in vivo. Cancer Res. 1994;54:2372–2377. [PubMed] [Google Scholar]
- 24.Pryor WA, Stone K. Oxidants in cigarette smoke. Radicals, hydrogen peroxide, peroxynitrate, and peroxynitrite. Ann NY Acad Sci. 1993;686:12–27. doi: 10.1111/j.1749-6632.1993.tb39148.x. [DOI] [PubMed] [Google Scholar]
- 25.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 26.Birrane G, Li H, Yang S, et al. Cigarette smoke induces nuclear translocation of heme oxygenase 1 (HO-1) in prostate cancer cells: nuclear HO-1 promotes vascular endothelial growth factor secretion. Int J Oncol. 2013;42:1919–1928. doi: 10.3892/ijo.2013.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aragon-Ching JB. Active surveillance for prostate cancer: has the time finally come? J Clin Oncol. 2010;28:e265–e266. doi: 10.1200/JCO.2010.28.1584. [DOI] [PubMed] [Google Scholar]
- 28.Dilly AK, Ekambaram P, Guo Y, et al. Platelet-type 12-lipoxygenase induces MMP9 expression and cellular invasion via activation of PI3K/Akt/NF-κB. Int J Cancer. 2013;133:1784–1791. doi: 10.1002/ijc.28165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian TV, Tomavo N, Huot L, et al. Identification of novel TMPRSS2:ERG mechanisms in prostate cancer metastasis: involvement of MMP9 and PLXNA2. Oncogene. 2014;24:2204–2214. doi: 10.1038/onc.2013.176. [DOI] [PubMed] [Google Scholar]
- 30.Delassus GS, Cho H, Hoang S, Eliceiri GL. Many new down- and up-regulatory signaling pathways, from known cancer progression suppressors to matrix metalloproteinases, differ widely in cells of various cancers. J Cell Physiol. 2010;224:549–558. doi: 10.1002/jcp.22157. [DOI] [PubMed] [Google Scholar]
- 31.Verslegers M, Lemmens K, Van Hove I, Moons L. Matrix metalloproteinase-2 and -9 as promising benefactors in development, plasticity and repair of the nervous system. Prog Neurobiol. 2013;105:60–78. doi: 10.1016/j.pneurobio.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Jinka R, Kapoor R, Sistla PG, Raj TA, Pande G. Alterations in cell-extracellular matrix interactions during progression of cancers. Int J Cell Biol. 2012;2012:219196. doi: 10.1155/2012/219196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo SY, Kwon SM. Angiogenesis and its therapeutic opportunities. Mediators Inflamm. 2013;2013:127170. doi: 10.1155/2013/127170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grässel S, Bauer RJ. Collagen XVI in health and disease. Matrix Biol. 2013;32:64–73. doi: 10.1016/j.matbio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Goel HL, Li J, Kogan S, Languino LR. Integrins in prostate cancer progression. Endocr Relat Cancer. 2008;15:657–664. doi: 10.1677/ERC-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao LJ, Lin P, Lin F, et al. ADAM17 targets MMP-2 and MMP-9 via EGFR-MEK-ERK pathway activation to promote prostate cancer cell invasion. Int J Oncol. 2012;40:1714–1724. doi: 10.3892/ijo.2011.1320. [DOI] [PubMed] [Google Scholar]
- 38.Ashida S, Nakagawa H, Katagiri T, et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res. 2004;64:5963–5972. doi: 10.1158/0008-5472.CAN-04-0020. [DOI] [PubMed] [Google Scholar]
- 39.Sathyanarayana UG, Padar A, Suzuki M, et al. Aberrant promoter methylation of laminin-5-encoding genes in prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2003;9:6395–6400. [PubMed] [Google Scholar]
- 40.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- 41.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/S0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 42.Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 1998;8:51–54. doi: 10.1016/S0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhen Y, Ye Y, Yu X, et al. Reduced CTGF expression promotes cell growth, migration, and invasion in nasopharyngeal carcinoma. PLoS One. 2013;8:e64976. doi: 10.1371/journal.pone.0064976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jinka R, Kapoor R, Sistla PG, Raj TA, Pande G. Alterations in cell-extracellular matrix interactions during progression of cancers. Int J Cell Biol. 2012;2012:219196. doi: 10.1155/2012/219196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okegawa T, Pong RC, Li Y, Hsieh JT. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim Pol. 2004;51:445–457. [PubMed] [Google Scholar]
- 46.Zhao Y, Zhou T, Li A, et al. A potential role of collagens expression in distinguishing between premalignant and malignant lesions in stomach. Anat Rec (Hoboken) 2009;292:692–700. doi: 10.1002/ar.20874. [DOI] [PubMed] [Google Scholar]
- 47.Fischer H, Salahshor S, Stenling R, et al. COL11A1 in FAP polyps and in sporadic colorectal tumors. BMC Cancer. 2001;1:17. doi: 10.1186/1471-2407-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu YH, Chang TH, Huang YF, et al. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33:3432–3440. doi: 10.1038/onc.2013.307. [DOI] [PubMed] [Google Scholar]
- 49.Nabha SM, dos Santos EB, Yamamoto HA, et al. Bone marrow stromal cells enhance prostate cancer cell invasion through type I collagen in an MMP-12 dependent manner. Int J Cancer. 2008;122:2482–2490. doi: 10.1002/ijc.23431. [DOI] [PMC free article] [PubMed] [Google Scholar]