Abstract
Mucin 1 (MUC1) is an oncogene that has a crucial role in the pathogenesis and progression of the majority of epithelial malignant tumors. Our previous study demonstrated that MUC1 gene silencing inhibited the growth of SMMC-7721 cells in vitro and in vivo, however, whether this growth inhibition is associated with apoptotic cell death remains to be elucidated. In the present study, it was found that MUC1 gene silencing not only resulted in the inhibition of SMMC-7721 cell growth, determined using a clone formation assay in vitro and a tumor xenograft mouse model with an in vivo imaging system, but also induced apoptotic alterations in SMMC-7721 cells, determined using Hoechst 33342 staining, flow cytometry with an Annexin V-PE staining and a DNA ladder assay. Further investigation using western blotting revealed that cytochrome c was released from the mitochondria into the cytoplasm, and caspase-8 and caspase-9 were activated in MUC1 gene-silenced SMMC-7721 cells. The pro-apoptotic protein Bcl-2-associated X protein (Bax) and the tumor suppressor p53 were increased, while the anti-apoptotic protein B-cell lymphoma 2 was decreased in MUC1 gene-silenced cells. In addition, results from the co-immunoprecipitation experiments demonstrated that the MUC1 cytoplasmic tail can bind directly to Bax or caspase-8 and these interactions were reduced upon MUC1 gene silencing in SMMC-7721 cells. The above results indicate that MUC1 gene silencing induces growth inhibition in SMMC-7721 cells through Bax-mediated mitochondrial and caspase-8-mediated death receptor apoptotic pathways.
Keywords: mucin 1, gene silencing, human hepatoma, apoptosis, Bcl-2-associated X protein, caspase-8
Introduction
Mucin 1 (MUC1) is a highly glycosylated transmembrane protein expressed on the apical surface of epithelial cells and is aberrantly overexpressed in the majority of malignant epithelial tumors, including breast, lung, ovarian, prostate and pancreatic cancer and several types of malignant hematological tumor (1). Previous studies have demonstrated that MUC1 is an oncoprotein that is involved in the regulation of carcinogenesis, tumor progression, invasion and metastasis of cancer (2). MUC1 consists of a large extracellular N-terminal subunit containing a variable number of tandem repeats region and a C-terminal subunit that resides on the cell surface as a heterodimeric complex via a strong noncovalent interaction. The C-terminal subunit is composed of a 58 amino acid extracellular domain, a 28 amino acid transmembrane domain and a 72 amino acid cytoplasmic tail (CT) (3). MUC1-CT is involved in numerous signaling pathways, including Wnt/β-catenin (4), c-Src (5), Grb2/Sos (6), glycogen synthase kinase 3 β (4), epidermal growth factor receptor (7,8) and nuclear factor-κB (9,10), which regulate the processes of cell survival and proliferation. By contrast, MUC1 interacts directly with several signaling molecules, including p53 (11,12), HSP70/90 (13,14) and caspase-8 (15) to regulate cell apoptosis.
Hepatocellular carcinoma (HCC) is one of the most common types of malignant tumor that severely threatens human health and quality of life. The occurrence of HCC is associated directly or indirectly with the abnormal expression of multiple genes, including B lymphoma Moloney murine leukemia virus insertion region 1 homolog (BMI1) (16), glypican-3 (17), heat shock protein 70 (18) and Sal-like protein 4 (19). Previous studies have demonstrated that MUC1 is also expressed in HCC cells and tissues (20–22), however, the mechanisms underlying the function MUC1 in the development of HCC remain to be elucidated. In order to determine the oncogenic role of MUC1 in HCC, MUC1 was knocked down in SMMC-7721 cells, using small interfering RNA (siRNA), to examine the effects and mechanisms of MUC1 gene silencing in HCC cells. Our previous study found that MUC1 gene silencing altered the phenotypic characteristics of the human hepatic cellular carcinoma cell line SMMC-7721 (23). In addition, cell proliferation was inhibited significantly in MUC1 gene-silenced SMMC-7721 cells. Furthermore, no tumor development was observed in mice injected with MUC1 gene-silenced SMMC-7721 cells. The results indicated that MUC1 gene silencing can inhibit the growth of SMMC-7721 cells.
The aim of the present study was to determine whether growth inhibition of MUC1 gene-silenced SMMC-7721 cells is associated with their apoptotic cell death. Furthermore, the aim was to elucidate the mechanisms and pathways of this apoptosis, which could provide a novel therapeutic target for the pathogenesis and gene therapy of HCC.
Materials and methods
Cell culture
The human HCC cell line, SMMC-7721, was purchased from the Cell Bank of the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Iscove's modified Dulbecco's medium (Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with heat-inactivated 10% fetal bovine serum (Invitrogen Life Technologies), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen Life Technologies), and maintained at 37°C and 5% CO2. MR1-C6 and MR1-D4, two stably transfected SMMC-7721 cell clones, were established using MUC1 gene specific siRNAs, as previously described (9). A clone stably transfected with a scramble siRNA was used as a negative control (NC). These transfected cells were maintained in the presence of 600 µg/ml G418.
Plate clone formation assay
Approximately 2,000 cells were plated per well in a 6-well plate and each group was analyzed in triplicate. Following incubation at 37°C and 5% CO2 for 3 weeks, the cells were gently washed twice in phosphate-buffered saline (PBS) and fixed in methanol for 15 min. Subsequently, the cell clones were stained with Giemsa staining solution for 30 min, followed by air drying. The visualized cell clones were observed and images were captured using a microscope (IX71; Olympus Corporation, Toyko, Japan).
Tumor xenograft mouse model and in vivo imaging
For the tumor xenograft mouse model construction, 15 BALB/c nude mice (4–6 weeks old; 18–20 g) were purchased from Beijing HFK Bioscience Co., Ltd. (Beijing, China). Animals were maintained in specific pathogen-free conditions and an environment with controlled conditions of light and humidity, and free access to water and a standard laboratory diet. Animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Bethesda, MD, USA). The present study was approved by the ethics committee of the Scientific Investigation Board of Science and Technology of Jilin Province (Changchun, China). Mice were randomly divided into four groups (five animals per group), including the SMMC-7721 group, the NC group, the MR1-D4 group and the MR1-D9 group. Cells (2×106) were subcutaneously injected into the right flank of each mouse. For real-time near-infrared (NIR) imaging, the mice were anesthetized and imaged on day 21 post-injection, and in vivo NIR images were obtained using a Xenogen IVIS Spectrum system (PerkinElmer, Inc., Waltham, MA, USA) using Living Image software version 3.0 (Caliper Life Sciences, Alameda, CA, USA). All image analysis and NIR fluorescent signal quantification was performed using the region of interest (ROI) function of Living Image software (Caliper Life Sciences).
Hoechst 33342 staining
Cells at logarithmic growth stage were cultured in a 96-well plate for 48 h and stained with Hoechst 33342 dye (Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 0.1 µg/ml at 37°C for 15 min in the dark. Cells were observed with an inverted fluorescence microscope (IX71; Olympus Coproration).
Annexin V-PE staining and flow cytometry
Following culturing the cells for 48 h, cells were digested with 0.25% trypsin (EDTA free) and washed twice with PBS. Cells (5×105) were centrifuged (300 x g for 5 min) and the cell pellets were stained with Annexin V-PE fluorescence labeling solution (Annexin V-PE apoptosis assay kits purchased from Nanjing KeyGen Biotech. Co., Ltd., Nanjing, China). Following incubation in the dark for 5-15 min, flow cytometric analysis was performed on a FACS Calibur Flow cytometer (BD Biosciences, San Jose, CA, USA). Data acquisition and analysis were performed using CellQuest version 4.0 software (BD Biosciences).
DNA ladder assay
Cells were cultured for 48 h, 5×106 cells were harvested and the DNA was extracted using an Apoptotic DNA Ladder Extraction kit (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's instructions. Electrophoresis of the DNA was performed in a 1% agarose gel.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies) and reverse-transcribed into cDNA (Takara Bio, Inc., Shiga, Japan), according to the manufacturer's instructions. RT-qPCR was performed using FastStart Universal SYBR-Green Master (Rox; Roche Life Science, Indianapolis, IN, USA) in an Applied Biosystems 7300 system (Applied Biosystems, Foster City, CA, USA). All the primers that were used for RT-qPCR analysis were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). The primers used were as follows: MUC1, forward 5′-TGC CGC CGA AAG AAC TACG-3′ and reverse 5′-TGG GGT ACT CGC TCA TAG GAT-3′; p53, forward 5′-TTG CAA TAG GTG TGC GTC AGA-3′ and reverse 5′-AGT GCA GGC CAA CTT GTT CAG-3′; B-cell lymphoma 2 (Bcl-2), forward 5′-TCA GGG ACG GGG TGA ACT-3′ and reverse 5′-CAG GTG CCG GTT CAG GTA CTC-3′; Bcl-2-associated X protein (Bax), forward 5′-CGC CGT GGA CAC AGA CTC-3′ and reverse 5′-CCT CCC TTC AAC ACT TCC T-3′; β-actin, forward 5′-AGT TGC GTT ACA CCC TTTC-3′ and reverse 5′-CCT TCA CCG TTC CAG TTT-3′. The PCR assays were performed in triplicate on a step-one RT-qPCR system. Cycle threshold (Ct) values were normalized to β-actin. The relative level of the mRNAs was calculated by 2−Δct (Δct=cttarget mRNA − ctβ-actin).
Western blotting
Western blotting was performed as previously described (24). Briefly, cells were lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology) and the protein concentration in the cell lysates was measured using the BCA protein assay kit (Beyotime Institute of Biotechnology). Cytosolic and mitochondrial proteins were extracted using cytosolic and mitochondrial protein extraction kits (Nanjing KeyGen Biotech. Co., Ltd.), according to the manufacturer's instructions. Equivalent quantities of protein from the soluble fractions of the cell lysates were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Following blocking with 5% non-fat milk in Tris-buffered saline containing 0.05% Tween 20 for 1 h, the membranes were incubated with appropriately diluted primary antibodies at 4°C overnight. The following primary antibodies were used: Rabbit anti-human caspase-3 monoclonal antibody (1:1,000; cat. no. sc-7148; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), rabbit anti-human Bax (cat. no. 50599-2-Ig; Proteintech, Chicago, IL, USA) and p53 (cat. no. 10442-1-AP; Proteintech) polyclonal antibodies (1:1,000), rabbit anti-human β-actin (1:2,000; cat. no. 5779-1), Bcl-2 (cat. no. 1017-1), cytochrome c (cat. no. 1896-1), Cox IV (cat. no. 7001-1), caspase-8 (cat. no. 1007-1), caspase-9 (cat. no. 1023-1)and poly (ADP-ribose) polymerase (PARP) (cat. no. 1051-1) monoclonal antibodies (1:1,000; Epitomics, Burlingame, CA, USA). The blots were then probed with horseradish peroxidase (HRP)-conjugated secondary antibody (HRP-conjugated goat anti rabbit IgG antibody; cat. no. RABHRP2; Sigma-Aldrich) at 1:2,000 for 1 h at room temperature. Immunoreactive bands were detected by enhanced chemiluminescence (Thermo Fisher Scientific, Waltham, MA, USA). The intensities of the bands were quantified by densitometry using ImageJ 1.49 d software (National Institutes of Health).
Co-immunoprecipitation
Cell lysates were first pre-cleaned with protein G agarose beads (Promega, Madison, WI, USA) for 3 h at 4°C and, subsequently, equal quantities of sample lysates were incubated with either 1.0 µg of mouse IgG or anti-MUC1-CT antibody (hamster anti MUC1-CT monoclonal antibody; Neomarkers Inc., Fremont, CA, USA) for 16 h at 4°C, followed by precipitation with protein G agarose beads. The precipitated proteins from the cell lysates or the whole cell lysates were subjected to immunoblot analysis with anti-Bax and anti-caspase-8 antibodies.
Statistical analysis
The data are presented as the mean ± standard deviation from three independent experiments. Statistical analysis was performed using SPSS 11.0 (SPSS, Inc., Chicago, IL, USA). The differences between groups were compared using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
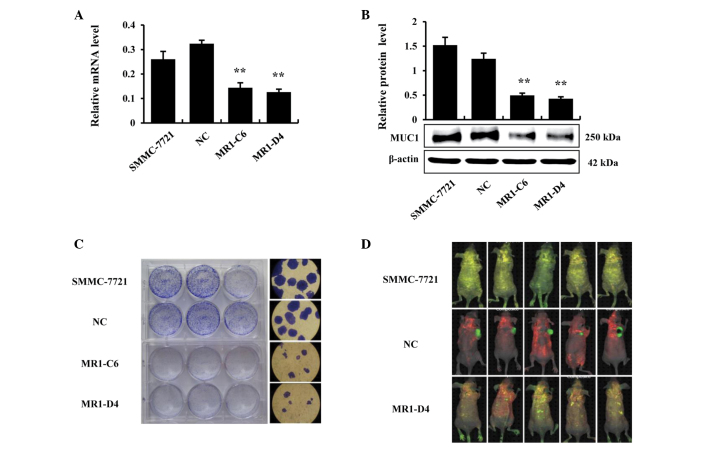
MUC1 gene silencing inhibits the growth of SMMC-7721 cells in vitro and in vivo
In our previous study, the expression of MUC1 was knocked down in the human HCC cell line SMMC-7721 by RNA interference. Two independent MUC1-knockdown clones were established, designated MR1-C6 and MR1-D4, and one negative control clone, which was designated NC, was established, as previously described (23). The effects of MUC1 gene silencing were validated by RT-qPCR and western blotting, and the results demonstrated that the expression of MUC1 in MR1-C6 and MR1-D4 cells was significantly reduced, compared with SMMC-7721 or NC cells (P<0.01; Fig. 1A and B). Subsequently, a clone formation assay was conducted to confirm that MUC1 gene silencing could inhibit the growth of SMMC-7721 cells in vitro. The results demonstrated that the clones in the MR1-C6 and MR1-D4 groups were significantly fewer in number and smaller in size, compared with those in the SMMC-7721 or NC groups (Fig. 1C). Furthermore, SMMC-7721 cells were implanted into BALB/c nude mice to investigate the inhibitory effects of MUC1 gene silencing on the growth of SMMC-7721 cells in vivo. By utilizing an in vivo imaging system, it was observed that the tumors in the MR1-D4 group were absent, compared with the NC group (Fig. 1D). Taken together, these results indicate that MUC1 gene silencing significantly inhibits the growth of SMMC-7721 cells in vitro and in vivo.
Figure 1.
MUC1 gene silencing inhibits the growth of SMMC-7721 cells in vitro and in vivo. (A) The mRNA level of MUC1 was detected by reverse transcription quantitative polymerase chain reaction and normalized to the internal control, β-actin. **P<0.01, compared with the NC. (B) The protein level of MUC1 was detected by western blotting and normalized to the internal control β-actin. Bars represent the relative protein level as calculated by the ratio of MUC1/β-actin. **P<0.01, compared with the NC. (C) A clone formation assay was conducted in a 6-well plate. Following incubation at 37°C and 5% CO2 for 3 weeks, images were captured. (D) SMMC-7721, NC and MR1-D4 cells (2×106 cells) were subcutaneously injected into BALB/c nude mice. The tumors of live mice were observed using an in vivo imaging system 5 weeks after injection. MUC1, mucin 1; NC, negative control.
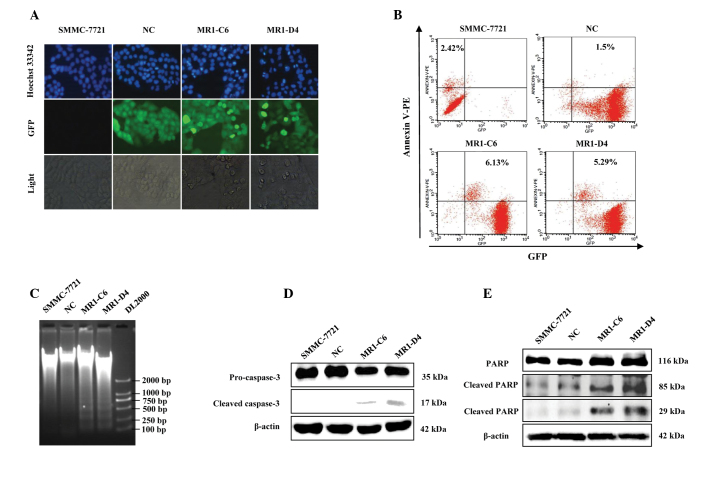
MUC1 gene silencing induces phenotypic alterations of apoptosis in SMMC-7721 cells
Subsequently, whether the growth inhibition of MUC1 gene-silenced SMMC-7721 cells was associated with their apoptotic death was determined. Using Hoechst 33342 staining, it was demonstrated that significantly more cells were undergoing characteristic apoptotic alterations, including nuclear condensation, nuclear fragmentation and darker dye staining in the MR1-C6 and MR1-D4 groups, however, these changes were not observed in the NC group (Fig. 2A). Annexin V is a calcium-dependent phospholipid binding protein and can bind to the plasma membrane in the early stages of apoptosis. It is one of a number of sensitive methods used to detect early apoptosis. Flow cytometric analysis demonstrated that the percentage of apoptotic cells in the MUC1 gene-silenced group (MR1-C6, 6.13%; MR1-D4, 5.29%) was markedly higher than that observed in the control group (SMMC-7721, 2.42%; NC, 1.5%; Fig. 2B). DNA fragmentation is a characteristic alteration observed during the late stage of apoptosis and can be detected using a DNA ladder assay. DNA Ladder extraction kits were used to extract the genomic DNA of SMMC-7721, NC, MR1-C6 and MR1-D4 cells. The DNA ladder was observed on 1% agarose electrophoresis gel in the MR1-C6 and MR1-D4 cells, but not in the SMMC-7721 or NC cells (Fig. 2C). Furthermore, in order to confirm that MUC1 gene silencing induced apoptosis of SMMC-7721 cells, the expression and activation of caspase-3, and its substrate PARP, were detected by western blotting. The results demonstrated that the cleaved products of caspase-3 and PARP were expressed in the MR1-C6 and MR1-D4 cells, but not in the SMMC-7721 or NC cells (Fig. 2D and E). The morphological and biochemical alterations observed in these cells indicate that MUC1 gene silencing can induce apoptosis of SMMC-7721 cells.
Figure 2.
MUC1 gene silencing induces apoptotic phenotypic alterations in SMMC-7721 cells. (A) Detection of cell nuclear condensation by immunofluorescence with Hoechst 33342 staining. GFP was expressed stably in NC, MR1-C6 and MR1-D4 cells. (B) Apoptotic cells were analyzed by flow cytometry with Annexin V-PE staining. (C) Agarose gel electrophoresis of cellular genomic DNA fragmentation. DL2000 was the DNA marker. (D) Expression of caspase-3 and its activating products was determined by western blotting. (E) Expression of PARP and its cleaved products was determined by western blotting. MUC1, mucin 1; NC, negative control; PARP, poly (ADP ribose) polymerase; GFP, green fluorescent protein.
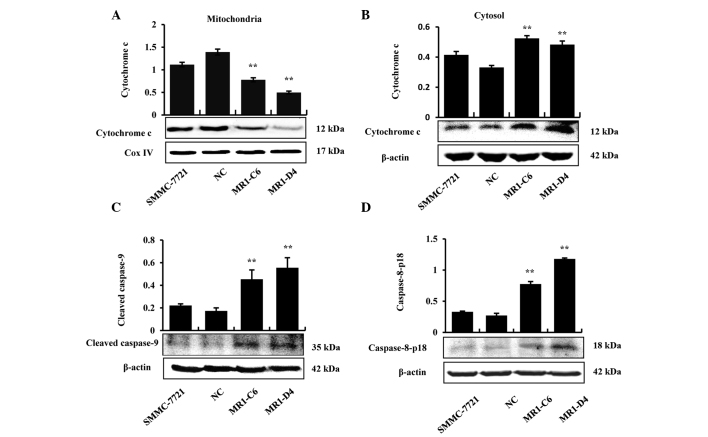
MUC1 gene silencing induces apoptosis of SMMC-7721 cells through mitochondrial and death receptor apoptotic pathways
Proteins were extracted from the mitochondria and cytoplasm to determine whether cytochrome c was released in MUC1 gene-silenced SMMC-7721 cells, and the results demonstrated that cytochrome c was only released from the mitochondria into the cytoplasm in the MR1-C6 and MR1-D4 groups (Fig. 3A and B). In addition, the results demonstrated that caspase-9, an initiator caspase, which is associated with the mitochondrial death pathway, was activated in the MR1-C6 and MR1-D4 cells (Fig. 3C). The results from western blotting demonstrated that pro-caspase-8 and cleaved caspase-8 increased in the MR1-C6 and MR1-D4 groups, compared with the control groups (Fig. 3D). The above results indicate that MUC1 gene silencing induces apoptosis in SMMC-7721 cells through the mitochondrial and death receptor apoptotic pathways.
Figure 3.
MUC1 gene silencing induces apoptosis of SMMC-7721 cells through mitochondrial and death receptor apoptotic pathways. (A) Mitochondrial extracts were assessed by western blotting to detect the levels of cytochrome c in the mitochondria. (B) Cytosol extracts were assessed by western blotting to detect the levels of cytoplasmic cytochrome c. (C) Western blotting for the expression of cleaved caspase-9. (D) Western blotting for the expression of caspase-8. Cox IV served as the mitochondrial loading control; β-actin was used as a protein loading control for whole cell lysis or cytoplasmic extracts. **P<0.01, compared with the NC. MUC1, mucin 1; NC, negative control.
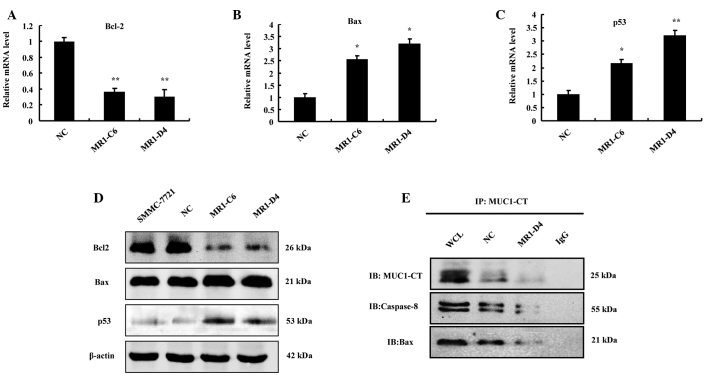
To further analyze the molecular mechanisms of apoptosis in SMMC-7721, as induced by MUC1 gene silencing, the expression of the pro-apoptotic protein Bax, the anti-apoptotic protein Bcl-2 and the tumor suppressor p53 was measured at the transcriptional and translational levels. RT-qPCR demonstrated that Bax and p53 significantly increased (P<0.05), while Bcl-2 significantly decreased (P<0.01) in the MR1-C6 and MR1-D4 groups, compared with the NC group (Fig. 4A–C). Western blotting revealed similar results to RT-qPCR (Fig. 4D). These results suggest that MUC1 gene silencing can regulate the expression and activation of proteins associated with the mitochondrial and death receptor apoptotic pathways.
Figure 4.
MUC1 gene silencing decreases MUC1-CT binding with Bax and caspase-8 directly to induce apoptosis of SMMC-7721 cells. (A-C) Relative mRNA levels of Bcl-2, Bax and p53 (fold change) compared with the NC, *P<0.05, **P<0.01. (D) Western blotting for the expression of p53, Bcl-2 and Bax. (E) Cell lysates were subjected to immunoprecipitation with the anti-MUC1-CT antibody or normal IgG and then immunoblotted with anti-caspase-8 and anti-Bax antibodies. Whole cell lysate was not subjected to immunoprecipitation. MUC1, mucin 1; NC, negative control; Bax, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma 2; CT, cytoplasmic tail; IgG, immunoglobulin G.
MUC1 gene silencing decreases MUC1-CT binding to Bax and caspase-8
To further investigate the mechanisms of SMMC-7721 cell apoptosis induced by MUC1 knockdown (15), co-immunoprecipitation experiments were conducted to determine the interaction between MUC1-CT and Bax or caspase-8 in SMMC-7721 cells. The results demonstrated that MUC1-CT can bind directly to Bax or caspase-8 and the interaction was reduced markedly in the MUC1 gene-silenced group (Fig. 4E). Taken together, these results suggest that MUC1 gene silencing can reduce the interaction of MUC1-CT with Bax or caspase-8, therefore resulting in activation of the mitochondrial and death receptor apoptotic pathways.
Discussion
Our previous study found that the knockdown of MUC1 in SMMC-7721 cells can significantly inhibit cell proliferation and induce cell cycle arrest (23). In the present study, a clone formation assay in vitro and a tumor xenograft mouse model with an in vivo imaging system were utilized and the growth inhibitory effects of MUC1 silencing in SMMC-7721 cells were further confirmed. Since MUC1 is an oncoprotein, it is not only involved in the regulation of carcinogenesis and tumor progression, invasion and metastasis (2), but it is also important in the inhibition of cell apoptosis (25-27). Consequently, the present study examined in depth whether the growth inhibition of MUC1 gene-silenced cells is associated with apoptotic cell death. Hoechst 33342 staining, flow cytometry with Annexin V-PE staining and a DNA Ladder assay were performed, and all results consistently demonstrated that MUC1 gene silencing induces cells to undergo apoptosis. These results are similar to results from a previous study, in which a MUC1-downregulated gastric carcinoma cell line was found to be more apoptotic compared with the controls (28). Since caspase activation has a key role in the process of apoptosis, caspase-3, in particular, is activated during early apoptosis. Once activated, caspase-3 can increase its substrate, PARP, which is an indicator of apoptosis. The expression of caspase-3 and PARP was detected by western blotting and the results demonstrated that cleaved caspase-3 and PARP were present, suggesting that caspase-3 was activated in MUC1 gene-silenced cells. Taken together, these results indicate that MUC1 gene silencing can not only inhibit the growth of SMMC-7721 cells but can also induce apoptosis of these cells.
Generally, cell apoptosis involves two major signaling pathways, including the mitochondrial apoptotic pathway and the death receptor apoptotic pathway (29). The intrinsic mitochondria apoptotic pathway is often characterized by the release of cytochrome c from the mitochondrial intermembrane space into the cytoplasm and activation of caspase-9. Bcl-2 is a major anti-apoptotic protein and Bax is a pro-apoptotic protein. These two proteins have been identified as major regulators in the mitochondrial apoptotic pathway. The tumor suppressor p53 can directly activate Bax and downregulate Bcl-2, in the absence of other proteins, to permeabilize mitochondria and engage the apoptotic program (30,31). In the present study, it was demonstrated that MUC1 gene silencing in SMMC-7721 cells could induce apoptosis, and further experiments revealed that MUC1 gene silencing induced cytochrome c release from the mitochondria to the cytoplasm and that caspase-9 was also activated. In parallel, the expression of Bax and p53 were also significantly increased, while the expression of Bcl-2 was markedly reduced, as compared with the control group, suggesting that the intrinsic mitochondrial apoptotic pathway was activated in MUC1 gene-silenced cell lines. It is well established that caspase-8, an initiator caspase, is required to activate the membrane bound receptor-mediated extrinsic apoptotic signaling pathway. To investigate whether the receptor-mediated apoptotic pathway is simultaneously activated with the mitochondrial apoptotic pathway when cell apoptosis occurs, the expression of caspase-8 was examined. The results demonstrated that caspase-8 was activated in MUC1 gene-silenced cell lines, suggesting that the receptor apoptotic pathway is also involved in MUC1 gene silencing-induced cell apoptosis. Ahmad et al reported that MUC1-CT could bind directly to Bax or caspase-8 in HCT116 cells and in breast cancer MCF7 cells to prevent Bax from localizing to the mitochondria (32) or inhibit the FAS-associated death domain protein from recruiting caspase-8 (15), thus suppressing the activation of proteins that are associated with the mitochondrial or receptor apoptotic pathways. In the present study, co-immunoprecipitation was also conducted to determine whether MUC1-CT could bind to Bax or caspase-8 directly. The results were positive and consistent with previous results, demonstrating that MUC1 gene silencing may reduce the interaction of MUC1-CT with Bax or caspase-8, and thus lead to activation of the mitochondrial or death receptor apoptotic pathway, ultimately resulting in SMMC-7721 cell apoptosis.
In conclusion, the present study demonstrated that MUC1 gene silencing induces the growth inhibition of SMMC-7721 cells via the mitochondrial and death receptor apoptotic pathways, and combined with our previous results, further suggests that MUC1 is important in the progression of HCC development, and thus MUC1 is a potential target for liver cancer therapy.
Acknowledgments
The authors would like to thank Dr O.J. Finn for the pcDNA3-MUC1 plasmid, which was used to transfect the SMMC-7721 cell line. This study was supported by grants from the Double Tenth Engineering of Major Research Project of Jilin Provincial Science and Technology Department (grant no. 20140201012YY) and the Major Development Programs for New Drugs of the Chinese Academy of Sciences during the 12th Five-Year Plan Period (grant no. 2011ZX09102-001-36).
References
- 1.Kufe DW. Functional targeting of the MUC1 oncogene in human cancers. Cancer Biol Ther. 2009;8:1197–1203. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nath S, Mukherjee P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kufe DW. Targeting the human MUC1 oncoprotein: A tale of two proteins. Cancer Biol Ther. 2008;7:81–84. doi: 10.4161/cbt.7.1.5631. [DOI] [PubMed] [Google Scholar]
- 4.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin. J Biol Chem. 2001;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- 6.Pandey P, Kharbanda S, Kufe D. Association of the DF3/MUC1 breast cancer antigen with Grb2 and the Sos/Ras exchange protein. Cancer Res. 1995;55:4000–4003. [PubMed] [Google Scholar]
- 7.Lau SK, Shields DJ, Murphy EA, Desgrosellier JS, Anand S, Huang M, Kato S, Lim ST, Weis SM, Stupack DG, et al. EGFR-mediated carcinoma cell metastasis mediated by integrin αvβ5 depends on activation of c-Src and cleavage of MUC1. PLoS One. 2012;7:e36753. doi: 10.1371/journal.pone.0036753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, Kufe D. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad R, Raina D, Joshi MD, Kawano T, Ren J, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, Gendler S, Bennett EP, Hollingsworth MA. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–26995. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X, Xu H, Kufe D. Human mucin 1 oncoprotein represses transcription of the p53 tumor suppressor gene. Cancer Res. 2007;67:1853–1858. doi: 10.1158/0008-5472.CAN-06-3063. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee S, Mujumdar N, Dudeja V, Mackenzie T, Krosch TK, Sangwan V, Vickers SM, Saluja AK. MUC1c regulates cell survival in pancreatic cancer by preventing lysosomal permeabilization. PLoS One. 2012;7:e43020. doi: 10.1371/journal.pone.0043020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- 15.Agata N, Ahmad R, Kawano T, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–6144. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonemitsu Y, Imazeki F, Chiba T, Fukai K, Nagai Y, Miyagi S, Arai M, Aoki R, Miyazaki M, Nakatani Y, Iwama A, Yokosuka O. Distinct expression of polycomb group proteins EZH2 and BMI1 in hepatocellular carcinoma. Hum Pathol. 2009;40:1304–1311. doi: 10.1016/j.humpath.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2013;125:89–97. doi: 10.1016/S0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 18.Jego G, Hazoumé A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer Lett. 2013;332:275–285. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, Reid LM, Nakauchi H. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bozkaya G, Korhan P, Cokaklı M, Erdal E, Sağol O, Karademir S, Korch C, Atabey N. Cooperative interaction of MUC1 with the HGF/c-Met pathway during hepatocarcinogenesis. Mol Cancer. 2012;11:64. doi: 10.1186/1476-4598-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan SF, Li KZ, Wang L, Dou KF, Yan Z, Han W, Zhang YQ. Expression of MUC1 and its significance in hepatocellular and cholangiocarcinoma tissue. World J Gastroenterol. 2005;11:4661–4666. doi: 10.3748/wjg.v11.i30.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gad A, Tanaka E, Matsumoto A, Wahab MA, Serwah Ael-H, Attia F, Ali K, Hassouba H, el-Deeb Ael-R, Ichijyo T, et al. Assessment of KL-6 as a tumor marker in patients with hepato-cellular carcinoma. World J Gastroenterol. 2005;11:6607–6612. doi: 10.3748/wjg.v11.i42.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li QS, Wang FL, Liu GM, Yuan HY, Chen TX, Wang J, Xie F, Zhai RP, Wang F, Guo YY, et al. Impact of Mucin1 knockdown on the phenotypic characteristics of the human hepatocellular carcinoma cell line SMMC-7721. Oncol Rep. 2014;31:2811–2819. doi: 10.3892/or.2014.3136. [DOI] [PubMed] [Google Scholar]
- 24.Wang FL, Li QS, Ni WH, Fang F, Sun XX, Xie F, Wang J, Wang F, Gao SJ, Tai GX. Expression of human full-length MUC1 inhibits the proliferation and migration of a B16 mouse melanoma cell line. Oncol Rep. 2013;30:260–268. doi: 10.3892/or.2013.2440. [DOI] [PubMed] [Google Scholar]
- 25.Castorina A, Giunta S. Mucin 1 (MUC1) signalling contributes to increase the resistance to cell death in human bronchial epithelial cells exposed to nickel acetate. Biometals. 2014;27:1149–1158. doi: 10.1007/s10534-014-9776-x. [DOI] [PubMed] [Google Scholar]
- 26.Yin L, Kufe T, Avigan D, Kufe D. Targeting MUC1-C is synergistic with bortezomib in downregulating TIGAR and inducing ROS-mediated myeloma cell death. Blood. 2014;123:2997–3006. doi: 10.1182/blood-2013-11-539395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, Li D, Ren J, Li C, Xiao Z. MUC1 activates JNK1 and inhibits apoptosis under genotoxic stress. Biochem Biophys Res Commun. 2013;440:179–183. doi: 10.1016/j.bbrc.2013.09.055. [DOI] [PubMed] [Google Scholar]
- 28.Costa NR, Paulo P, Caffrey T, Hollingsworth MA, Santos-Silva F. Impact of MUC1 mucin downregulation in the phenotypic characteristics of MKN45 gastric carcinoma cell line. PloS One. 2011;6:e26970. doi: 10.1371/journal.pone.0026970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmermann KC, Green DR. How cells die: Apoptosis pathways. J Allergy Clin Immunol. 2001;108(4 Suppl):S99–S103. doi: 10.1067/mai.2001.117819. [DOI] [PubMed] [Google Scholar]
- 30.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 31.Srinivas G, Kusumakumary P, Nair MK, Panicker KR, Pillai MR. Mutant p53 protein, Bcl-2/Bax ratios and apoptosis in paediatric acute lymphoblastic leukaemia. J Cancer Res Clin Oncol. 2000;126:62–67. doi: 10.1007/s004320050010. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad R, Alam M, Rajabi H, Kufe D. The MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX protein and blocks BAX function. J Biol Chem. 2012;287:20866–20875. doi: 10.1074/jbc.M112.357293. [DOI] [PMC free article] [PubMed] [Google Scholar]