Abstract
Schizophrenia (SCZ) is one of the most complex mental illnesses affecting ~1% of the population worldwide. SCZ pathogenesis is considered to be a result of genetic as well as epigenetic alterations. Previous studies have aimed to identify the causative genes of SCZ. However, DNA methylation of long non-coding RNAs (lncRNAs) involved in SCZ has not been fully elucidated. In the present study, a comprehensive genome-wide analysis of DNA methylation was conducted using samples from two male patients with paranoid and undifferentiated SCZ, respectively. Methyl-CpG binding domain protein-enriched genome sequencing was used. In the two patients with paranoid and undifferentiated SCZ, 1,397 and 1,437 peaks were identified, respectively. Bioinformatic analysis demonstrated that peaks were enriched in protein-coding genes, which exhibited nervous system and brain functions. A number of these peaks in gene promoter regions may affect gene expression and, therefore, influence SCZ-associated pathways. Furthermore, 7 and 20 lncRNAs, respectively, in the Refseq database were hypermethylated. According to the lncRNA dataset in the NONCODE database, ~30% of intergenic peaks overlapped with novel lncRNA loci. The results of the present study demonstrated that aberrant hypermethylation of lncRNA genes may be an important epigenetic factor associated with SCZ. However, further studies using larger sample sizes are required.
Keywords: schizophrenia, DNA methylation, protein coding gene, long non-coding RNA
Introduction
Schizophrenia (SCZ) is considered a heritable disorder that affects brain responses to external stimuli (1,2). This abnormality may cause certain psychological disorders and cause a patient to withdraw from society. The precise etiological factors associated with SCZ remain unknown. In clinical practice, SCZ diagnosis predominantly depends on symptoms that are numerous and debilitating (3). A number of SCZ sub-types are distinguishable according to the symptoms that a patient exhibits. Paranoid and undifferentiated SCZ are the most common SCZ sub-types (4). However, the causes of these two SCZ sub-types have remain elusive.
Studies have demonstrated that a number of susceptibility genes may be associated with specific illness sub-types (4). For example, the dystrobrevin-binding protein 1 gene has been shown to be associated with negative and cognitive symptoms in patients with SCZ (4). SCZ is a complex disorder that may be caused by genetic and epigenetic factors (5). The symptoms associated with SCZ are variable. Therefore, there is a requirement to identify biomarkers that may indicate a patients' genetic pre-disposition to SCZ and the possible environmental exposure risks associated with SCZ.
DNA methylation, one of the most common and important epigenetic factors, is influenced by environmental factors and regulates gene expression (6). Numerous studies have demonstrated that DNA methylation is involved in SCZ (7–18). For example, Reelin is a protein which is responsible for neuronal connectivity and synaptic plasticity (19). Abnormal expression of Reelin may result in cognitive deficits in SCZ (19). In brain samples from patients with SCZ, mRNA and protein levels of Reelin were shown to be reduced by ~50% in response to Reelin promoter hypermethylatation compared to those in control samples (8). However, to the best of our knowledge, studies on DNA methylation in SCZ have predominantly focussed on protein-coding genes. Few studies have investigated the involvement of long non-coding RNAs (lncRNAs) in SCZ (20,21).
lncRNA is a type of non-coding RNA that is >200 nucleotides long. Sequence characteristics are similar to those of mRNAs; however, they lack encoding capacity. In mammals, a number lncRNAs have been shown to be associated with SCZ (22,23). The functions of the majority of lncRNAs are unknown. However, studies have demonstrated that lncRNAs are regulators of expression, sub-cellular location and activity of certain protein-coding genes. lncRNAs are associated with a number of biological processes via protein-coding gene regulation, including genomic imprinting control, cell differentiation, immune responses, human diseases and tumorigenesis (24–26). Therefore, alterations in lncRNA gene methylation may result in differential expression patterns and, therefore, affect the expression of target protein-coding genes. Usually, these lncRNAs are anti-sense transcripts and their promoters are located in the intronic region of the corresponding protein coding genes. DNA methylation in lncRNA gene promotor regions may affect the expression of their corresponding target genes by influencing histone modification, which is mediated by binding to histone modification proteins, including polycomb repressive complex 2 and protein regulator of cytokinesis 1 (27). DNA methylation patterns of lncRNA genes have been investigated in developmental biology and in diseases, such as cancer (28,29). However, to the best of our knowledge, no studies on DNA methylation patterns of lncRNA genes in SCZ have been preformed. In the present study, it was hypothesized that epigenetic alterations of lncRNAs may be associated with SCZ.
In the present study, methylated DNA-binding domain-sequencing (MBD-Seq) was used in order to identify DNA hyper-methylated regions in male patients with paranoid or undifferentiated SCZ. DNA methylation levels of protein-coding genes and lncRNAs were measured in peripheral blood samples in order to explore epigenetic biomarkers for diagnosis and provide a foundation for the development of novel anti-psychotic drugs for SCZ.
Materials and methods
Sample collection
Peripheral blood samples were obtained from Ningbo Kangning Hospital (Zhejiang, China) in 2011. Patients were clinically diagnosed by trained psychiatrists using the Diagnostic and Statistical Manual of Mental Disorders, 4th Ed. (30). Case 1 refers to a male patient with paranoid SCZ and case 2 refers to a male patient with undifferentiated SCZ. Patients were verified to be without any serious or unstable medical illness. A healthy male individual was used as a negative control. Table I lists the demographic and clinical characteristics of the subjects in the present study. The negative control male and the supervisors of SCZ patients provided their written informed consent for the study. The present study was approved by the Human Research Ethics Committees of Ningbo University and Ningbo Kangning Hospital (Zhejiang, China).
Table I.
Demographic and clinical data.
| Characteristic | Control | Case 1 | Case 2 |
|---|---|---|---|
| Gender | Male | Male | Male |
| Age (years) | 25 | 34 | 22 |
| Diagnostic subtype | NA | Paranoid | Undifferentiated |
| Age at onset | NA | 19 | 11 |
| Family history | NA | Negative | Positive |
| Antipsychotic drug | NA | Quetiapine | Clozapine |
| Psychotic trauma | NA | Negative | Negative |
MBD enrichment and next-generation sequencing
Methods for MBD-seq were similar to those described in a previous study (31). Genomic DNA was isolated from peripheral blood samples of the three individuals using Qiagen Puregene kit (Qiagen, Hiden, Germany). DNA was then sonicated into 200- to 400-bp fragments by 20 cycles of 30 sec sonication, using a Bioruptor sonicator (Diagenode, Liège, Belgium). Subsequently, the active motif methyl collector kit (Qiagen) was used to complete MBD2 enrichment. According to the manufacturer's protocol, ~1 mg of sonicated genomic DNA was incubated with MBD2-His-conjugated protein and magnetic beads. DNA was purified for each biological replicate, and then pooled post-enrichment. After enrichment, DNA purification columns (Qiagen) were used to purify both the methylated fraction and supernatant fractions, as reported previously (31). Finally, MBD2-enriched genomic DNA from each sample was submitted for high-throughput sequencing analysis. Libraries were sequenced twice using an Illumina Hiseq 2000 platform (Illumina, San Diego, CA, USA), obtaining two datasets of 51- and 101-bp single-end reads, which were used separately in order to identify the MBD2-bound fraction of the human genome, respectively.
Data processing of MBD-seq
MBD-seq reads were filtered using FASTX-Toolkit software with a cutoff of <Q20 in order to obtain high-quality reads. Remaining reads were then mapped to the human genome (hg19; ensembl) using bowtie and <2 mismatches (32). DNA methylation peaks for each sample were identified using MACS software (Miltenyi Biotech, Bergisch Gladbach, Germany) (33) with a threshold of P<10–5. Reads from the healthy sample were used as a negative control for the experiments.
Annotation of DNA methylation peaks
Genomic regions of each peak were compared with the known genes in the hg19 reference genome, including coding and lncRNAs, using the RefSeq database (http://www.ncbi.nlm.nih.gov/refseq/). The regions were assigned to one of six classes associated with known genes using HOMER software (http://homer.salk.edu/homer/microarray/) (34) The six classes of genome regions were intergenic, 3′ untranslated regions (UTR), intron, exon, 5′UTR, promoter-transcript start site (promoter-TSS) and transcript termination site (TTS). Promoter-TSS were identified from −1000 bp to +100 bp, while TTS was identified from −100 bp to +1000 bp. Subsequently, intergenic peaks identified using the RefSeq database were compared with genomic regions of lncRNAs from NONCODE version 4.0 database (http://www.noncode.org/). Images of peak visualization were obtained using Drompa 2.3.1 version software (35).
CpG island analysis
Genomic locations of CpG islands were downloaded from UCSC (https://genome.ucsc.edu/) (36). Genomic regions of each peak were compared with those of CpG islands in order to identify peaks overlapping with or containing CpG islands. The number of CG sites was counted and compared with that in random genomic regions with the same length distribution.
Gene ontology (GO) and pathway enrichment analysis
GO terms and their corresponding depth levels in the GO tree were obtained from the GO database (http://geneontology.org/page/go-database). For each GO term, the P-value was calculated using a hyper-geometric test and adjusted with false rate discovery using the 'multtest' package in R statistics. Enriched GO terms with adjusted P-values <0.05 were then obtained. Enriched Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) pathways were obtained.
Results
Statistical information from the MBD-seq dataset
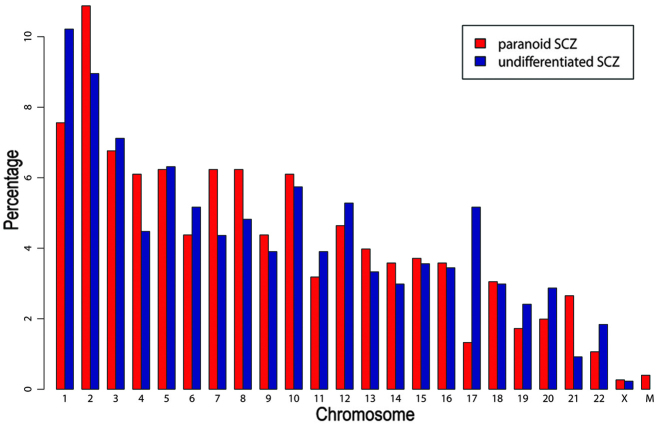
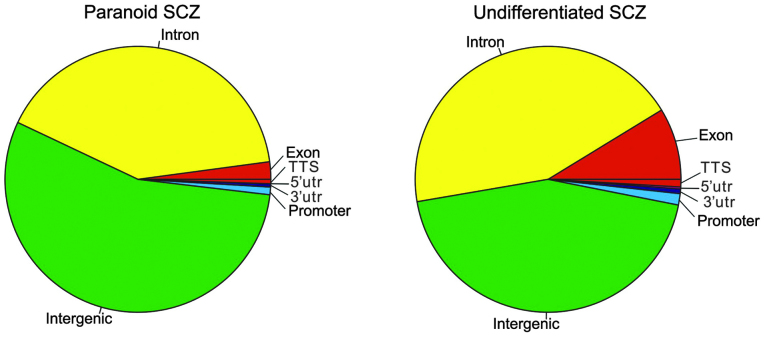
Sequencing was repeated twice and two datasets of MBD-seq were obtained for each SCZ sample. Sample reads were filtered using a cutoff of Q20, which resulted in 40–75% of the original reads, of which 65–90% matched with hg19 human genome reads (Table II). Using MACS software for peak calling, 1,236 and 1,552 peaks were obtained in the first dataset from patients with paranoid and undifferentiated SCZ, respectively. In the second dataset, 1,397 and 1,437 peaks were obtained from patients with paranoid and undifferentiated SCZ, respectively. The results of two datasets were compared by HOMER software with default parameters. Compared with the second dataset, there were 12.14 and 12.37% different peaks in the first dataset of paranoid and undifferentiated SCZ, respectively. Peak regions from the two datasets were then merged and peaks shared by the two datasets were selected using HOMER software. Finally, 757 and 871 peaks were obtained from patients with paranoid and undifferentiated SCZ, respectively. The average peak length was 288 and 431 bp in patients with paranoid SCZ and undifferentiated SCZ, respectively. As demonstrated in Fig. 1, chromosome distributions in the two SCZ subtypes were similar (pearson correlation test: r=0.84, P<0.01). G+C contents and CG percentages were similar in the patient with paranoid SCZ (G+C=49.0%; CG=1.9%) and that with undifferentiated SCZ (G+C=51.5%; CG=2.7%). By contrast, G+C and CG contents of random sequences with the same length distribution were markedly lower in the patients with paranoid SCZ (G+C=37.3%; CG=0.8%) and undifferentiated SCZ (G+C=38.4%; CG=0.9%), compared with those in the control patient. Three and 19 peaks overlapped with CpG islands in the paranoid and the undifferentiated SCZ sample. Among them, four peaks in the undifferentiated SCZ sample were located in the gene promotor regions, including tripartite motif containing 45 (TRIM45), major histocompatibility complex, class II, DP α 1 (HLA-DPA1), thioesterase super-family member 6 (THEM6) and chromosome 6 open reading frame 123 (C6orf123). As demonstrated in Fig 2, the intergenic region was the most enriched (paranoid SCZ, 54.96%; undifferentiated SCZ, 44.20%), followed by the intron region (paranoid SCZ, 40.69%; undifferentiated SCZ, 43.97%).
Table II.
Methylated DNA-binding domain-sequencing datasets.
| Data | Control | Paranoid SCZ | Undifferentiated SCZ |
|---|---|---|---|
| Total reads (n) | 30,901,427 | 8,720,205 | 20,384,242 |
| Reads post-Q20 filter, n (% of total) | 13,800,627 (45) | 4,017,085 (47) | 9,003,647 (45) |
| Read/match ratio (%) | 75.69 | 81.43 | 76.34 |
| Unique read/match ratio (%) | 66.48 | 72.83 | 67.61 |
| Peaks (n) | 1,236 | 1,552 |
SCZ, schizophrenia.
Figure 1.
Chromosomal distribution of DNA hyper-methylation peaks. SCZ, schizophrenia; M, mitochondria.
Figure 2.
Distribution of gene function classes for the DNA hypermethylation peaks. UTR, untranslated region; TTS, transcript termination site; SCZ, schizophrenia.
Hypermethylation peaks in SCZ are predominantly enriched in genes characterized by neuron function
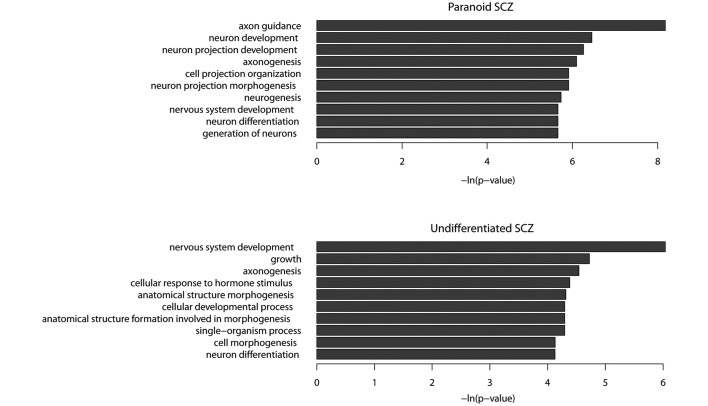
There were 291 coding genes, 18 non-coding genes and 1 pseudogene in the paranoid SCZ sample, whilst 411 coding genes, 21 non-coding genes and 10 pseudogenes were observed in the undifferentiated SCZ sample. GO term enrichment analysis demonstrated that the functions of protein-coding genes in the paranoid SCZ sample were predominantly associated with the nervous system. The five most enriched GO terms were axon guidance, neuron development, neuron projection development, axonogenesis and neuron projection morphogenesis (P<0.01; Fig. 3A). A number of these GO terms are associated with SCZ. For example, contactin-associated protein-like 2 has been shown to mediate cell-cell interactions in the nervous system and is associated with SCZ (37,38) and synapsin III, a member of the synaptic vesicle-associated protein family, was shown to be a candidate gene for SCZ (39). Furthermore, hyper-methylated genes have been shown to be enriched in several important pathways, including the Wnt signaling pathway (40), calcium signaling pathway (41) and a number of pathways associated with cancer (P<0.05).
Figure 3.
Most enriched gene ontology terms of protein-coding genes exhibiting DNA hypermethylation. SCZ, schizophrenia.
The functions of hypermethylated protein-coding genes observed in the undifferentiated SCZ sample were associated with the nervous system, including nervous system development, axonogenesis, neuron differentiation and cell morphogenesis (Fig. 3B). There were 32 hypermethylated genes shared by paranoid and undifferentiated SCZ data-sets (Table III). GO term enrichment analysis demonstrated that these shared hypermethylated genes were enriched in the following functions: Axon guidance, axonogenesis, neuron projection morphogenesis and neuron projection development. The results of the present study suggested that SCZ may be caused by aberrant hypermethylation of a number of genes associated with the nervous system.
Table III.
Hypermethylation genes shared between paranoid and undifferentiated SCZ samples.
| Gene ID | Gene name | Gene definition |
|---|---|---|
| 2139 | EYA2 | Eyes absent homolog 2 (Drosophila) |
| 2272 | FHIT | Fragile histidine triad |
| 11240 | PADI2 | Peptidyl arginine deiminase, type II |
| 100507421 | TMEM178B | Transmembrane protein 178B |
| 5332 | PLCB4 | Phospholipase C, β 4 |
| 5184 | PEPD | Peptidase D |
| 84691 | FAM71F1 | Family with sequence similarity 71, member F1 |
| 400941 | LINC00487 | Long intergenic non-protein coding RNA 487 |
| 23261 | CAMTA1 | Calmodulin binding transcription activator 1 |
| 84131 | CEP78 | Centrosomal protein 78kDa |
| 29994 | BAZ2B | Bromodomain adjacent to zinc finger domain, 2B |
| 121256 | TMEM132D | Transmembrane protein 132D |
| 57221 | KIAA1244 | KIAA1244 |
| 3680 | ITGA9 | Integrin, α 9 |
| 643650 | LOC643650 | Uncharacterized LOC643650 |
| 3899 | AFF3 | AF4/FMR2 family, member 3 |
| 10395 | DLC1 | Deleted in liver cancer 1 |
| 3786 | KCNQ3 | Potassium voltage-gated channel, KQT-like subfamily, member 3 |
| 29119 | CTNNA3 | Catenin (cadherin-associated protein), α 3 |
| 1002 | CDH4 | Cadherin 4, type 1, R-cadherin (retinal) |
| 54715 | RBFOX1 | RNA binding protein, fox-1 homolog (Caenorhabditis elegans) 1 |
| 27086 | FOXP1 | Forkhead box P1 |
| 169044 | COL22A1 | Collagen, type XXII, α 1 |
| 256380 | SCML4 | Sex comb on midleg-like 4 (Drosophila) |
| 25771 | TBC1D22A | TBC1 domain family, member 22A |
| 10426 | TUBGCP3 | Tubulin, gamma complex associated protein 3 |
| 124540 | MSI2 | Musashi homolog 2 (Drosophila) |
| 51151 | SLC45A2 | Solute carrier family 45, member 2 |
| 10404 | CPQ | Carboxypeptidase Q |
| 125336 | LOXHD1 | Lipoxygenase homology domains 1 |
| 80216 | ALPK1 | α-kinase 1 |
| 92293 | TMEM132C | Transmembrane protein 132C |
All genes are protein-coding with the exception of LINC00487 and LOC643650. SCZ, schizophrenia.
Protein-coding genes with hypermethylated promoters
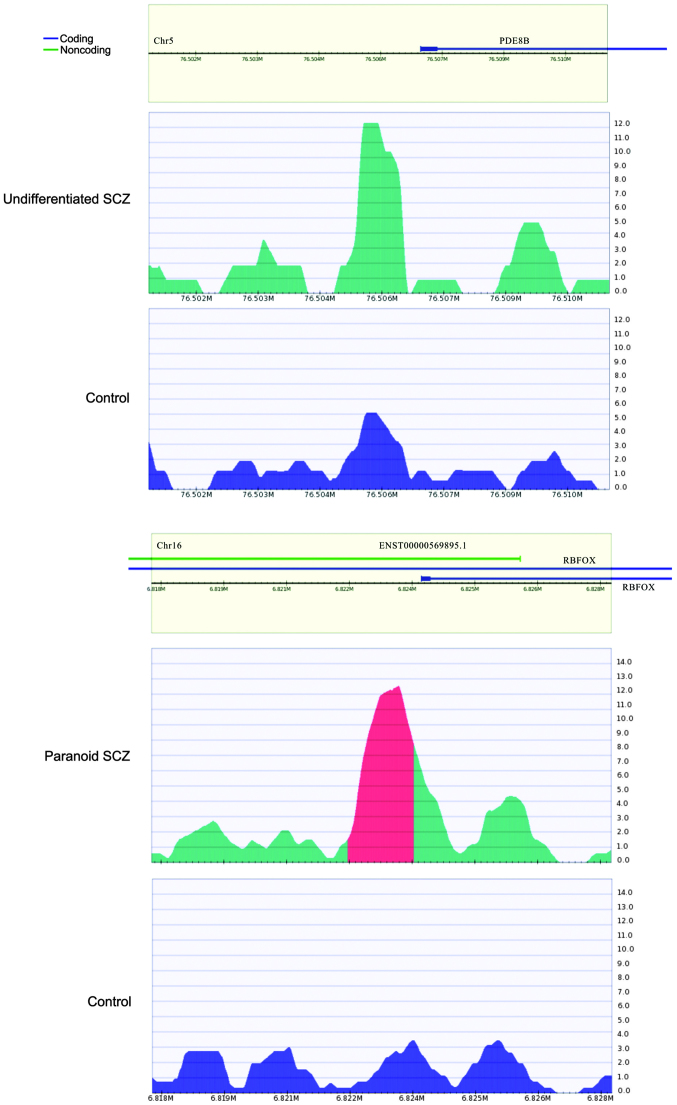
There were 7 (0.92%) and 12 (1.38%) peaks observed in the promoter regions in the paranoid and undifferentiated SCZ samples, respectively. The 7 genes in the paranoid SCZ sample were zinc finger protein 641, RNA binding protein, fox-1 homolog (RBFOX1), solute carrier family 45 member 2, ubiquitin-conjugating enzyme E2Q family-like 1, cluster of differentiation 8a, retinoic acid early transcript 1K pseudogene and GRAM domain containing 3. RBFOX1 (Fig. 4) is a type of RNA binding protein that is capable of regulating alternative splicing. RBFOX1 binds with ataxin-2, which causes familial neurodegenerative diseases. Therefore, RBFOX1 is involved in neuron development (42). It was hypothesized that the hypermethylated RBFOX1 promoter may affect RBFOX1 expression, thereby regulating biological processes involved in SCZ.
Figure 4.
Hypermethylated promoter of (top) the PED8B gene in the undifferentiated SCZ samples and the negative control and (bottom) RBFOX1 gene in the paranoid SCZ samples and the negative control. Methylation levels surrounding the peaks from −5,000 to +50,000 bp; the methylation peaks are located in the center. PDE8B, phosphodiesterase 8B; RBFOX, RNA-binding protein, fox-1 homolog; SCZ, schizophrenia.
For the undifferentiated SCZ sample, genes with hypermethylated promoters included claudin 5, prostaglandin reductase 1, HLA-DPA1, C6ORF123, THEM6, phosphodiesterase 8B (PDE8B), zinc finger protein 622, TRIM45, C11ORF16, polymerase (RNA) I polypeptide E, tensin like C1 domain containing phosphatase and potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2. Among these, PDE8B (Fig. 4) has been reported to be associated with SCZ (43). A study has demonstrated that PDE8B expression is upregulated following treatment with olanzapine and down-regulated following treatment with lithium, which are two drugs that are used to treat schizoaffective disorders (44).
lncRNA genes and DNA hypermethylation
A total of 17 hyper-methylated lncRNAs were identified in the paranoid SCZ sample. Among them, 3 were shown to be associated with cancer (disrupted in renal carcinoma 3, cancer susceptibility candidate 2 and plasmacytoma variant translocation 1; data available on request). SCZ patients are more likely to suffer from cancer compared with the general population. Therefore, SCZ may lead to alterations in epigenetic modifications for a number of cancer-associated genes and, therefore, increase the risk of cancer (45). In the undifferentiated SCZ sample, 20 lncRNAs were identified, including 5 anti-sense RNAs to protein-coding genes, which were shown to be associated with SCZ. Sodium leak channel non-selective-anti-sense RNA 1 (NALCN-AS1; data available on request) is the anti-sense of NALCN that has been reported as a SCZ candidate gene (46–48). Microtubule-associated protein tau-AS1 (MAPT-AS1; data available on request) is the anti-sense lncRNA gene of MAPT that has been shown to be associated with neurodegenerative disorders and SCZ (49). Guanine nucleotide binding protein (G protein) α stimulating activity polypeptide 1-AS (GNAS-AS; data available on request) is the anti-sense lncRNA gene of GNAS1 that may influence a patients' susceptibility to SCZ (50). POU domain, class 4, transcription factor 1-AS1 (POU4F1-AS1; data available on request) is the anti-sense of POU4F1 that exhibits functions associated with the nervous system (51).
The development of DNA sequencing technology has enabled the identification of an increasing number of human lncRNAs. Although their functions are currently unknown, lncRNAs are likely to be transcribed and be involved in a number of biological processes. The present study has identified a number of peaks in the intergenic regions that may correspond to novel lncRNAs. Therefore, lncRNAs were obtained from NONCODE v4.0 and their genomic regions were compared with the peaks of the intergenic regions identified in the present study. The results identified 131 out of 416 (31.5%) and 114 out of 385 (29.6%) intergenic peaks in the paranoid and undifferentiated SCZ, respectively, corresponding to 434 and 320 lncRNAs from NONCODE database for paranoid and undifferentiated SCZ, respectively. The results of the present study suggested that intergenic hypermethylation may influence lncRNA expression and function. The results of the present study showed that lncRNA n373859 was hypermethylated in the promoter region in paranoid SCZ. As shown in the NONCODE database, lncRNA n373859 is highly expressed in brain tissue. Further study is warranted to investigate whether the aberrant promoter hypermethylation of lncRNA n373859 may lead to downregulation in patients with paranoid SCZ.
Discussion
DNA methylation is associated with epigenetic modifications and may be an important factor for SCZ. Aberrant hypermethylation may contribute to abnormal gene expression and influence the development of SCZ. Although a number of studies have reported the effects of numerous important coding genes on DNA methylation in SCZ, including reelin (8,9), catechol-O-methyltransferase (10,11), sex determining region Y (SRY)-box 10 (12), brain-derived neurotrophic factor (16), dopamine receptor D4 (17) and cytotoxic T-lymphocyte-associated protein 4 (17), there are limited studies on lncRNAs. In the present study, a number of hypermethylated loci were identified in protein-coding genes and lncRNAs in male patients with SCZ. The results have provided novel insights for the understanding of SCZ etiology.
In the present study, male patients were selected in order to investigate candidate hypermethylated lncRNAs associated with SCZ. Gender differences are common for a number of aspects of SCZ, such as incidence and prevalence, the presentation and course of the illness, and the various effects on brain development (52). The onset of SCZ is delayed in females and there is typically a second peak in female patients at >45 years of age (53). Estrogen level reduction is considered to be associated with this process (54). Males typically suffer from SCZ more severely and are less responsive to medication than females (52). A number of reasons to explain the early onset in men compared with women have been proposed, including slower maturation and differences in lateralization of the brain, greater incidence of head trauma and the lack of neuroprotective effects from female hormones (52). Therefore, males are more susceptible to SCZ than females (52,55–57). Studies of hormones in female mammals have revealed that psychotic symptoms may vary throughout the menstrual cycle, and that estrogen may enhance neurocognitive performance and improves symptoms of SCZ (58,59). Due to the gender differences associated with SCZ, gender-specific characteristics and endocrine gland side effects require further research in order to develop anti-psychotic treatments. Advances in epigenetic research in SCZ will provide novel insights into SCZ pathogenesis and the development of medication for patients with SCZ.
SCZ pathophysiology research is often based on individual genes and the environment (60). An epidemiological study suggested that interactions between genes and the environment are involved in the molecular mechanisms underlying the onset of SCZ (60). Epigenetic factors, including DNA methylation, genomic imprinting, histone modifications and non-coding RNA expression are involved in SCZ pathogenesis (60–63). These factors may contribute to the mechanisms associated with gender-specific genes and gender dimorphism in SCZ (61).
DNA extracted from peripheral blood samples has been shown to be informative for SCZ research. Dysregulation levels of a number of SCZ candidate genes in peripheral blood samples are associated with a number of clinical symptoms of SCZ (63,64). In the present study, which used genome-wide methylation analyses, DNA methylation levels of protein-coding genes and lncRNAs in peripheral blood samples were investigated. Epigenetic biomarkers were identified that may be used for the diagnosis and development of anti-psychotic drugs for patients with SCZ.
The number of peaks that exhibited DNA hypermethylation were similar between the different SCZ subtypes (paranoid and undifferentiated). Hypermethylated genes were shown to exhibit functions associated with the nervous system in paranoid and undifferentiated SCZ. This suggested that DNA methylation may affect the activity or expression of neuron-associated genes, and thereby influence the development of SCZ. Hypermethylation of promoters may affect the transcription of corresponding genes. In the present study, 7 and 12 peaks were identified in the promoter regions of Refseq genes in paranoid and undifferentiated SCZ samples, respectively. The expression of these genes may be affected by changes to DNA methylation and, therefore, influence biological processes associated with SCZ. With the exception of the peaks corresponding to coding genes, a number of peaks were shown to correspond with known lncRNAs. Furthermore, ~30% of the intergenic peaks that were identified corresponded with novel lncRNAs, according to the RefSeq database. These results suggested that lncRNAs may represent an important regulator for SCZ pathogenesis.
In conclusion, hypermethylated coding and non-coding genes were identified in samples from males with paranoid and undifferentiated SCZ. The results of the present study may clarify the mechanisms underlying SCZ pathogenesis.
Acknowledgments
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 31100919, 81371469 and 31301084), Natural Science Foundation of Zhejiang Province (grant nos. LR13H020003 and LQ13C060002), Natural Science Foundation of Ningbo (grant no. 2013A610249), K.C. Wong Magna Fund in Ningbo University, and Ningbo social development research projects (grant no. 2012C50032).
References
- 1.Hall MH, Schulze K, Rijsdijk F, Picchioni M, Ettinger U, Bramon E, Freedman R, Murray RM, Sham P. Heritability and reliability of P300, P50 and duration mismatch negativity. Behav Genet. 2006;36:845–857. doi: 10.1007/s10519-006-9091-6. [DOI] [PubMed] [Google Scholar]
- 2.Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11:100–113. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- 3.Tamminga CA, Holcomb HH. Phenotype of schizophrenia: A review and formulation. Mol Psychiatry. 2005;10:27–39. doi: 10.1038/sj.mp.4001563. [DOI] [PubMed] [Google Scholar]
- 4.Fanous AH, Kendler KS. Genetics of clinical features and subtypes of schizophrenia: A review of the recent literature. Curr Psychiatry Rep. 2008;10:164–170. doi: 10.1007/s11920-008-0028-z. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors O, Mortensen PB, et al. Genetic Risk and Outcome in Psychosis (GROUP): Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies W. Genomic imprinting on the X chromosome: Implications for brain and behavioral phenotypes. Ann NY Acad Sci. 2010;1204(Suppl):E14–E19. doi: 10.1111/j.1749-6632.2010.05567.x. [DOI] [PubMed] [Google Scholar]
- 7.Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: Analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: A preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 10.Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari MR, Ahmadkhaniha H, Thiagalingam S, Abdolmaleky HM. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res. 2011;45:1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, Kato T. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25:5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolosa A, Sanjuán J, Dagnall AM, Moltó MD, Herrero N, de Frutos R. FOXP2 gene and language impairment in schizophrenia: Association and epigenetic studies. BMC Med Genet. 2010;11:114. doi: 10.1186/1471-2350-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pun FW, Zhao C, Lo WS, Ng SK, Tsang SY, Nimgaonkar V, Chung WS, Ungvari GS, Xue H. Imprinting in the schizophrenia candidate gene GABRB2 encoding GABA(A) receptor β(2) subunit. Mol Psychiatry. 2011;16:557–568. doi: 10.1038/mp.2010.47. [DOI] [PubMed] [Google Scholar]
- 15.Abdolmaleky HM, Yaqubi S, Papageorgis P, Lambert AW, Ozturk S, Sivaraman V, Thiagalingam S. Epigenetic dysregulation of HTR2A in the brain of patients with schizophrenia and bipolar disorder. Schizophr Res. 2011;129:183–190. doi: 10.1016/j.schres.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Ikegame T, Bundo M, Sunaga F, Asai T, Nishimura F, Yoshikawa A, Kawamura Y, Hibino H, Tochigi M, Kakiuchi C, et al. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. 2013;77:208–214. doi: 10.1016/j.neures.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Kordi-Tamandani DM, Vaziri S, Dahmardeh N, Torkamanzehi A. Evaluation of polymorphism, hypermeth-ylation and expression pattern of CTLA4 gene in a sample of Iranian patients with schizophrenia. Mol Biol Rep. 2013;40:5123–5128. doi: 10.1007/s11033-013-2614-3. [DOI] [PubMed] [Google Scholar]
- 18.Cheng J, Wang Y, Zhou K, Wang L, Li J, Zhuang Q, Xu X, Xu L, Zhang K, Dai D, et al. Male-specific association between dopamine receptor D4 gene methylation and schizophrenia. PLoS One. 2014;9:e89128. doi: 10.1371/journal.pone.0089128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folsom TD, Fatemi SH. The involvement of Reelin in neuro-developmental disorders. Neuropharmacology. 2013;68:122–135. doi: 10.1016/j.neuropharm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vučićević D, Schrewe H, Orom UA. Molecular mechanisms of long ncRNAs in neurological disorders. Front Genet. 2014;5:48. doi: 10.3389/fgene.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Pyschiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 22.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 23.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 27.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Ibala-Romdhane S, Al-Khtib M, Khoueiry R, Blachère T, Guérin JF, Lefèvre A. Analysis of H19 methylation in control and abnormal human embryos, sperm and oocytes. Eur J Hum Genet. 2011;19:1138–1143. doi: 10.1038/ejhg.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiesa N, De Crescenzo A, Mishra K, Perone L, Carella M, Palumbo O, Mussa A, Sparago A, Cerrato F, Russo S, et al. The KCNQ1OT1 imprinting control region and non-coding RNA: New properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet. 2012;21:10–25. doi: 10.1093/hmg/ddr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.American Psychiatric Association . Diagnostic and statistical manual of mental disorders, text revision (DSM-IV-TR) 4th Edition. American Psychiatric Association; 2000. [Google Scholar]
- 31.Hogart A, Lichtenberg J, Ajay SS, Anderson S, Margulies EH. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22:1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakato R, Itoh T, Shirahige K. DROMPA: Easy-to-handle peak calling and visualization software for the computational analysis and validation of ChIP-seq data. Genes Cells. 2013;18:589–601. doi: 10.1111/gtc.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji W, Li T, Pan Y, Tao H, Ju K, Wen Z, Fu Y, An Z, Zhao Q, Wang T, et al. CNTNAP2 is significantly associated with schizophrenia and major depression in the Han Chinese population. Psychiatry Res. 2013;207:225–228. doi: 10.1016/j.psychres.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 38.Rodenas-Cuadrado P, Ho J, Vernes SC. Shining a light on CNTNAP2: Complex functions to complex disorders. Eur J Hum Genet. 2014;22:171–178. doi: 10.1038/ejhg.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao HT, Porton B, Czernik AJ, Feng J, Yiu G, Häring M, Benfenati F, Greengard P. A third member of the synapsin gene family. Proc Natl Acad Sci USA. 1998;95:4667–4672. doi: 10.1073/pnas.95.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bousman CA, Glatt SJ, Chandler SD, Lohr J, Kremen WS, Tsuang MT, Everall IP. Negative symptoms of psychosis correlate with gene expression of the Wnt/β-catenin signaling pathway in peripheral blood. Psychiatry J. 2013;2013:852930. doi: 10.1155/2013/852930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berridge MJ. Dysregulation of neural calcium signaling in Alzheimer disease, bipolar disorder and schizophrenia. Prion. 2013;7:2–13. doi: 10.4161/pri.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fatemi SH, Folsom TD, Reutiman TJ, Vazquez G. Phosphodiesterase signaling system is disrupted in the cerebella of subjects with schizophrenia, bipolar disorder, and major depression. Schizophr Res. 2010;119:266–267. doi: 10.1016/j.schres.2010.02.1055. [DOI] [PubMed] [Google Scholar]
- 44.Fatemi SH, Folsom TD, Reutimann TJ, Braun NN, Lavergne LG. Levels of phosphodiesterase 4A and 4B are altered by chronic treatment with psychotropic medications in rat frontal cortex. Synapse. 2010;64:550–555. doi: 10.1002/syn.20762. [DOI] [PubMed] [Google Scholar]
- 45.Irwin KE, Henderson DC, Knight HP, Pirl WF. Cancer care for individuals with schizophrenia. Cancer. 2014;120:323–334. doi: 10.1002/cncr.28431. [DOI] [PubMed] [Google Scholar]
- 46.Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124:192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Teo C, Zai C, Borlido C, Tomasetti C, Strauss J, Shinkai T, Le Foll B, Wong A, Kennedy JL, De Luca V. Analysis of treatment-resistant schizophrenia and 384 markers from candidate genes. Pharmacogenet Genomics. 2012;22:807–811. doi: 10.1097/FPC.0b013e3283586c04. [DOI] [PubMed] [Google Scholar]
- 48.Al-Sayed MD, Al-Zaidan H, Albakheet A, Hakami H, Kenana R, Al-Yafee Y, Al-Dosary M, Qari A, Al-Sheddi T, Al-Muheiza M, et al. Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am J Hum Genet. 2013;93:721–726. doi: 10.1016/j.ajhg.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobson-Stone C, Polly P, Korgaonkar MS, Williams LM, Gordon E, Schofield PR, Mather K, Armstrong NJ, Wen W, Sachdev PS, Kwok JB. GSK3B and MAPT polymorphisms are associated with grey matter and intracranial volume in healthy individuals. PLoS One. 2013;8:e71750. doi: 10.1371/journal.pone.0071750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minoretti P, Politi P, Coen E, Di Vito C, Bertona M, Bianchi M, Emanuele E. The T393C polymorphism of the GNAS1 gene is associated with deficit schizophrenia in an Italian population sample. Neurosci Lett. 2006;397:159–163. doi: 10.1016/j.neulet.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 51.Zou M, Li S, Klein WH, Xiang M. Brn3a/Pou4f1 regulates dorsal root ganglion sensory neuron specification and axonal projection into the spinal cord. Dev Biol. 2012;364:114–127. doi: 10.1016/j.ydbio.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abel KM, Drake R, Goldstein JM. Sex differences in schizophrenia. Int Rev Psychiatry. 2010;22:417–428. doi: 10.3109/09540261.2010.515205. [DOI] [PubMed] [Google Scholar]
- 53.Lindamer LA, Lohr JB, Harris MJ, Jeste DV. Gender, estrogen, and schizophrenia. Psychopharmacol Bull. 1997;33:221–228. [PubMed] [Google Scholar]
- 54.Mellios N, Galdzicka M, Ginns E, Baker SP, Rogaev E, Xu J, Akbarian S. Gender-specific reduction of estrogen-sensitive small RNA, miR-30b, in subjects with schizophrenia. Schizophr Bull. 2012;38:433–443. doi: 10.1093/schbul/sbq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boksa P. Animal models of obstetric complications in relation to schizophrenia. Brain Res Brain Res Rev. 2004;45:1–17. doi: 10.1016/j.brainresrev.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Missios S, Harris BT, Dodge CP, Simoni MK, Costine BA, Lee YL, Quebada PB, Hillier SC, Adams LB, Duhaime AC. Scaled cortical impact in immature swine: Effect of age and gender on lesion volume. J Neurotrauma. 2009;26:1943–1951. doi: 10.1089/neu.2009.0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson C, Terry AV., Jr Neurodevelopmental animal models of schizophrenia: Role in novel drug discovery and development. Clin Schizophr Relat Psychoses. 2010;4:124–137. doi: 10.3371/CSRP.4.2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cosimo Melcangi R, Garcia-Segura LM. Sex-specific therapeutic strategies based on neuroactive steroids: In search for innovative tools for neuroprotection. Horm Behav. 2010;57:2–11. doi: 10.1016/j.yhbeh.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 59.Kulkarni J, Gurvich C, Lee SJ, Gilbert H, Gavrilidis E, de Castella A, Berk M, Dodd S, Fitzgerald PB, Davis SR. Piloting the effective therapeutic dose of adjunctive selective estrogen receptor modulator treatment in postmenopausal women with schizophrenia. Psychoneuroendocrinology. 2010;35:1142–1147. doi: 10.1016/j.psyneuen.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 60.Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS. Evidence for X-chromosomal schizophrenia associated with microRNA alterations. PLoS One. 2009;4:e6121. doi: 10.1371/journal.pone.0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 63.Melas PA, Rogdaki M, Ösby U, Schalling M, Lavebratt C, Ekström TJ. Epigenetic aberrations in leukocytes of patients with schizophrenia: Association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 2012;26:2712–2718. doi: 10.1096/fj.11-202069. [DOI] [PubMed] [Google Scholar]
- 64.Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, Huang YH, Hsiao PC, Hsiao CK, Liu CM. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]