Abstract
Over the last few decades, the epithelial-to-mesenchymal transition (EMT) has been identified as being involved in a number of aspects of physiological processes and various pathological events, including embryonic development and renal fibrosis. Transforming growth factor-β receptor 2 (TGFβR2) is a widely studied gene, which fulfils a vital role in the TGFβ signaling pathway and exerts a crucial function in the progression of EMT. Previous studies demonstrated that the dysregulation of microRNAs (miRNAs) is considered to be associated with the EMT process. However, the precise functional involvement of miRNAs in EMT remains to be fully elucidated. In the present study, the level of miR-590 was decreased in an EMT model in vitro and in vivo. Furthermore, the overexpression of miR-590 inhibited EMT by upregulating the epithelial marker, E-cadherin, and downregulating the mesenchymal markers, laminin, α-smooth muscle actin (α-SMA) and collagen, in the human kidney 2 (HK2) cell line. Furthermore, TGFβR2 was negatively regulated by miR-590. In addition, performing a knockdown of TGFβR2 with small-interfering RNA had an effect similar to miR-590 on EMT in the HK2 cell line, whereas the transfection of pCMV-tag2B-TGFβR2 reversed the effect of miR-590 on EMT in HK2 cells. Taken together, the present study demonstrated that miR-590 is a novel EMT-suppressive microRNA, which targets TGFβR2.
Keywords: miR-590, epithelial-to-mesenchymal transition, Transforming growth factor-β receptor 2, unilateral ureteral obstruction, kidney, HK2
Introduction
The epithelial-to-mesenchymal transition (EMT) is considered to be an essential biological process, which comprises a series of epithelial plasticity alterations between the epithelial and mesenchymal states, and has an important role in physiological and pathological processes (1,2). During the progression of EMT, epithelial cells, which form monolayers that exist in body tissues and organs, lose their cell-cell adhesiveness, attain an elongated morphology and exhibit an increased motility and invasiveness (1). In addition to these phenotypic changes, the cells may also exhibit alterations in their gene expression, including an upregulation of a variety of transcription factors, including Snail, a downregulation of epithelial markers, including E-cadherin, and an increased expression of mesenchymal markers, such as α-smooth muscle actin (α-SMA) (1). Concomitantly with the changes observed in the EMT markers, the further progression through the stages of EMT is marked by the stimulation of epithelial cells, which are transformed into myofibroblasts. EMT is considered to contribute to fibrogenesis by providing a source of myofibro-blasts, and also by activating a paracrine signaling pathway between the epithelial and stromal cells (3–5). Furthermore, a number of reviews reported on the role of EMT in epithelial-mesenchymal interactions in the development of fibrotic diseases (6,7). These findings provide substantial evidence for an important role for the epithelium in fibrogenesis, as a source of myofibroblasts, and as a mediator influencing the development of myofibroblasts from epithelia cells.
MicroRNAS (miRNAs) form a class of small, single-stranded, non-coding RNAs, which are 19–22 nucleotides in length and negatively regulate gene expression by base-pairing with the 3′ untranslated region (UTR) of cognate mRNAs, which have the conserved seed sequence (8). These non-coding RNAs are considered to exert a critical role in physiological and pathological processes, ranging from embryonic develop ment to tumorigenesis, depending on their specific gene targets (9,10). An accumulating body of evidence supports that miRNAs exert a crucial role in EMT through the modulation of EMT-associated genes. In the cell model of EMT induced by transforming growth factor-β (TGFβ1), previous studies have demonstrated that all five members of the miR-200 family and miR-205 are capable of repressing EMT by targeting the transcription factors, zinc finger E-box binding homeobox 1 and 2 (ZEB1/2) (11–13). A further study revealed that miR-155 exerts an essential role in TGFβ1-induced EMT through targeting the protein, RhoA (14). Taken together, these results suggested that miRNAs may be crucially important in EMT, however, the biological functions of the majority of the miRNAs in EMT remain to be fully elucidated.
In the present study, ample evidence is provided to support a role for miR-590 in EMT, based on studies performed in TGFβ1-treated human kidney 2 (HK2) cells and a unilateral ureteral obstruction (UUO) kidney model. The study aimed to investigate whether the overexpression of miR-590 exerted any influence on EMT in HK2 cells and in human renal tubular epithelial cells. Furthermore, the results of the present study are discussed with respect to potential effects on epithelium homeostasis, and the consequences of these effects in kidney fibrosis.
Materials and methods
Prediction of miRNAs which target TGFβ receptor 2 (R2)
The miRNAs targeting TGFβR2 were predicted using the prediction programs: miRDB (http://www.microrna.org/), PicTar(http://pictar.mdc-berlin.de/) and TargetScan (http://www.targetscan.org/). Only those miRNAs that were predicted by all three algorithms were selected as putative regulators of TGFβR2, and were suggested for further experimental identification.
Cell culture and transfection
Human embryonic kidney 293 (HEK293T) and HK2 cells were cultivated in Dulbecco's modified Eagle's medium and RPMI-1640 media (Invitrogen Life Technologies, Carlsbad, CA, USA), respectively, supple mented with 10% fetal bovine serum (Invitrogen Life Technologies), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in an atmosphere of 5% CO2. The HEK293T and the HK2 cells were purchased from the American Tissue Culture Collection (Manassas, VA, USA). For the transfection experiments, when the cells had reached 60% confluence, the HK2 cells were transfected with pCDNA3.1 (800 ng), pCDNA3.1-mir-590 (800 ng), small-interfering (si) RNA-TGFβR2 (20 nM; sense and antisense sequences, 5′-GUGCCUGUAACAUGGAAGATT-3′ and 5′-UCUUCCAUGUUACAGGCACTT-3′, respectively) or siRNA-control (20 nM; sense and antisense sequences, 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′, respectively) using Lipofectamine™ 2000 transfection reagent (Invitrogen Life Technologies), according to the manufacturer's instructions.
Plasmid construction
To overexpress the miR-590, the 81 base pairs (bp) genomic sequence, which encodes mature miR-590, was cloned into the PUC57 vector (manufactured by Genewiz, Inc., South Plainfield, NJ, USA). The sequence used was: 5′-AGTCAGAAATGAGCTTATTCATAAAAGTGCAGTATGGTGAAGTCAATCTGTAATTTTATGTATAAGCTAGTCTCTGATTGA-3′. Subsequently, the premature sequence was transferred into pcDNA3.1 (Invitrogen Life Technologies) downstream of the cytomegalovirus (CMV) promoter between the restriction sites HindIII and EcoRI. For the luciferase reporter plasmids, the 3′ UTR of the mouse TGFβR2, or its mutant variations, was amplified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) from the mouse genome, and inserted downstream of the luciferase coding sequence of the pcDNA3.1-luciferase reporter plasmid between the restriction sites BamHI and EcoRI. For the overexpression of TGFβR2, the coding sequence was cloned to generate the plasmid pCMV-tag-2B-TGFβR2 using the restriction sites of BamHI and Xhol, and (forward: 5′-CGCGGATCCCTGTCCACTTGCGACAACCAG-3′ and reverse: 5′-GGCCTCGAGTTTGGTAGTGTTCAGCGAGC-3′). All the plasmids used in the present study were verified by sequencing.
Generation of the EMT model in vitro
EMT was induced in the HK2 cells in the presence of TGFβ1. Cells that had reached 50% confluence were treated with recombinant human TGFβ1 (10 ng/ml; R&D Systems, Minneapolis, MN, USA) in RPMI-1640 medium for 48 h.
Generation of the UUO kidney model
The UUO kidney model was generated in male C57BL/6 mice (12–16 weeks old, weighing 24–28 g), which were purchased from the Animal Center of Chongqing Medical University. Animals were treated according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The experimental procedures were approved by the Animal Experimental Ethics Committee of Chongqing Medical University. The animals were operated on by left ureteral ligation, as described previously (15,16). Briefly, following anesthetization with sodium pentobarbital (50 mg/kg), the middle portion of the left ureter was ligated completely by surgical suture, whereas the control group underwent an identical process without ligation. The mice were sacrificed on day 14 following the ligation surgery, and the kidneys were harvested for subsequent studies. These experiments were approved by the Medicine Animal Care Committee of the Chongqing Medical University.
Histochemical analysis
The kidneys were dissected, fixed with 10% neutral formalin for 48 h, and embedded in paraffin. Briefly, tissues were immersed in acetone at 4°C, transferred to a freezer at −20°C, and fixed overnight. Then the tissues were dehydrated in acetone at 4°C for 15 min and in acetone for another 15 min at room temperature. Thereafter the tissues were cleared twice in methyl benzoate for 15 min, and twice in xylene for 15 min at room temperature. Finally they were penetrated with paraffin (melting point, 58–60°C; Junsei Chemical Co., Ltd., Tokyo, Japan) at 60°C for 2–3 h in a vacuum-evaporating embedder and embedded in paraffin. Subsequently, 6-mm thick sections were selected and stained with Masson's stain for the determination of the tubulointerstitial index, as previously described (17–19).
TGFβR2 3′ UTR-luciferase reporter assay
The luciferase reporter analyses were performed on HEK293T cells. For the transfection assay, the HEK293T cells were seeded (1×105 cells/well) in 24-well plates. The plasmids pcDNA3.1-Luc-TTGFβR2WT, pcDNA3.1-Luc-TGFβR2Mut and pcDNA3.1-Luc were transfected into HEK293T cells, which stably expressed pcDNA3.1-miR-590 or pcDNA3.1, using Lipofectamine™ 2000 in OptiMEM medium (Thermo Fisher, Waltham, MA, USA). At 48 h later, the cells were harvested with PBS, and lysed with radioimmunoprecipitation assay (RIPA) buffer. The dual-luciferase reporter assay system (Promega Corp., Madison, WI, USA) was used for the analysis, and the data are shown as the ratio of luciferase activity normalized against that of Renilla.
RT-qPCR
RT-qPCR was used to determine the mRNA levels. The total RNA was isolated from cells or kidneys using Trizol® (Invitrogen Life Technologies), and 5 µg total RNA was selected to be reverse-transcribed using the First-Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Aliquots of 0.5 µl transcription product served as the template for the qPCR reactions with UltraSYBR mixture (CWBio, Inc., Beijing, China) or UltraSYBR mixture (Takara Bio, Inc., Otsu, Japan). The mRNA expression levels of E-cadherin, α-SMA and collagen were normalized to 18S RNA, whereas the expression level of miR-590 was normalized to U6 RNA. Primers for the detection of the miRNAs were as follows. The primers for miR-590 were provided by Ruibo (Guangzhou, China); the U6 primers were 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The primers for the detection of the mRNAs were as follows: 18S primers were 5′-GTAACCCGTTGAACCCCATT-3′ (forward) and 5′-CCATCCAATCGGTAGTAGCG-3′ (reverse); the mouse E-cadherin primers, 5′-GTCAACACCTACAACGCTGC-3′ (forward) and 5′-ACGTGCTTGGGTTGAAGA-3′ CA (reverse); the mouse α-SMA primers, 5′-GCTATGCCCTGCCTCATGC-3′ (forward) and 5′-TCACGGACAATCTCACGCTC-3′ (reverse); the mouse collagen primers, 5′-AACTTTGCTTCCCAGATGTCCTATG-3′ (forward) and 5′-GCTTCCCCATCATCTCCATTCTTGC-3′ (reverse); the human E-cadherin primers, 5′-GGTGCTCTTCCAGGAACCTC-3′ (forward) and 5′-GGAAACTCTCTCGGTCCAGC-3′ (reverse); the human α-SMA primers, 5′-GGGGTGATGGTGGGAATG-3′ (forward) and 5′-GCAGGGTGGGATGCTCTT-3′ (reverse); and the human collagen primers were 5′-AAGGTGTTGTGCGATGACG-3′ (forward) and 5′-TGGTCGGTGGGTGACTCTG-3′ (reverse). All the experiments were performed in triplicate. The expression levels were quantified using the 2−ΔCt method. ΔCt was calculated either as Ct (E-cadherin/ α-SMA/collagen) - Ct (i8s) or as Ct (miR-590) - Ct (U6).
Protein isolation and western blotting
The HK2 cells were washed three times with 100 µl ice-cold PBS buffer and lysed by adding 200 µl RIPA buffer/well immediately, with a 30-min incubation period. For the kidney tissue, the kidneys were homogenized in lysis buffer (RIPA) containing 1 mg/l protease cocktail (BioTool, Jupiter, FL, USA; containing 104 mM AEBSF, 80 µM aprotinin, 5 mM bestatin, 1.5 mM E-64, 2 mM leupeptin, 1.5 mM pepstatin A), prior to sonication Xo-1200D (Heng Long Instrument Co., Ltd., Changzhou, China) at 4°C for 60 sec. The total protein concentration of each sample was measured using the bicinchoninic acid protein assay with a microplate procedure (Beyotime Institute of Biotechnology, Haimen, China), prior to western blotting. Samples of 30 µg total protein were subjected to 12% SDS-PAGE, and the protein bands were subsequently transferred onto polyvinylidene fluoride membranes (Millipore Corp., Bedford, MA, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline (TBS)/Tween 20 (0.1%) at room temperature for 1 h. Subsequently, the membranes were immersed in primary antibody solution [rabbit polyclonal anti-E-cadherin, 1:3,000 (20648-1-AP; Proteintech Group Inc., Chicago, IL, USA); rabbit polyclonal anti-laminin, 1:2,500 (sc-20145; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); rabbit polyclonal anti-TGFβR2, 1:2,000 (our laboratory); or mouse monoclonal anti-β-tubulin, 1:5,000 (HC101-01 TransGen, Beijing, China)] at 4°C for an overnight incubation. After washing three times with TBS/Tween 20 (0.1%; 5 min each wash), incubation with horseradish peroxidase-conjugated goat anti-rabbit and mouse secondary antibody (1:5,000, Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing, China) secondary antibodies was performed at room temperature for 1 h (1:5,000 dilution). The immunoreactivity was developed using chemiluminescent horseradish peroxidase substrate reagent (Millipore Corp.), and the signal was detected using the Bio-Rad ChemiDoc XRS+imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). β-tubulin was used as a protein loading control, and western blot analyses were performed in triplicate.
Immunofluorescent analysis
For the detection of immunofluorescence, HK2 cells were cultured with sterile glass coverslips in six-well plates. The glass slides were treated with 95% alcohol for 15 min at room temperature prior to cell culture. Subsequently, the cell coverslips were washed three times with 500 µl PBS and permeabilized with 0.1% Triton X-100 for 30 min. Following blocking with 10% normal goat serum for 1 h at room temperature, the cells were incubated with rabbit anti-laminin primary antibody (1:100; Santa Cruz Biotechnology, Inc.) for 3 h at room temperature. The cover-slips were subsequently washed three times with PBS, prior to incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglubulin G at room temperature for 1 h. The slides were examined under a Nikon (Melville, NY, USA) Eclipse TE300 fluorescence microscope, and images were captured with a SPOT Diagnostic (Sterling Heights, MI, USA) charged-couple device camera.
Statistical analysis
GraphPad Prism 5 (San Diego, CA, USA) software was used to analyze the experimental data. The statistical differences between groups, including the bar graph of the luciferase assay and the results from the RT-qPCR experiments, were analyzed by one-way analysis of variance. All the experiments were performed at least in triplicate, and the statistical errors of averaged data are presented as the mean ± standard error of the mean. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-590 is decreased during EMT in vitro and in vivo
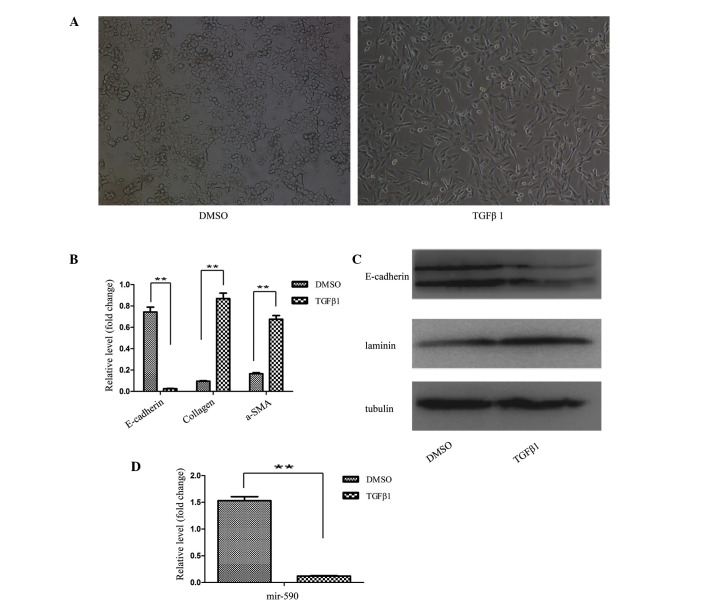
To investigate whether miR-590 has a direct function on EMT, TGFβ1-induced epithelial cells were used as a cell model, and UUO mice were an animal model, for modulating EMT in vitro and in vivo, respectively. TGFβ1-induced EMT experiments were performed in HK2 cells according to the method described previously (20,21), whereas the UUO model was generated in mice by left ureter ligation (22). Following treatment with TGFβ1 for 48 h, the HK2 cells were transformed from the epithelial state, characterized by polygonal morphology and tight connections, into a spindle-like mesenchymal morphology, which appeared as the HK2 cells underwent the process of EMT (Fig. 1A). Subsequently, the EMT process at the molecular level was identified by determining the levels of the marker genes. The results from the RT-qPCR analysis revealed decreased mRNA expression levels of the epithelial marker (E-cadherin), and increased expression levels of the mesenchymal markers, collagen and α-SMA, following TGFβ1 treatment of the HK2 cells (Fig. 1B). Western blot analysis also confirmed the EMT process.
Figure 1.
mRNA and protein expression levels of miR-590 were decreased during EMT in vitro. (A) The EMT model with TGFβ1-induced HK2 cells. The morphological changes of the HK2 cells in the presence of TGFβ1 (10 ng/ml) were observed under a phase-contrast microscope (magnification, ×100). (B) The regulation of collagen, α-SMA and E-cadherin in the cell EMT model. The mRNA expression levels were detected using RT-qPCR. (C) The regulation of laminin and E-cadherin in the cell EMT model. The protein expression levels were detected by western blotting. (D) The downregulation of miR-590 expression in the cell EMT model. The expression level was examined by RT-qPCR. The results are shown as the fold change compared with the non-treated control. Data are presented as the mean ± standard deviation of three independent experiments, **P<0.01 compared with the control. DMSO, dimethyl sulfoxide; TGFβ1, transforming growth factor β1; a-SMA, α-smooth muscle actin.
As shown in Fig. 1C, downregulation of the protein expression level of E-cadherin, and an upregulation of the protein expression level of laminin, were observed in TGFβ1-treated HK2 cells. Furthermore, the expression level of miR-590 in HK2 cells was significantly decreased following TGFβ1 treatment, as revealed by the RT-qPCR analysis (Fig. 1D). These results indicated an inverse correlation between the expression of EMT-associated genes and miR-590. To investigate the association between miR-590 and the EMT process in renal fibrosis, the identical experiments were performed in the UUO kidney. Prior to the experiments, the efficiency of the UUO model was confirmed by Masson staining.
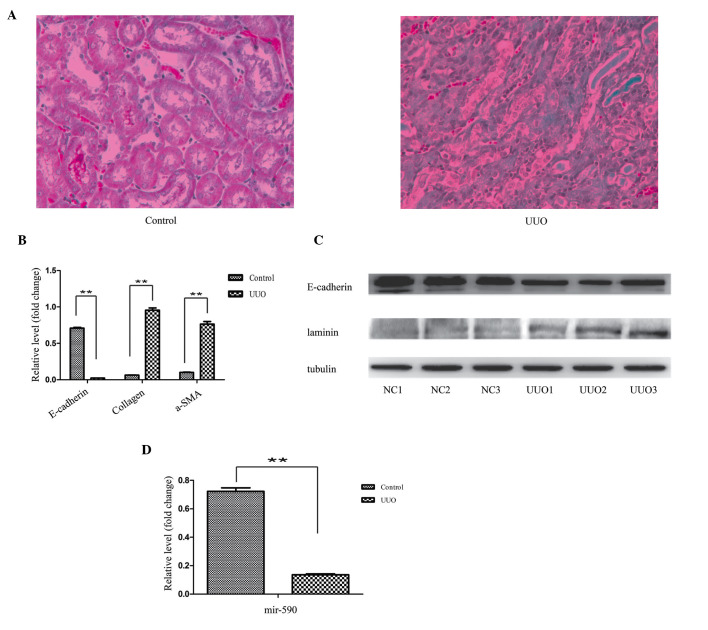
The histopathological results revealed substantial structural alterations, induced by left ureter ligation. In the control specimens, the kidney tissues were well aligned and surrounded by only a small quantity of interstitial tissue, whereas the section of the kidney from the UUO-induced mice featured a large proportion of fibres (Fig. 2A). For the EMT analysis, consistently with the cell model, a decreased mRNA expression level of E-cadherin, and an increased mRNA expression level of collagen and α-SMA, were observed in the RT-qPCR experiment (Fig. 2B). The western blot analysis revealed a decreased protein expression level of E-cadherin, and an increased protein expression level of laminin, in the UUO kidney (Fig. 2C). Similarly, the expression level of miR-590 was significantly decreased in the UUO animals compared with the control (Fig. 2D).
Figure 2.
mRNA expression levels of miR-590 were decreased during EMT in vivo. (A) The EMT model with UUO kidneys. The UUO kidney section exhibited a large proportion of fibrotic tissues (revealed by the blue coloration). Magnification, ×100. (B) Regulation of the mRNA expression levels of collagen, α-SMA and E-cadherin in a kidney EMT model, as detected by RT-qPCR. (C) Regulation of the protein expression levels of laminin and E-cadherin in the kidney EMT model, as determined by western blot analysis. β-tubulin was used as a loading control. (D) Downregulation of the expression level of miR-590 in the kidney EMT model, as determined by RT-PCR. The results are shown as the fold change compared with the control. The data are expressed as the mean ± standard deviation of three independent experiments. **P<0.01, compared with the control. a-SMA, α-smooth muscle actin; UUO, unilateral ureteral obstruction.
mir-590 downregulates TGFβR2 by targeting its 3′ UTR
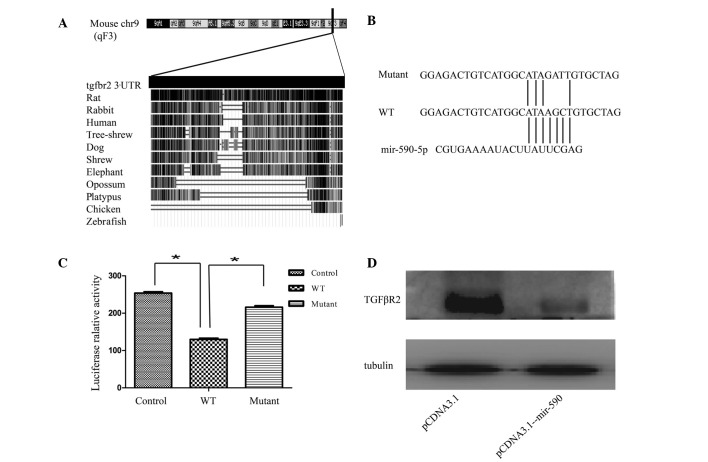
Since a pronounced downregulation of miR-590 was revealed in the EMT model, to investigate the role exerted by miR-590 in EMT regulation, a search of the prediction software programs, TargetScan, miRanda and PicTar, was performed. It was observed that the 3′ UTR of the TGFβR2 mRNA contained a potential site for miR-590 (23). Subsequently, conservation of the 3′ UTR of TGFβR2 was analyzed by blasting the sequences between different species, according to the University of California Santa Cruz genome database (Santa Cruz). As shown in Fig. 3A, the entire 3′ UTR of TGFβR2 was 2,698 bp long, and was highly conserved among different species, including humans, rats, rabbits, shrews, tree shrews and dogs. Subsequently, the validation of the association between miR-590 and TGFβR2 was assessed. Given that it was considered to be one of the most effective miRNAs in inhibiting the luciferase activity, the plasmid pCDNA-3.1-miR-590 was used in the present study. To examine whether miR-590 regulates TGFβR2 expression by directly binding to the predicted 3′ UTR sequence, luciferase reporter plasmids containing the wild-type or the mutated binding site for miR-590 were constructed (Fig. 3B). These results revealed that the overexpression of miR-590 significantly suppressed the luciferase activity of the wild-type TGFβR2 3′ UTR reporter, although not that of the mutant reporter containing a mutation in the miR-590-binding site (Fig. 3C). To examine the physiological importance of miR-590 in regulating TGFβR2 production, the effect of miR-590 on TGFβR2 production in HK2 cells was assessed. The western blot analysis revealed that the transfection of HK2 cells with pCDNA-3.1-miR-590 markedly reduced the protein expression level of TGFβR2 (Fig. 3D). These results demonstrated that miR-590 is able to negatively regulate the expression of TGFβR2 by directly targeting its 3′ UTR.
Figure 3.
mir-590 downregulates TGFβR2 by targeting its 3′ UTR. (A) Bioinformatics analysis of the 3′ UTR of TGFβR2. The 3′ UTR of TGFβR2 is 2,698 bp in length, and is highly conserved among mammals, as revealed by the University of California, Santa Cruz genome database. (B and C) Analysis of the luciferase activity in HEK293T cells. HEK293T cells were transiently co-transfected with the luciferase reporter plasmid, containing the wild-type TGFβR2 3′ UTR, or mutant variations, in the presence of pCDNA3.1-mir-590 or pCDNA3.1. (D) Effects of miR-590 on the endogenous TGFβR2 level in HK2 cells. The expression level was analyzed by western blotting. The data are presented as fold changes compared with the control cells. Data are presented as the mean ± standard deviation, and are representative of at least three independent experiments. *P<0.05, compared with the control. chr9, chromosome 9; WT, wild-type; UTR, untranslated region; TGFβR2, transforming growth factor receptor β2.
Effects of miR-590 and TGFβR2 on EMT in HK2 cells
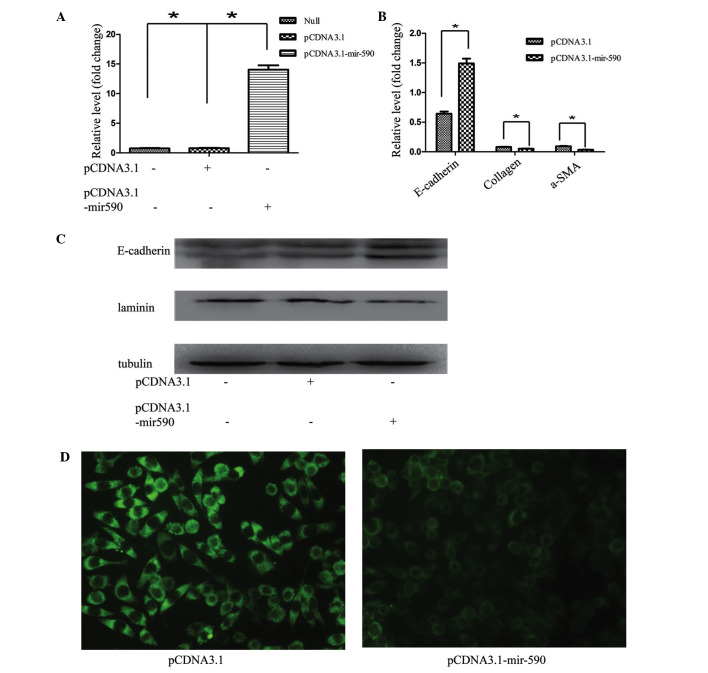
A consistent correlation between the expression levels of miR-590 and E-cadherin was observed, and a marked downregulation of miR-590 was identified in the EMT model. Furthermore, the expression of endogenous miR-590 was higher in HK2 cells compared with L929 cells (mice embryonic myofibroblasts; data not shown) indicating that the downregulation of miR-590 may explain phenotypic stabilization of mesenchymal features in the L929 cell line. To investigate the role of miR-590 in EMT, HK2 cells were transfected with pCDNA3.1-mir-590, and the expression level of mir-590 was significantly increased (Fig. 4A). The induction of miR-590 markedly inhibited EMT in HK2 cells, as revealed by epithelial marker changes. The RT-qPCR results demonstrated that the expression of the miR-590 plasmid in HK2 cells also resulted in a decrease in the mRNA expression of other known fibroblast markers, including α-SMA and collagen I, and an increased mRNA expression of E-cadherin (Fig. 4B). Furthermore, the western blotting and immunofluorescence results revealed that transfection of the miR-590 plasmid markedly reduced the expression of laminin, and upregulated E-cadherin expression in HK2 cells (Fig. 4C and D). A previous study in our laboratory revealed that TGFβ1 induced EMT in vitro, accompanied by the upregulation of laminin, and the downregulation of E-cadherin, at the mRNA and protein levels in HK2 cells, whereas the transfection of exogenous miR-590 markedly attenuated the EMT process in HK2 cells.
Figure 4.
Effects of miR-590 on EMT in HK2 cells. (A) Validation of the transfection efficiency in HK2 cells with pCDNA3.1-mir-590 or pCDNA3.1. (B) The mRNA expression levels of E-cadherin, collagen and α-SMA in HK2 cells 48 h following transfection with pCDNA3.1-mir-590 or pCDNA3.1. (C) The protein expression levels of laminin and E-cadherin 48 h following transfection with pCDNA3.1-mir-590 or pCDNA3.1. (D) The altered expression levels in HK2 cells following miR-590 overexpression, as measured by fluorescence. Magnification, ×400. The data are shown as fold changes compared with control cells. The data are presented as the mean ± standard deviation, and the results are representative of at least three independent experiments. *P<0.05, compared with the control. a-SMA, α-smooth muscle actin.
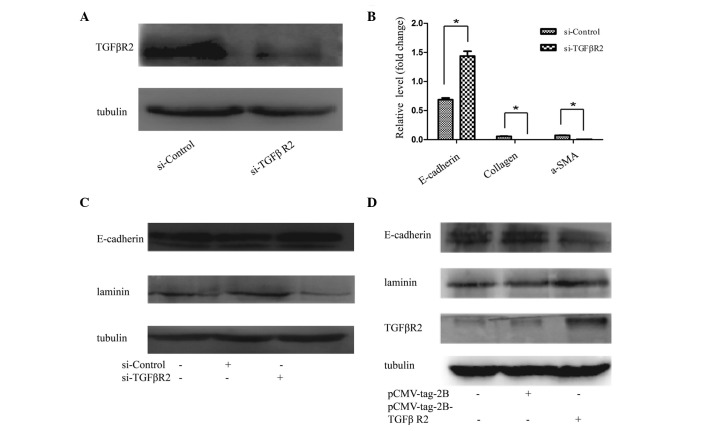
The TGFβR2 3′ UTR has a functional site for miR-590, and it is well known that TGFβR2 exerts an important role in the process of TGFβ-mediated signal transduction by linking TGFβ1 to its receptor (TGFβR2) and causing the activation of TGFβ receptor type I (TGFβR1) kinase, leading to phosphorylation of the Smad2 and -3 complexes (24). To examine the effect of TGFβR2 on EMT, TGFβR2 siRNA or scrambled siRNA was introduced into HK2 cells. As shown in Fig. 5A, TGFβR2 siRNA effectively suppressed the protein levels of TGFβR2, and prevent EMT in HK2 cells. The results revealed that E-cadherin expression was induced, whereas the mRNA expression levels of collagen and α-SMA were reduced (Fig. 5B). HK2 cells transfected with TGFβR2-specific siRNA exhibited increased protein levels of E-cadherin and a reduced protein expression of laminin, compared with the negative controls (Fig. 5C). By contrast, the upregulation of TGFβR2 with pcmv-tag2b-TGFβR2 in the HK2 cells was able to reverse the EMT-associated gene expression, as demonstrated by the increased protein expression levels of laminin, and reduced protein expression levels of E-cadherin, revealed in the western blot analysis of cells transfected with pcmv-tag2b-TGFβR2 compared with transfected pcmv-tag2b vector (Fig. 5D).
Figure 5.
Effect of TGFβR2 on EMT. (A) The expression of TGFβR2 in HK2 cells was successfully suppressed by TGFβR2 siRNA. (B) The relative mRNA levels of E-cadherin, collagen and α-SMA in HK2 cells, following TGFβR2 siRNA treatment for 48 h. (C) Western blot analysis of the alterations of E-cadherin and laminin in HK2 cells treated with TGFβR2 siRNA for 48 h. (D) The overexpression of TGFβR2 with pCMV-tag2B-TGFβR2 promoted EMT in HK2 cells. The data are shown as the fold changes compared with cells transfected with the control. The data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, compared with the control. TGFβR2, transforming growth factor receptor β2; a-SMA, α-smooth muscle actin.
Discussion
Regardless of the type of kidney disease, fibrosis is unavoidably the outcome, resulting in the marked destruction of the kidney structure, with consequential functional deterioration (24). Myofibroblasts contribute to the accumulation of the extracellular matrix observed in fibrotic disease, and it was demonstrated that myofibroblasts may originate from renal residential fibroblasts (25,26). In addition to the residential fibroblasts, other sources of fibroblasts have been studied, including fibrocytes and pericytes. The EMT of mature tubular epithelial cells is another source of fibroblasts, which are fundamentally linked to the process of renal fibrosis (27,28). As revealed in the present study, the mRNA expression level of E-cadherin in the epithelial cells was markedly decreased, whereas the mesenchymal markers, collagen I and α-SMA, were increased in the EMT model in vitro and in vivo. Consistent with the results of the RT-qPCR analysis, the western blotting data further confirmed that the EMT process existed in TGFβ1-treated HK2 cells and the UUO model, as demonstrated by the lower expression levels of E-cadherin and higher expression levels of laminin. Furthermore, previous studies revealed that a proportion of the fibroblasts, which are predominantly the functional cells in this process, originate from the tubular epithelial cells in the diseased kidney by EMT (28–30), which has an important role in the process of kidney disease. Therefore, understanding the regulatory mechanisms of EMT may help to elucidate the events involved in the process of fibrosis, leading to the development of novel therapeutics for treating kidney disease.
miRNAs are endogenously encoded, non-coding RNAs which predominantly post-transcriptionally regulate gene expression through either translational suppressors or mRNA degradation (8). miRNAs, a class of newly identified regulators, were demonstrated to be involved in the process of EMT. In vitro and animal studies revealed that these miRNAs (miR-200a, miR-200b and miR-205) suppress EMT by targeting ZEB1 and ZEB2 (11–13). Other studies demonstrated that miR-655 and miR-34a are associated with EMT-suppressive miRNAs (31,32). The present study has revealed that miR-590 causes EMT, as demonstrated by the evidence that, to the best of our knowledge, for the first time, miR-590 affects EMT in HK2 cells. Transfection of the HK2 cells with pCDNA3.1-mir-590 resulted in an inhibition of a number of the fibrotic changes, and the promotion of various epithelial characteristics. Therefore, miR-590 appears to exert a key role in EMT, and may promote fibrosis.
Previous studies revealed that miR-590 has multiple, experimentally validated target genes, including TGFβR2, protein polybromo-1, S100A10 and the cell adhesion molecule L1-like gene (33–36). Among these targets, TGFβR2 was revealed to be associated with EMT in epithelial cells. To investigate the mechanism underlying the involvement of miR-590 in EMT, the present study has revealed that miR590 directly targets TGFβR2, as demonstrated by the luciferase reporter activity assays and western blot analyses. Protein levels of TGFβR2 were markedly decreased following miR-590 overexpression. Additionally, E-cadherin expression was upregulated and the expression of laminin was decreased. TGFβR2 was identified as a functional target of miR-590, and this target is well established as a component of the TGFβ signaling pathway. The TGFβ signaling pathway consists of TGFβ, TGFβR, the Smad protein family and several other important transcriptional regulatory factors. The TGFβR family comprises three subtypes, TGFβR1, TGFβR2 and TGFβR3, although only TGFβR2 may accept TGFβ1 as a protein partner independently of the other two receptors, which are unable to function in the absence of TGFβR2 (37). Furthermore, a previous study demonstrated that mir-655 is an EMT-suppressive miRNA, which targets TGFβR2 (32). In the present study, it was also revealed that the knockdown of TGFβR2 with siRNA elicited a similar pattern of alteration of the EMT-associated genes, whereas the transfection of pcmv-tag2B-TGFβR2 reversed the protein expression, results which are consistent with the previous hypothesis. Therefore, mir-590 was identified as an EMT-suppressive miRNA in the present study.
In conclusion, the results reported in the present study provide the first evidence, to the best of our knowledge, that miR-590 is involved in the process of EMT in tubular epithelial cells. The overexpression of miR-590 markedly increased E-cadherin expression and suppressed laminin expression in HK2 cells, clearly indicating that this miRNA is an inhibitor of EMT. Furthermore, TGFβR2, which is a core component of the TGFβ signaling pathway, was characterized as a direct target of miR-590. The present results suggest that the EMT-suppressor miR-590, targeting TGFβR2, has the potential to serve as a biomarker of, and a potential therapeutic target for, fibrosis.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 31271563), the National Basic Research Program of China (no. 2011CB944002) and the Chongqing Application Development Project (no. cstc2014yykfB10003).
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y. Epithelial to mesenchymal transition in renal fibrogenesis: Pathologic significance, molecular mechanism and therapeutic intervention. J Am Soc Nephrol. 2004;15:1–12. doi: 10.1097/01.ASN.0000106015.29070.E7. [DOI] [PubMed] [Google Scholar]
- 3.Schrimpf C, Duffield JS. Mechanisms of fibrosis: the role of the pericyte. Curr Opin Nephrol Hypertens. 2011;20:297–305. doi: 10.1097/MNH.0b013e328344c3d4. [DOI] [PubMed] [Google Scholar]
- 4.Lin SL, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cell sin obstructive fibrosis of the kidney. Am J Pathol. 2008;173:1617–1627. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31:653–662. doi: 10.1007/s10555-012-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akkina S, Becker BN. MicroRNAs in kidney function and disease. Transl Res. 2011;157:236–240. doi: 10.1016/j.trsl.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 12.Paterson EL, Kolesnikoff N, Gregory PA, Bert AG, Khew-Goodall Y, Goodall GJ. The microRNA-200 family regulates epithelial to mesenchymal transition. Scientific World Journal. 2008;8:901–904. doi: 10.1100/tsw.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YT, Chang FC, Wu CF, Chou YH, Hsu HL, Chiang WC, Shen J, Chen YM, Wu KD, Tsai TJ, Duffield JS, Lin SL. Platelet-derived growth factor receptor signaling activates pericyte myofibroblast transition in obstructive and post-ischemic kidney fibrosis. Kidney Int. 2011;80:1170–1181. doi: 10.1038/ki.2011.208. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Shultz RW, Mars WM, Wegner RE, Li Y, Dai C, Nejak K, Liu Y. Disruption of tissue type plasminogen activator gene in mice reduces renal interstitial fibrosis in obstructive nephropathy. J Clin Invest. 2002;110:1525–1538. doi: 10.1172/JCI0216219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales C, González GE, Rodríguez M, Bertolasi CA, Gelpi RJ. Histopathologic time course of myocardial infarct in rabbit hearts. Cardiovasc Pathol. 2002;11:339–345. doi: 10.1016/S1054-8807(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 18.Junqueira LC, Bignolas G, Brentani RR. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem J. 1979;11:447–455. doi: 10.1007/BF01002772. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Faulhaber-Walter R, Wen Y, Huang Y, Mizel D, Chen M, Sequeira Lopez ML, Weinstein LS, Gomez RA, Briggs JP, Schnermann J. Renal failure in mice with Gsalpha deletion in juxtaglomerular cells. Am J Nephrol. 2010;32:83–94. doi: 10.1159/000314635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaimori A, Potter J, Kaimori JY, Wang C, Mezey E, Koteish A. Transforming growth factor-beta 1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro. J Biol Chem. 2007;282:22089–22101. doi: 10.1074/jbc.M700998200. [DOI] [PubMed] [Google Scholar]
- 21.Aresu L, Rastaldi MP, Scanziani E, Baily J, Radaelli E, Pregel P, Valenza F. Epithelial-mesenchymal transition (EMT)of renal tubular cells in canine glomerulonephritis. Virchows Arch. 2007;451:937–942. doi: 10.1007/s00428-007-0482-8. [DOI] [PubMed] [Google Scholar]
- 22.Huang XR, Chung AC, Wang XJ, Lai KN, Lan HY. Mice overexpressing latent TGF-beta 1 are protected against renal fibrosis in obstructive kidney disease. Am J Physiol Renal Physiol. 2008;295:F118–F127. doi: 10.1152/ajprenal.00021.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Zhang H, Liu J, Tu X, Zang Y, Zhu J, Chen J, Dong L, Zhang J. MiR-30 inhibits TGF-b1-induced epithelial-to-mesenchymal transition in hepatocyte by targeting Snail1. Biochem Biophys Res Commun. 2012;417:1100–1105. doi: 10.1016/j.bbrc.2011.12.121. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat Rev Nephrol. 2011;7:684–696. doi: 10.1038/nrneph.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hills CE, Squires PE. The role of TGF-β and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev. 2011;22:131–139. doi: 10.1016/j.cytogfr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Barnes JL, Gorin Y. Myofibroblast differentiation during fibrosis: Role of NAD(P)H oxidases. Kidney Int. 2011;79:944–956. doi: 10.1038/ki.2010.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan HY. Tubular epithelial-myofibroblast transdifferentiation mechanisms in proximal tubule cells. Curr Opin Nephrol Hypertens. 2003;12:25–29. doi: 10.1097/00041552-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 28.Zeisberg M, Duffield JS. Resolved: EMT produces fibroblasts in the kidney. J Am Soc Nephrol. 2010;21:1247–1253. doi: 10.1681/ASN.2010060616. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Liu Y. Blockage of tubular epithelial to myofibroblast transition by hepatocyte growth factor prevents renal interstitial fibrosis. J Am Soc Nephrol. 2002;13:96–107. doi: 10.1681/ASN.V13196. [DOI] [PubMed] [Google Scholar]
- 30.Iwano M. EMT and TGF-beta in renal fibrosis. Front Biosci (Schol Ed) 2010;2:229–238. doi: 10.2741/s60. [DOI] [PubMed] [Google Scholar]
- 31.Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, Wang H, Huang C, Sun S. Hypoxia-induced downregulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS One. 2012;7:e30771. doi: 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K. MiR-655 is an EMT- suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu Y, Ouyang Y, Wang F, Zheng A, Bai L, Han L, Chen Y, Wang H. MicroRNA-590 promotes cervical cancer cell growth and invasion by targeting CHL1. J Cell Biochem. 2014;115:847–853. doi: 10.1002/jcb.24726. [DOI] [PubMed] [Google Scholar]
- 34.Xiao X, Tang C, Xiao S, Fu C, Yu P. Enhancement of proliferation and invasion by MicroRNA-590-5p via targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res. 2013;20:537–544. doi: 10.3727/096504013X13775486749335. [DOI] [PubMed] [Google Scholar]
- 35.Shan X, Miao Y, Fan R, Qian H, Chen P, Liu H, Yan X, Li J, Zhou F. MiR-590-5P inhibits growth of HepG2 cells via decrease of S100A10 expression and inhibition of the Wnt pathway. Int J Mol Sci. 2013;14:8556–8569. doi: 10.3390/ijms14048556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, Xiang G, Wang Y, Zhang L, Yang X, Cao L, Peng H, Xue P, Chen D. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-β RII. Mol Cells. 2012;33:545–551. doi: 10.1007/s10059-012-2267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]