Abstract
The hepatitis B virus (HBV) infection is a major risk factor in the development of chronic hepatitis (CH) and hepa-tocellular carcinoma (HCC). The activation-induced cytidine deaminase (AID)/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family of cytidine deaminases is significant in innate immunity, as it restricts numerous viruses, including HBV, through hypermutation-dependent and -independent mechanisms. It is important to induce covalently closed circular (ccc)DNA degradation by interferon-α without causing side effects in the infected host cell. Furthermore, organisms possess multiple mechanisms to regulate the expression of AID/APOBECs, control their enzymatic activity and restrict their access to DNA or RNA substrates. Therefore, the AID/APOBECs present promising targets for preventing and treating viral infections. In addition, gene polymorphisms of the AID/APOBEC family may alter host susceptibility to HBV acquisition and CH disease progression. Through G-to-A hypermutation, AID/APOBECs also edit HBV DNA and facilitate the mutation of HBV DNA, which may assist the virus to evolve and potentially escape from the immune responses. The AID/APOBEC family and their associated editing patterns may also exert oncogenic activity. Understanding the effects of cytidine deaminases in CH virus-induced hepatocarcinogenesis may aid with developing efficient prophylactic and therapeutic strategies against HCC.
Keywords: hepatitis B virus, activation-induced cytidine deaminase, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like, polymorphism, hepatocellular carcinoma
1. Introduction
The hepatitis B virus (HBV) infection is a prevalent type of infectious disease that is causing a global concern for public health (1). Although there has been considerable improvement in understanding the molecular virology and pathogenesis of the HBV infection over the past decade and effective therapeutic measures have been developed for its treatment, there are currently 240 million individuals globally who are chronic HBV carriers, and ~620,000 succumb per year due to late sequelae of liver cirrhosis or hepatocellular carcinoma (HCC) (1). The treatment of chronic hepatitis (CH) B is currently limited; predominantly consisting of interferons (IFNs) and nucleoside analogs (NUCs) (2). NUCs are efficient antiviral agents, however, NUCs only control rather than cure HBV infections due to persistent viral covalently closed circular (ccc)DNA. Therefore, a long-term treatment is required, which is expensive and may lead to concomitant resistance (2). IFN therapy is associated with side effects, and treatment with this cytokine can result in viral clearance in a small proportion of patients (3). Thus, it is important to continue conducting research in order to identify therapeutic targets to stimulate the development of novel antiviral agents and immunotherapies.
The activation-induced cytidine deaminase (AID)/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family, which was first described for inhibition of human immunodeficiency virus type 1 (HIV-1), is important in the innate immune system, as it defends against viruses, including HBV through hypermutation-dependent and -independent mechanisms. Nine of 11 APOBEC3 family members have been identified to exert varying levels of activity against HBV under experimental conditions (Table I). APOBEC-3A and APOBEC-3B are essential for cccDNA degradation by IFN-α or the lymphotoxin-β receptor-agonist without damaging the infected host cells (4). Thus, targeting the formation and subsequent processing of viral cccDNA may be more rational approaches.
Table I.
Reported hyperediting and restriction activity of AID/APOBECs against HBV DNA.
| Cytidine deaminases | Hyperediting activity | Dinucleotide preference | Restriction activity against HBV DNA | Reference |
|---|---|---|---|---|
| AID | Deaminates each viral RNA and HBV minus-strand DNA | GpC, ApC | Active | 25,27 |
| A1 | Deaminates HBV minus-strand DNA | TpC | Active | 19,21,25 |
| A2 | NI | NI | Not active | 25 |
| A3A | Deaminates each strand of HBV DNA | Weak bias | Active/not activea | 4,17,25 |
| A3B | Deaminates each strand of HBV DNA | GpC, ApC | Active | 4,16,24–26,28,37 |
| A3C | Deaminates HBV minus-strand DNA | No bias | Active but weak/not activea | 16–18,20,26,37 |
| A3DE | Not active | N/A | NI | 17 |
| A3F | Deaminates each strand of HBV DNA | TpC, GpC | Active | 16–18,20,25,26,37 |
| A3G | Deaminates each strand of HBV DNA | CpC | Active | 16–18,20,26,37,75 |
| A3H | Deaminates each strand of HBV DNA | TpC | NI | 17,20 |
Controversial experimental results exist in the article. NI, no information found; N/A, not applicable; AID, activation-induced cytidine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; HBV, hepatitis B virus.
The aim of the present study was to review the rapidly accumulating evidence for the involvement of AID/APOBEC cytidine deaminase expression and HBV replication, and to summarize the current knowledge surrounding the role of editing in HBV-associated liver disease as a consequence of its actions in host cells.
2. AID/APOBEC family of deaminases
AID/APOBEC family members
All members of the AID/APOBEC family possess one or two catalytic domains that deaminate cytidine in RNA and DNA. The deaminases mediate the hydrolytic removal of an amino group at the C4 position of a cytidine (C) or deoxycytidine (dC) generating a uridine (U) or deoxyuridine (dU), respectively (5). The presence of the enzyme in cells producing RNA virus results in C-to-U conversion of minus strand reverse transcripts and G-to-A in plus strand DNA. The binding to the target DNA creates a U-G mismatch, which generates a C-to-T transition in minus strand DNA and a G-to-A transition in plus strand DNA during general DNA replication without repair pathways (Fig. 1) (6). In humans, the family comprises 11 members with distinct functions, including AID, APOBEC1 (A1), APOBEC2 (A2), APOBEC4 (A4) and APOBEC3 (A3) subgroups. The A3 group consists of seven proteins: A3A, A3B, A3C, A3DE, A3F, A3G and A3H (7–10). The seven A3 genes are arranged in a tandem gene cluster on chromosome 22 in humans (11). The presence of the AID/APOBEC family is restricted to vertebrates. AID and A2 are likely to be the ancestral members, while A1 and A3 are later evolutionary arrivals (12), A3s are restricted to placental mammals, and their gene copy number is species-specific. For example, mice only possess a single A3 gene, pigs have two, sheep and cattle have three, cats have four, horses have six and primates have at least seven A3 genes (13). The rapid expansion of the A3 locus in humans indicates an important role in the host genome defense against exogenous viruses and endogenous retroelements (12–14). The role of the AID/APOBEC family in the inhibition of viral infection was initially described for HIV-1. Various studies have shown that the genome of hepadnaviruses is hyperedited by cytidine deaminases (15–21). Recent reports demonstrated that HBV DNA replication is restricted by A1, AID, A3A, A3B, A3C, A3G, A3F, but not A3DE (4,15,17,22–26), specifically via the degradation of HBV cccDNA (4), which was investigated in an experimental setting through deaminase-independent and -dependent mechanisms (27,28).
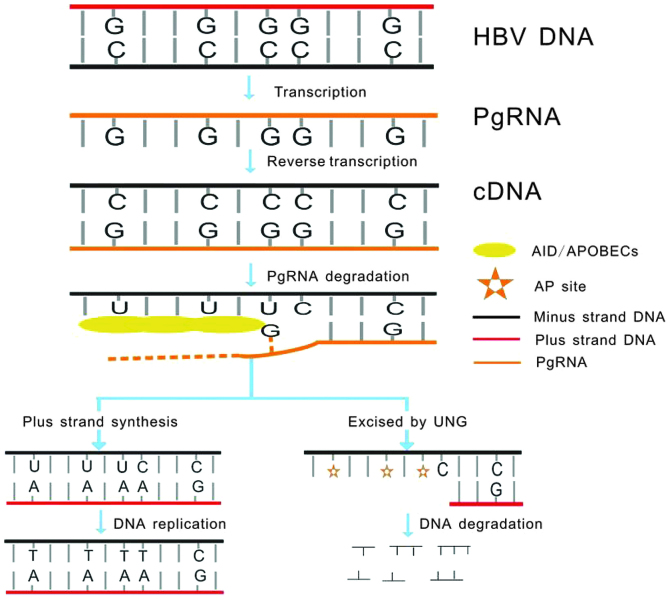
Figure 1.
HBV DNA is transcribed to pgRNA as a replicative RNA intermediate, according to which the minus DNA strand (cDNA) forms. ssDNA is formed as the pgRNA is degraded. AID/APOBECs catalyze cytosine deamination of HBV DNA on the cDNA, producing uracil during reverse transcription. Uracils in DNA (including cccDNA) are recognized and excised by UNG leading to formation of AP sites. These AP sites are recognized by cellular AP endonucleases leading to DNA digestion. The HBV DNAs that do not undergo degradation, generate C-to-T transitions in minus strand DNA and G-to-A transitions in plus strand DNA during general DNA replication. HBV, hepatitis B virus; pgRNA, pregenomic RNA; ssDNA, single stranded DNA; AID, activation-induced cytidine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; cccDNA, covalently closed circular DNA; UNG, uracil DNA glycosylases; AP, apurinic/apyrimidinic.
AID/APOBEC family gene expression
A1 is primarily expressed in the gastrointestinal compartment and catalyzes the expression of a truncated form of apolipoprotein (Apo) B (ApoB48) with the distinct biological function of ApoB100 (10). AID is expressed in germinal center B cells and randomly edits dC residues to dU in the variable region of immunoglobulin gene loci, which is essential for the process of antibody diversification (29,30). As expected, AID and A1 were not detected in the HCC cell lines and liver tissue (20,25). A2 is widely expressed in muscle; predominantly in cardiac and skeletal muscle, thus, is exclusively associated with the development of cardiac and skeletal muscle, as well as early embryogenesis (9,31–33). A4 was discovered by a computational homology search and was found to be expressed in the testicles, although its function remains undetermined (8). A3 (particularly A3G and A3F) is widely expressed in human tissues, where the mRNA levels broadly correlate with the lymphoid cell content (with the exception of gonadal tissues) (34). In the healthy human liver, low to moderate APOBEC expression levels have been observed (11,20,26,35,36), with A3G expression levels identified to be the highest, followed by A3C and A3H expression (18). Positive transcription and expression has been identified for A3G, A3C and A3H in HepG2 cells, although not for A3B and A3F. Amplification experiments with A2- and A3A-specific primers resulted in particularly faint signals, and A1- and A3DE-specific sequences were not detected (18).
Onset of liver disease results in changes to the levels of these deaminases. For example, in hepatitis/cirrhotic samples, five of seven A3 genes were significantly upregulated in the following order: HCV ± HBV > HBV > alcoholic cirrhosis. A3C and A3D were upregulated for all groups, while IFN inducible A3G was overexpressed in virus-associated cirrhotic patients, and AID was present in 50% of the HBV/HCV samples (21). The expression levels of A1, A2, and A4 were not identified to be significantly different between cirrhotic liver and normal tissue samples (21). Furthermore, the A3B cytidine deaminase was widely upregulated in HCC tumor tissue samples (37).
Upregulation by IFNs and other cytokines
The expression of APOBEC genes is induced by IFNs, as well as other cytokines. IFN-α has been observed to trigger the expression of A3 in numerous reports. For example, treatment with IFN-α in primary human hepatocytes (PHHs) and HCC cell lines resulted in induction of A3-associated genes, particularly A3G; however, no changes were triggered in the expression of A1, A2 or AID (25,26,36). In addition, A3A was demonstrated to be significantly expressed in peripheral blood mononuclear cells, HBV-infected differentiated HepaRG cells and PHHs that were treated with IFN-α (4,36,38). IFN-α enhances A3 expression levels in human macrophages (39), phagocytes (38) and peripheral plasmacytoid dendritic cells (40). This indicates that A3 expression may contribute to the antiviral effects of IFN-α against HBV DNA. In addition to IFN-α, other IFN family members, such as IFN-γ and IFN-β, directly upregulate A3G expression in macrophages (39,41). IFN-γ is known to control HBV in a dose-dependent reduction manner, which was consistent with the observation of a dose-dependent increase in A3G and A3F protein expression following IFN-γ therapy (42). In addition, the expression of AID is induced in response to tumor necrosis factor (TNF)-α. Interleukin (IL)-1β stimulation in cultured human hepatocytes and pro-inflammatory cytokine-mediated expression was meditated by nuclear factor-κB signaling pathways (43,44) and by IL-4 and -13 through STAT6 (45). Transforming growth factor-β is a cytokine that induces AID in B and HepG2 cells (46), whereas TNF-α and IL-1β stimulate the expression of A2 in human hepatocytes (44).
3. Specific regulation of editing by AID/APOBECs
Mechanism of regulation
The mutagenic activity resulting from AID/APOBEC-mediated deamination is toxic for retroviruses and beneficial to organisms in host cell defense (13). However, its excessive activity and off-target mutations within the cellular genome are also toxic and oncogenic to the host. Thus, organisms possess multiple mechanisms to regulate the expression of AID/APOBECs; for example, controlling their enzymatic activity and restricting their access to DNA or RNA substrates (47). A1 is the sole family member capable of recognizing and using mRNA as a substrate. Editing only occurs on cytidines 5′ of the mooring sequence (48). Although RNA editing occurs in the cytoplasm when A1 is overexpressed (49), cytoplasmic A1 editing activity is suppressed under normal physiological conditions. In addition, RNA editing is restricted to the cell nucleus within a temporal and spatial window that occurs subsequent to pre-mRNA splicing and prior to mRNA nuclear export, only when interacting with cofactor, APOBEC-1 complementation factor (50,51). The sequence requirements for AID/APOBEC deamination are not stringent, however, deamination generally occurs within transcribed or single-stranded regions of DNA. Therefore, single-stranded (ss)DNA is always recognized as a substrate by the AID/APOBEC family. The deaminase activity of AID is responsible for a variety of point mutations and DNA breaks. This mutagenic activity leads to somatic hypermutation (SHM) and class switch recombination (52). Notably, the expression of AID is restricted to activated B cells under physiological conditions, and only targets variable and switch regions of immunoglobulin genes for mutagenesis. A2 is essential for muscle development with its expression being largely restricted to striated muscle. However, in vitro (32,53) A2 was identified to lack autonomous deaminase activity. The above-mentioned processes are regulated in a tissue-specific manner during development and in response to metabolic regulation. Furthermore, subcellular localization and RNA binding to AID/APOBECs regulate their deaminase activity. Furthermore, AID is regulated by nuclear-cytoplasm transport and is predominantly located in the cytoplasm. Controlling the abundance of AID exclusion from the nucleus in steady state is one of the major regulatory mechanisms restricting its contact with genomic material (54,55). A3G is strongly retained in the cytoplasm and excluded from chromosomes based on the specialized property of a novel cytoplasmic retention signal (56). A3G is present in two distinct molecular forms within the cell: A low molecular mass form and a high molecular mass (HMM) complex, which contains one or more inhibitory RNAs that inactivate ssDNA deaminase activity (57–59). A3G-RNA complexes in viral particles have been found to be inactive until RNase H activity of reverse transcriptase (RT) degrades the RNA of DNA-RNA hybrids (60). A3C, A3F and A3H all form intracellular HMM complexes (61–63) in addition to A3G. AID exerts no measurable deaminase activity on ssDNA unless the AID is pretreated with RNase to remove the inhibitory RNA, which is bound to the AID (29).
4. Role of AID/APOBECs in HBV-associated disease progression
Promotion of the evolution of HBV virus
The encapsidated HBV genome consists of a 3.2-kb partially double-stranded relaxed circular (rc)DNA molecule. Upon translocation to the nucleus, the rcDNA genome is converted into a cccDNA by cellular repair factors and remains an episomal minichromo-some, which transcribes all viral RNAs, including pregenomic (pg)RNA as a replicative RNA intermediate. The pgRNA is translated to form the core protein and internal translation initiation synthesizes the DNA polymerase. The pgRNA, viral core, and polymerase proteins are assembled into the nucleocapsid in the cytoplasm (1). Reverse transcription starts to synthesize the (minus) DNA strand via reverse transcription activity of the viral polymerase. Following degradation of the pgRNA by the viral RNase H, the plus strand is synthesized to form mature core particles, which interact with the preS domain of the membrane-associated viral surface proteins and acquire the viral envelope (64). Approximately 1011 viral particles are released per day into the circulation of individuals with the chronic HBV infection and HBV particles are cleared from the plasma with a half-life of ~1.0 day (65,66). In the initial immunotolerant phase of chronic HBV infection, the immune pressure is weak. With the progression of chronic infection, especially during Hepatitis B e antigen (HBeAg) seroconversion, HBV mutations gradually occur (67). The mutation rate of HBV DNA caused by AID/APOBECs in chronic HBV infection patients has been shown to be higher than that of acute HBV infection patients. In the chronic HBV infection group, the frequency of hypermutated genomes was found to be higher in HBeAg-negative individuals when compared with seropositive cases (68,69), and the degree of mutation of the HBV DNA was significantly associated with the extent of fibrosis (68). This may have been due to the fact that HBeAg-negative patients had undergone HBeAg seroconversion, resulting in an already elevated immune response (70). Although mutations in A3 that result in altered editing can be highly deleterious, lightly edited genomes may facilitate the virus to evolve and even escape from the immune responses (21). Non-resolving inflammation has been shown to be indispensable for immune-selection of the AID/APOBECs-dependent HBV mutations (6).
5. Inhibition of HBV by AID/APOBEC proteins
A3s are effective inhibitors of HBV DNA replication
A3s are effective restrictive factors for various types of virus with ssDNA, including HBV. It has been shown that A3 catalyzes cytosine deamination of HBV DNA produced during reverse transcription as a template, resulting in the presence of uracil in cDNA. Uracils in DNA (including cccDNA) are recognized and excised by cellular uracil DNA glycosylases (UNGs) leading to formation of apurinic/apyrimidinic (AP) sites, which are then recognized by cellular AP endonucleases resulting in DNA digestion (Fig. 1) (4,71–73). Certain studies describe contradictory results and report that the loss of UNG does not restore the levels of viral reverse transcripts, which had been decreased by A3G (74). A previous study found that HBV replication was inhibited by A3G (75). However, the G-to-A hypermutation was not observed in HBV DNA. Further studies using a 3D-polymerase chain reaction (PCR) method showed that A3B, A3C, A3F and A3G extensively deami-nated cytidine residues in minus strand DNA. Unexpectedly, three of the four enzymes (A3B, A3F and A3G) deaminated HBV plus strand DNA in human HCC cell lines; however, the underlying mechanism remained undetermined (16). In another study, Henry et al (17) compared the HBV editing by all seven enzymes in a quail cell line, which did not produce any endogenous DNA cytidine deaminase activity. The study identified that all of the A3s, not including A3DE, deaminated the HBV DNA levels from 10−2 to 10−5 in vitro, with A3A proving to be the most efficient editor (17). The hyperediting ability of A3A was confirmed by Lucifora et al (4). Although A3C was weaker at inhibiting HBV replication compared with A3G, its capacity for DNA hyperediting was higher than A3G and A3H (18,20). A3G demonstrated a strong preference for CpC, whereas A3H demonstrated a preference for TpC (19). These findings indicate that human A3 enzymes impact HBV replication by cytidine deamination. However, the induction of hypermutations is not sufficient for full inhibition of HBV replication.
A cytidine deaminase null mutant of A3B (E255A) was also found to inhibit HBV RNA production, although it was unable to edit HBV (28). It was proposed that only the carboxy (C)-terminal deaminase domain of A3B catalyzes HBV hypermutations (24,26). By investigating a series of A3B point mutants, it was found that the N- and C-terminal cytosine deaminase domains of A3B (24) and A3G (76) exerted inhibitory effects on HBV replication. By contrast, a truncated splice variant of A3B that lacked the C-terminal deaminase domain exerted no effect on HBV replication (26). However, A3G, which was defective for deaminase activity, was found to inhibit HBV replication (75). From the above-mentioned reports, it is hypothesized that the inhibition of HBV may be mediated by additional, different mechanisms.
Possible editing-independent mechanisms
In principle, a decrease in HBV DNA may result from either accelerated DNA degradation or decreased synthesis. A previous report suggested that A3G decreased the viral DNA levels by inhibiting pgRNA packaging (75). Subsequently, Rösler et al (15) revealed that the early stages of HBV DNA morphogenesis, including RNA and protein synthesis, binding of pgRNA to the core protein, and self-assembly of the viral core protein, were unaffected. However, A3G rendered HBV core protein-associated full-length pgRNA nuclease sensitive. Nguyen et al (27) analyzed the mechanisms of deamination-independent suppression of HBV replication by A3G and did not identify enhanced DNA degradation by A3G, either in vivo or in vitro. A3G appeared to inhibit the very early steps in viral reverse transcription and blocked DNA strand elongation. Furthermore, the deamination-independent antiviral function of A3G targets DNA-RNA hybrids (27). The A3B protein inhibits the binding of heterogeneous nuclear ribonucleoprotein K to the enhancer II of HBV and represses the activity of HBV. In addition, A3B directly suppresses HBV S-promoter activity (23). The process is presented schematically in Fig. 2.
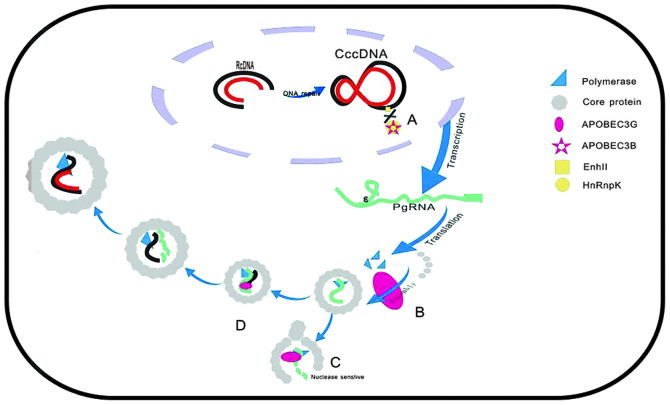
Figure 2.
The rcDNA genome is converted into cccDNA by cellular repair factors. Then, the cccDNA is transcribed to the pgRNA and subgenomic mRNAs (not shown). The mRNAs are transported to the cytoplasm. The pgRNA is translated in the cytosol to form HBV core protein and the viral polymerase. These three components assemble to form the core particle. The first (minus) DNA strand forms within the core particles via reverse transcription of the pgRNA to DNA; the pgRNA is degraded by viral RNase H as the plus strand is synthesized. (A) A3B inhibits the binding of HnRnp K to the Enh II of HBV; (B) A3G may inhibit pgRNA packaging; (C) A3G renders HBV core protein-associated full-length pgRNA nuclease-sensitive; (D) A3G blocks DNA strand elongation and targets a DNA-RNA hybrid. rcDNA, relaxed circular DNA; cccDNA, covalently closed circular DNA; pgRNA, pregenomic RNA; HBV, hepatitis B virus; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; HnRnp K, heterogeneous nuclear ribonucleoprotein K; Enh II, enhancer II.
Inhibition of HBV by additional APOBEC proteins
AID and A1 also inhibit HBV replication. AID was significantly upregulated in ~50% of HBV and HCV cirrhotic samples indicating that ectopic expression of AID was also a feature of HBV liver disease. A1 was markedly expressed in one sample (although normalization of the expression levels was not conducted), A2 transcripts were detected in certain samples, but A4 was not detected in any sample. AID was identified to be as effective as A3G at hyperediting HBV DNA (21). Mutation matrices and editing frequencies in the HBV minus strand DNA were comparable, for example, the editing frequency for A1 was 32%, and for A3G was 34% (18). C-to-T (U) hypermutations were observed in rcDNA and nucleocapsid viral RNA indicating that AID deaminates viral RNA and ssDNA (77). A1 exhibited a strong preference for TpC and a strong aversion to GpC (19) and AID editing of the HBV target was concentrated at the GpC and ApC sites (21). Furthermore, the low expression levels of A1 and AID in Huh7 cells was consistently associated with decreased levels of core-associated HBV DNA by quantitative PCR. However, A2 did not exert any effect on HBV replication (25).
AID/APOBECs incorporation into viral particles
Research on HBV reveals that the antiviral activity of A3G requires incorporation into assembling viral particles to inhibit reverse transcription. Factors required for incorporation of the antiviral deaminase protein, A3G into HBV nucleocapsids continues to be investigated. It has been demonstrated that A3G and A3C bind to the HBV core protein in immunoprecipitation experiments (78,79). Such binding is essential for reverse transcription following infection. The binding of A3G to the HBV core protein was only indirectly demonstrated with coexpression of RT and pgRNA, however, not with core protein alone (15,18). The results are consistent with the findings of Nguyen et al (80) that A3G was specifically incorporated into replication-competent HBV nucleocapsids by interacting with viral RT and RNA packaging signals. However, by fluorescence resonance energy transfer (FRET) and acceptor photobleaching experiments, Zhao et al (81) revealed that A3G directly binds to core proteins. Similarly, direct interaction of HBV core protein and A3A was confirmed by proximity ligation assay and FRET analysis. Deletion analysis was used to confirm that the central region of the HBV core protein (between aa 77 and 149) was involved in the interaction with A3A (4). Additionally, the A3B, A3C and A3F enzymes were also found to be associated with the HBV capsid by interaction with the core protein (16). Similar to A3G, AID was co-immunoprecipitated with the nucleocapsid core protein. The assumption was made that AID formed a ribonucleopro-tein complex with the HBV core proteins and RNA during nucleocapsid assembly in which AID deaminated cytosines of the viral RNA, including pgRNA and ssDNA (77).
6. APOBEC polymorphisms in HBV infections
The APOBEC family is considered to be significant in innate cellular immunity, particularly in HBV hyperediting and infection inhibition. It was of interest to elucidate whether gene polymorphisms alter host susceptibility to HBV progression of CH. A3B is an efficient editor and inhibitor in the process of HBV infection. A 29.5-kb deletion between exon 5 in A3A and exon 8 in an A3B gene cluster resulted in complete removal of the A3B coding region (82,83). A hybrid gene containing the coding region of A3A and the 3′-untranslated region of A3B was initially discovered by mapping end-sequence pairs from a human fosmid library against the human genome reference sequence assembly (82). This was confirmed by dense single nucleotide polymorphism marker mapping (83). A report on the frequency of the deletion allele conducted using 1,277 diverse human samples revealed that its expression is rare in African and European individuals (frequency of 0.9 and 6%, respectively), more common in East Asian and Amerindian individuals (36.9 and 57.7%, respectively) and almost universal in Oceanic populations (92.9%) (84). Analyzing the association between this polymorphism and chronic HBV infection may clarify the effect of A3B on the establishment of chronic HBV carrier state. Abe et al (85) initially investigated the association between this polymorphism and chronic HBV infection. No association was identified between the chronic HBV carrier state and deletion polymorphisms; however, the extent of liver fibrosis was found to be associated with insertion homozygosity (85). Similarly, another report demonstrated that there was no significant difference in the frequencies of deleted A3B alleles or genotypes between patients with CH B and control subjects. By contrast, subjects bearing a deleted genotype experienced a more rapid progression of liver disease when compared with those exhibiting an insertion genotype (86). This result indicated that in the A3B deletion polymorphism, A3 cytidine deaminases do not predispose an individual to chronicity, but may modulate the course of persistent HBV infection. These results are contradictory to a study, which revealed that a 29.5-kb deletion obliterated the production of A3B, and was significantly associated with increased susceptibility to persistent HBV infection in an ethnic Han Chinese population (28).
Seven haplotypes (designated with Roman numerals, I-VII) were reported to be present in various human populations based on the five single amino acid polymorphisms, N15Δ, R18L, G105R, K121D and E178D (87). The N15Δ and R105G mutations may independently cause a marked decrease in A3H expression. Only the haplotypes II, IV and VII were without these two mutations, and were stably expressed and found to efficiently inhibit HIV-1 replication (88). In addition, Wang et al (87) found that E140K, H54R, E56Q, C85S and C88S, as well as the two mutants, W115A and W115L, disrupted A3H expression. The A3G H186R polymorphisms in HBV-infected patients were found to be different to those of healthy Moroccan individuals, although the difference was not significant (86). The outcomes of a selection of studies are summarized in Table II. The association between AID/APOBEC polymorphisms and liver disease requires further, in-depth investigation.
Table II.
Summary of linkage studies between clinical indicators and AID/APOBEC.
| A, APOBEC3 polymorphism studies
| ||
|---|---|---|
| Study (ref.) | Cohort description | Association with APOBEC3 polymorphisms |
| Abe et al, 2009 (85) | 724 patients with chronic HBV infection and 469 healthy control subjects. | No significant association between A3B deletion polymorphism and chronic HBV carrier state. A3B gene deletion homozygosity was associated with mild liver fibrosis. |
| Zhang et al, 2013 (28) | 1,124 patients with HBV-associated HCC, 510 individuals with persistent HBV infection and 826 healthy control subjects. Population, Han Chinese. |
At least one A3B deletion allele increased the risk for persistent HBV infection and HCC development. |
| Ezzikouri et al, 2013 (86) | 179 HBV chronic carriers and 216 healthy control subjects. Population, Moroccan. |
No significant difference in the frequencies of deleted A3B alleles or genotypes between the two groups. Patients with deleted genotypes experienced a faster progression of liver disease than those with insertion genotypes. A3B deletion exhibited significantly lower viral loads than patients with the wild-type. A3G H186R polymorphism R/R genotype frequencies were not significantly different in the HBV-infected patients and healthy subjects. |
| B, Hypermutation studies
| ||
|---|---|---|
| Study (ref.) | Cohort description | Association with hypermutation |
| Beggel et al, 2013 (68) | 80 treatment-naïve HBV infection patients (47 HBeAg-positive and 33 HBeAg-negative). | Hypermutation rates for HBeAg-negative patients were >10-fold higher than those of HBeAg-positive patients. HBeAg-negative patients higher hypermutation rates were significantly associated with the degree of fibrosis. |
| Noguchi et al, 2005 (69) | 8 Japanese adult patients with acute HBV infection, 10 patients were chronic carriers. | Hypermutated HBV DNA was detected in 1/8 patients with acute HBV infection and 4/10 patients with chronic HBV infection. In the latter group, hypermutated genomes were found only in eAb-positive patients. |
| C, APOBEC3 expression studies
| ||
|---|---|---|
| Study (ref.) | Cohort description | Association with APOBEC3 expression |
| Vartanian et al, 2010 (21) | 41 cirrhotic samples (10 alcoholic cirrhosis, 10 HBV+, 11 HBV+HCV+, 10 HCV+, 4 normal livers). | 5/7 A3 genes were significantly upregulated in the order: HCV ± HBV > HBV > alcoholic cirrhosis. A3C and A3D were up regulated for all groups. Interferon inducible A3G was overexpressed in virus-associated cirrhosis, as was AID in 50% of these HBV/HCV samples. |
| Xu et al, 2007 (37) | 29 pairs of HCC and surrounding non-cancerous tissue samples. | A3B transcripts were significantly elevated in 24/29 HCC. |
| Kou et al, 2007 (46) | 51 HCC patients (14 HBV+, 30 HCV+) with 25 CH and 26 LC patients, and 6 normal livers. | AID was significantly upregulated in HCC and surrounding non-cancerous liver tissues underlying CH or LC. |
AID, activation-induced cytidine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; HBV, hepatitis B virus; CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HBeAg, hepatitis B e antigen.
7. Emerging role of the AID/APOBEC family in the development of cancer
The AID/APOBEC family and their associated editing patterns may be involved in oncogenesis. Recent analyses of the mutations have implicated APOBEC cytidine deaminases as significant factors in the mutagenesis of human cancer genomes (89,90). Within cancer genomes, APOBEC mutagenesis has been found to be pervasive and correlated with APOBEC mRNA levels (89–91). Roberts et al (89) identified that tumor samples, from 14 types of cancer, contained hundreds of APOBEC-signature mutations, constituting ≤68% of mutations in the exome. In at least six distinct types of cancer, similar results have revealed that A3B is upregulated, and its preferred target sequence was frequently mutated and clustered (91).
AID is critical in class-switch recombination and SHM of the immunoglobulin gene in B lymphocytes (92). Animal models have demonstrated that the aberrant expression of AID contributes to HCC tumorigenesis (93,94). A positive correlation between somatic mutation frequencies and AID expression levels was observed in the p53 gene in liver tissues with AID upregulation, as downregulation and somatic mutations in the p53 gene have been well characterized in human hepatocarcinogenesis (46). These data indicate that AID/APOBEC-catalyzed deamination may be important in generating somatic mutations during the progression of virus-associated HCC.
Transgenic overexpression of A1 in rabbit and mouse livers resulted in liver dysplasia and HCC through excessive editing of hepatic mRNAs, such as tyrosine kinase (95). Furthermore, hyperediting of the novel A1 target no 1 mRNA that encoded a tumor suppressor gene, created stop codons and truncated protein products, which are linked to liver cancer (96). Aberrant A2 expression resulted in nucleotide alterations in the transcripts of a specific target gene and may be involved in the development of human HCC via hepatic inflammation (97). A3 deaminases generate C-terminally truncated HBx mutants, which enhance the colony-forming ability and proliferative capacity of neoplastic cells. Notably, A3B was observed to be widely upregulated in HCC tissues, and promoted the growth of neoplastic human HepG2 liver cells and upregulated heat shock transcription factor 1 expression levels (37). The expression of A3A may lead to induction of DNA breaks and activation of damage responses in a deaminase-dependent manner. Consistent with the above-mentioned observations, A3A expression was found to affect genomic integrity by inducing cell-cycle arrest (98). The outcomes of a selection of studies are summarized in Table III.
Table III.
Summary of linkage studies between HCC and AID/APOBECs.
| Protein | Experimental targets | Indication | Reference |
|---|---|---|---|
| A1 | TM, TR | All of the TM and one TR had liver dysplasia. 8/35 TM developed HCC. | 95 |
| A1 | TM, TR | The aberrant editing markedly reduced levels of protein expression by the tumor suppressor gene, NAT1. | 96 |
| A2 | TM | HCC developed in 2/20 A2 TM at 72 weeks of age. Significantly high frequencies of nucleotide alterations in EIF4G2 and PTEN genes were observed in hepatocytes. | 97 |
| A3 | CL, HCCT | C-terminally truncated HBx mutants generated by A3 enhanced the colony forming ability and proliferative capacity of neoplastic cells. A3B upregulated HSF1. | 37 |
| A3A | CL | A3A led to induction of cellular DNA breaks and activation of damage responses in a deaminase-dependent manner. A3A expression induced cell cycle arrest. | 98 |
| AID | CL, HCCT | The majority of liver tissues with AID upregulation contained multiple genetic changes in the p53 gene. Aberrant activation of AID in hepatocytes resulted in accumulation of multiple genetic alterations in the p53 gene. | 46 |
| AID | TM | HCC developed in 27% of tissue-nonspecific alkaline phosphatase-AID TM at the age of 90 weeks. The HCC expressed α-fetoprotein and possessed deleterious mutations in the tumor suppressor gene, TRP53. | 94 |
AID, activation-induced cytidine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; TM, transgenic mice; TR, transgenic rabbit; CL, cell line; CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HCCT, HCC tissue; NAT1, novel A1 target no 1; EIF4G2, eukaryotic translation initiation factor 4 γ, 2; PTEN, phosphatase and tensin homolog; HBx, hepatitis B virus X; HSF1, heat shock transcription factor 1; TRP53, tumor protein P53.
8. Conclusion
Innate immunity mechanisms are the first line of defense against invading viruses. The AID/APOBEC family of cytidine deaminases is significant in the regulation of tissue development, responding to metabolic regulation and facilitating innate immunity by restricting numerous types of virus, including HBV. Unresolved issues are the mechanisms of AID/APOBEC-dependent specific recognition of HBV DNA, degradation of cccDNA, and the security and availability of experimental models, which are required for further investigation. Recent analyses of the mutations have indicated that AID/APOBEC cytidine deaminases are significant factors in the mutagenesis of human cancer genomes. A3B, which is only localized to the nucleus, is proposed to be responsible for a large proportion of dispersed and clustered mutations in multiple distinct cancers, including HCC. It is essential to elucidate the mutational processes underlying the development of cancer, and its potential implications on cancer etiology, prevention and therapy.
Abbreviations
- AID
activation-induced cytidine deaminase
- APOBEC
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like
- HCC
hepatocellular carcinoma
- cccDNA
covalently closed circular DNA
- ApoB48
apolipoprotein B
- IFN-α
interferon-α
- PHH
primary human hepatocytes
- ssDNA
single stranded DNA
- SHM
somatic hypermutation
- HMM
high molecular mass
- rcDNA
relaxed circular DNA
- pgRNA
pregenomic RNA
- UNG
uracil DNA glycosylases
- RT
reverse transcriptase
- FRET
fluorescence resonance energy transfer
- CH
chronic hepatitis
References
- 1.Gerlich WH. Medical virology of hepatitis B: How it began and where we are now. Virol J. 2013;10:239. doi: 10.1186/1743-422X-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoulim F. Hepatitis B virus resistance to antiviral drugs: Where are we going? Liver Int. 2011;31(Suppl 1):111–116. doi: 10.1111/j.1478-3231.2010.02399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M, et al. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology. 2006;44:675–684. doi: 10.1002/hep.21282. [DOI] [PubMed] [Google Scholar]
- 4.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: New members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 6.Deng Y, Du Y, Zhang Q, Han X, Cao G. Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation and hepatocellular carcinoma. Cancer Lett. 2014;343:161–171. doi: 10.1016/j.canlet.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 7.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 9.Liao W, Hong SH, Chan BH, Rudolph FB, Clark SC, Chan L. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem Biophys Res Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- 10.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 11.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 12.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 13.Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013;2013:683095. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severi F, Chicca A, Conticello SG. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol Biol Evol. 2011;28:1125–1129. doi: 10.1093/molbev/msq338. [DOI] [PubMed] [Google Scholar]
- 15.Rösler C, Köck J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsäcker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 16.Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry M, Guétard D, Suspène R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3 G. PLoS One. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumert TF, Rösler C, Malim MH, von Weizsäcker F. Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology. 2007;46:682–689. doi: 10.1002/hep.21733. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez MC, Suspène R, Henry M, Guétard D, Wain-Hobson S, Vartanian JP. Human APOBEC1 cytidine deaminase edits HBV DNA. Retrovirology. 2009;6:96. doi: 10.1186/1742-4690-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köck J, Blum HE. Hypermutation of hepatitis B virus genomes by APOBEC3 G, APOBEC3C and APOBEC3H. J Gen Virol. 2008;89:1184–1191. doi: 10.1099/vir.0.83507-0. [DOI] [PubMed] [Google Scholar]
- 21.Vartanian JP, Henry M, Marchio A, Suspène R, Aynaud MM, Guétard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P, et al. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010;6:e1000928. doi: 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Liu J, Kang F, Guan W, Gao X, Wang Y, Sun D. Core-APOBEC3C chimerical protein inhibits hepatitis B virus replication. J Biochem. 2011;150:371–374. doi: 10.1093/jb/mvr086. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Zhang X, Tian C, Wang T, Sarkis PT, Fang Y, Zheng S, Yu XF, Xu R. Cytidine deaminase APOBEC3B interacts with heterogeneous nuclear ribonucleoprotein K and suppresses hepatitis B virus expression. Cell Microbiol. 2008;10:112–121. doi: 10.1111/j.1462-5822.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 24.Bonvin M, Greeve J. Effects of point mutations in the cytidine deaminase domains of APOBEC3B on replication and hypermu-tation of hepatitis B virus in vitro. J Gen Virol. 2007;88:3270–3274. doi: 10.1099/vir.0.83149-0. [DOI] [PubMed] [Google Scholar]
- 25.Jost S, Turelli P, Mangeat B, Protzer U, Trono D. Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J Virol. 2007;81:10588–10596. doi: 10.1128/JVI.02489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen DH, Gummuluru S, Hu J. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol. 2007;81:4465–4472. doi: 10.1128/JVI.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T, Cai J, Chang J, Yu D, Wu C, Yan T, Zhai K, Bi X, Zhao H, Xu J, et al. Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet. 2013;22:1262–1269. doi: 10.1093/hmg/dds513. [DOI] [PubMed] [Google Scholar]
- 29.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxy-cytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 31.Vonica A, Rosa A, Arduini BL, Brivanlou AH. APOBEC2, a selective inhibitor of TGFβ signaling, regulates left-right axis specification during early embryogenesis. Dev Biol. 2011;350:13–23. doi: 10.1016/j.ydbio.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato Y, Probst HC, Tatsumi R, Ikeuchi Y, Neuberger MS, Rada C. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass and myopathy. J Biol Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etard C, Roostalu U, Strahle U. Lack of Apobec2-related proteins causes a dystrophic muscle phenotype in zebrafish embryos. J Cell Biol. 2010;189:527–539. doi: 10.1083/jcb.200912125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka Y, Marusawa H, Seno H, Matsumoto Y, Ueda Y, Kodama Y, Endo Y, Yamauchi J, Matsumoto T, Takaori-Kondo A, et al. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem Biophys Res Commun. 2006;341:314–319. doi: 10.1016/j.bbrc.2005.12.192. [DOI] [PubMed] [Google Scholar]
- 36.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: Implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu XF. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46:1810–1820. doi: 10.1002/hep.21893. [DOI] [PubMed] [Google Scholar]
- 38.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang FX, Huang J, Zhang H, Ma X, Zhang H. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008;89:722–730. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- 41.Stopak KS, Chiu YL, Kropp J, Grant RM, Greene WC. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages and dendritic cells. J Biol Chem. 2007;282:3539–3546. doi: 10.1074/jbc.M610138200. [DOI] [PubMed] [Google Scholar]
- 42.Argyris EG, Acheampong E, Wang F, Huang J, Chen K, Mukhtar M, Zhang H. The interferon-induced expression of APOBEC3G in human blood-brain barrier exerts a potent intrinsic immunity to block HIV-1 entry to central nervous system. Virology. 2007;367:440–451. doi: 10.1016/j.virol.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endo Y, Marusawa H, Kinoshita K, Morisawa T, Sakurai T, Okazaki IM, Watashi K, Shimotohno K, Honjo T, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26:5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto T, Marusawa H, Endo Y, Ueda Y, Matsumoto Y, Chiba T. Expression of APOBEC2 is transcriptionally regulated by NF-kappaB in human hepatocytes. FEBS Lett. 2006;580:731–735. doi: 10.1016/j.febslet.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 45.Endo Y, Marusawa H, Kou T, Nakase H, Fujii S, Fujimori T, Kinoshita K, Honjo T, Chiba T. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 2008;135:889–898. 898 e1–e3. doi: 10.1053/j.gastro.2008.06.091. [DOI] [PubMed] [Google Scholar]
- 46.Kou T, Marusawa H, Kinoshita K, Endo Y, Okazaki IM, Ueda Y, Kodama Y, Haga H, Ikai I, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int J Cancer. 2007;120:469–476. doi: 10.1002/ijc.22292. [DOI] [PubMed] [Google Scholar]
- 47.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol. 2012;23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Backus JW, Schock D, Smith HC. Only cytidines 5′ of the apolipoprotein B mRNA mooring sequence are edited. Biochim Biophys Acta. 1994;1219:1–14. doi: 10.1016/0167-4781(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, Sowden MP, Smith HC. Induction of cytidine to uridine editing on cytoplasmic apolipoprotein B mRNA by overexpressing APOBEC-1. J Biol Chem. 2000;275:22663–22669. doi: 10.1074/jbc.M910406199. [DOI] [PubMed] [Google Scholar]
- 50.Lau PP, Xiong WJ, Zhu HJ, Chen SH, Chan L. Apolipoprotein B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation. J Biol Chem. 1991;266:20550–20554. [PubMed] [Google Scholar]
- 51.Sowden MP, Smith HC. Commitment of apolipoprotein B RNA to the splicing pathway regulates cytidine-to-uridine editing-site utilization. Biochem J. 2001;359:697–705. doi: 10.1042/bj3590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes: Merging mechanisms for genetic diversity. Cell. 2002;109(Suppl):S35–S44. doi: 10.1016/S0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 53.Lada AG, Krick CF, Kozmin SG, Mayorov VI, Karpova TS, Rogozin IB, Pavlov YI. Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry (Mosc) 2011;76:131–146. doi: 10.1134/S0006297911010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stavnezer J. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 2011;32:194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patenaude AM, Di Noia JM. The mechanisms regulating the subcellular localization of AID. Nucleus. 2010;1:325–331. doi: 10.4161/nucl.1.4.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennett RP, Presnyak V, Wedekind JE, Smith HC. Nuclear Exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J Biol Chem. 2008;283:7320–7327. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 57.Smith HC. APOBEC3G: A double agent in defense. Trends Biochem Sci. 2011;36:239–244. doi: 10.1016/j.tibs.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 59.McDougall WM, Smith HC. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochem Biophys Res Commun. 2011;412:612–617. doi: 10.1016/j.bbrc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Dolan PT, Dang Y, Zheng YH. Biochemical differentiation of APOBEC3F and APOBEC3G proteins associated with HIV-1 life cycle. J Biol Chem. 2007;282:1585–1594. doi: 10.1074/jbc.M610150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol. 2007;81:9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewin SR, Ribeiro RM, Walters T, Lau GK, Bowden S, Locarnini S, Perelson AS. Analysis of hepatitis B viral load decline under potent therapy: Complex decay profiles observed. Hepatology. 2001;34:1012–1020. doi: 10.1053/jhep.2001.28509. [DOI] [PubMed] [Google Scholar]
- 66.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tran A, Kremsdorf D, Capel F, Housset C, Dauguet C, Petit MA, Brechot C. Emergence of and takeover by hepatitis B virus (HBV) with rearrangements in the pre-S/S and pre-C/C genes during chronic HBV infection. J Virol. 1991;65:3566–3574. doi: 10.1128/jvi.65.7.3566-3574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beggel B, Münk C, Däumer M, Hauck K, Häussinger D, Lengauer T, Erhardt A. Full genome ultra-deep pyrosequencing associates G-to-A hypermutation of the hepatitis B virus genome with the natural progression of hepatitis B. J Viral Hepat. 2013;20:882–889. doi: 10.1111/jvh.12110. [DOI] [PubMed] [Google Scholar]
- 69.Noguchi C, Ishino H, Tsuge M, Fujimoto Y, Imamura M, Takahashi S, Chayama K. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626–633. doi: 10.1002/hep.20580. [DOI] [PubMed] [Google Scholar]
- 70.Hannoun C, Horal P, Lindh M. Long-term mutation rates in the hepatitis B virus genome. J Gen Virol. 2000;81:75–83. doi: 10.1099/0022-1317-81-1-75. [DOI] [PubMed] [Google Scholar]
- 71.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 72.Har ris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 73.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 76.Lei YC, Tian YJ, Ding HH, Wang BJ, Yang Y, Hao YH, Zhao XP, Lu MJ, Gong FL, Yang DL. N-terminal and C-terminal cytosine deaminase domain of APOBEC3G inhibit hepatitis B virus replication. World J Gastroenterol. 2006;12:7488–7496. doi: 10.3748/wjg.v12.i46.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang G, Kitamura K, Wang Z, Liu G, Chowdhury S, Fu W, Koura M, Wakae K, Honjo T, Muramatsu M. RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc Natl Acad Sci USA. 2013;110:2246–2251. doi: 10.1073/pnas.1221921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlicht HJ, Bartenschlager R, Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989;63:2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen DH, Hu J. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol. 2008;82:6852–6861. doi: 10.1128/JVI.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao D, Wang X, Lou G, Peng G, Li J, Zhu H, Chen F, Li S, Liu D, Chen Z, Yang Z. APOBEC3G directly binds Hepatitis B virus core protein in cell and cell free systems. Virus Res. 2010;151:213–219. doi: 10.1016/j.virusres.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 83.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 84.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abe H, Ochi H, Maekawa T, Hatakeyama T, Tsuge M, Kitamura S, Kimura T, Miki D, Mitsui F, Hiraga N, et al. Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection. Hepatol Res. 2009;39:1159–1168. doi: 10.1111/j.1872-034X.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- 86.Ezzikouri S, Kitab B, Rebbani K, Marchio A, Wain-Hobson S, Dejean A, Vartanian JP, Pineau P, Benjelloun S. Polymorphic APOBEC3 modulates chronic hepatitis B in Moroccan population. J Viral Hepat. 2013;20:678–686. doi: 10.1111/jvh.12042. [DOI] [PubMed] [Google Scholar]
- 87.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, III, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 93.Chiba T, Marusawa H. A novel mechanism for inflammation-associated carcinogenesis; an important role of activation-induced cytidine deaminase (AID) in mutation induction. J Mol Med (Berl) 2009;87:1023–1027. doi: 10.1007/s00109-009-0527-3. [DOI] [PubMed] [Google Scholar]
- 94.Takai A, Toyoshima T, Uemura M, Kitawaki Y, Marusawa H, Hiai H, Yamada S, Okazaki IM, Honjo T, Chiba T, Kinoshita K. A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53 mutations. Oncogene. 2009;28:469–478. doi: 10.1038/onc.2008.415. [DOI] [PubMed] [Google Scholar]
- 95.Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci USA. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 97.Okuyama S, Marusawa H, Matsumoto T, Ueda Y, Matsumoto Y, Endo Y, Takai A, Chiba T. Excessive activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide 2 (APOBEC2) contributes to liver and lung tumorigenesis. Int J Cancer. 2012;130:1294–1301. doi: 10.1002/ijc.26114. [DOI] [PubMed] [Google Scholar]
- 98.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]