Abstract
Long non-coding RNAs (lncRNAs) are a class of RNAs that do not code protein but are important in diverse biological processes. In previous years, with the application of high-throughput sequencing, a large number of lncRNAs associated with virus infections have been identified and intensively investigated, however, there are few studies examining the association between lncRNAs and HCV replication. Previous studies have demonstrated that signal transducer and activator of transcription 3 (STAT3) is activated by the hepatitis C virus (HCV) and in turn increases the replication of HCV. However, the detailed molecular mechanism is only partially understood. In the present study, using human lncRNA polymerase chain reaction (PCR) arrays, it was identified that lnc-IGF2-AS, lnc-7SK, lnc-SChLAP1 and lnc-SRA1 are upregulated by STAT3. In addition, among these four lncRNAs, only lnc-IGF2-AS and lnc-7SK were involved in HCV replication. Transfection of siRNA lnc-7SK and siRNA lnc-IGF2-AS partially inhibited the replication of HCV in Huh7 cells. Data also indicated that when transfected with siRNA lnc-7SK and siRNA lnc-IGF2-AS, the expression of phosphatidylinositol 4-phosphate (PI4P), which was identified to be associated with HCV replication, was reduced. Thus, the present study identified two new types of lncRNAs, lnc-IGF2-AS and lnc-7SK, which can be upregulated by STAT3 and are involved in HCV replication by regulating PI4P.
Keywords: long non-coding RNAs, signal transducer and activator of transcription 3, hepatitis C virus replication, phosphatidylinositol 4-phosphate
Introduction
Hepatitis C virus (HCV) is a hepatotropic RNA virus of the genus Hepacivirus in the Flaviviridae family. Since it was identified in 1989, HCV has become a globally spread virus, leading to a worldwide health problem, which requires significant resources for its prevention and control. HCV can cause acute and chronic hepatitis in humans and chimpanzees, and if untreated it may progress to cirrhosis and hepatocellular carcinoma (HCC) (1). HCV is a positive-sense, single-stranded enveloped RNA virus ~9,600 nucleotides in length (2). Signal transducer and activator of transcription (STATs) factors are a family of transcription factors that have essential roles in cell growth, development, proliferation, immune defense and differentiation. Seven members of the STAT family have been identified and characterized, including STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b and STAT6 (3). Among the STAT family, STAT3 has been identified as a critical regulator in tumor cells (4). The activation of STAT3 may be one of the mechanisms by which human papilloma virus (HPV) achieves oncogenic transformation. E2, an early protein of HPV16, potentiates tumor necrosis factor (TNF)-α-mediated nuclear factor-κB activation through activation of STAT3 in the presence of TNF-α. In addition, this results in the reversal of E2-induced apoptosis, leading to cell survival (5). Tacke et al found that extracellular HCV core protein activates STAT3 in human monocytes, macrophages and dendritic cells (DCs) through an interleukin (IL)-6 autocrine pathway (6). However, the specific mechanism underlying how STAT3 is involved in the regulation of HCV replication remains to be elucidated.
Non-coding RNAs (ncRNAs) are transcribed RNA molecules with little or none protein coding capacity. Among all types of ncRNAs, long non-coding RNAs (lncRNAs), a type of regulatory ncRNA, are crucial in almost every aspect of cellular processes (7). Zhang et al found that nuclear enriched abundant transcript 1 (NEAT1) is one of several lncRNAs whose expression is altered by human immunodeficiency virus type 1 (HIV-1) infection and the knockdown of NEAT1 enhanced HIV-1 production through increased nucleus-to-cytoplasm export of Rev-dependent instability element-containing HIV-1 mRNAs (8). According to a large number of studies, lncRNAs are also associated with the development and progression of HCV, which causes HCC (9–12). A number of previous studies have implicated phosphatidylinositol 4-phosphate (PI4P) and several interacting partners in HCV infection (13–16). It has been demonstrated that host phosphatidylinositol 4-kinase and its product PI4P are involved in HCV replication and exert their effect through the effector oxysterol-binding protein (17). However, there are no relevant studies investigating whether lncRNAs can affect HCV replication through regulating PI4P. Therefore, identifying the association between lncRNAs regulated by STAT3 and PI4P expression may provide a novel pathogenic mechanism and therapeutic target for HCV infection.
In order clarify the detailed molecular mechanism of how STAT3 promotes the replication of HCV, human lncRNA polymerase chain reaction (PCR) arrays were used to identify the lncRNAs regulated by STAT3 in the current study. Finally, the mechanism underlying the regulation of HCV replication by lnc-7SK and lnc-IGF2-AS was investigated.
Materials and methods
Cell culture and virus
Human hepatoma cells (Huh7; American Type Culture Collection, Manassas, VA, USA) are a cell line highly permissive for HCV replication (18). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) medium (HyClone Laboratories, Inc., Logan, UT, USA) with 10% fetal bovine serum (FBS; HyClone Laboratories, Inc.) and cell culture plates were all placed in a humidified 5% CO2 incubator at 37°C.
The genotype 2a HCV strain JFH-1 was produced and propagated as described by Wakita et al (19).
Lentiviral vector construction and transfection
The lentiviral vector system contains GV-166, pHelper1.0 and pHelper2.0. pHelper1.0 and pHelper2.0 contain essential elements for virus packaging. The STAT3 gene was amplified by PCR and then inserted at a unique BamHI site and AgeI site in the vector (GV-166) to construct the GV166-STAT3 plasmid using the following primers: STAT3, forward ATGGGACACTGGGTGAGAGT and reverse CGGCAGGTCAATGGTATTGC. E. coli was transformed with GV166-STAT3 and cultured overnight at 37°C. PCR of the connected product was performed and the construct was confirmed by DNA sequencing. Huh7 cells were transfected with GV166-STAT3, pHelper1.0 and pHelper2.0 using Lipofectamine™ 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA). The virus supernatant was collected after 48 h of cultivation and the viral titers were determined.
Stably transfected cells were transfected with the lentivirus (10 MOI) at 50–60% confluence in 6 well plates. The lentivirus vector with 2 mg/ml polybrene was added to the Opti-MEM medium and after 12–16 h, the culture medium containing lentivirus was removed and the cell was incubated in Opti-MEM medium with 10% FBS. The cells were selected with 2 µg/ml puromycin (Sigma-Aldrich, St. Louis, MO, USA) for 7–10 days to produce the GV166-STAT3 stable transfected clones. An empty GV166-control vector was used as a negative control. The expression of STAT3 was then identified by quantitative (q)PCR and western blot analysis.
Total RNA extraction
Total RNA from the cells was extracted using TRIzol. TRIzol (1 ml) reagent was added to the 6-well plates and then the homogenized samples were incubated for 10 min at room temperature. Chloroform (0.2 ml) was added to the samples and agitated vigorously for 15 sec. The samples were incubated for 5 min at room temperature and then centrifuged at 12,000 × g for 10 min at 4°C. The upper aqueous phase was transferred to new tubes and 0.5 ml isopropyl alcohol was added. The samples were incubated for 10 min at room temperature and were then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant liquid was carefully removed without disturbing the pellet containing the RNA. The RNA pellet was washed with 1 ml of 75% ethanol three times and then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was carefully removed and the RNA pellet was dried for 10–15 min. Subsequently, the RNA pellet was redissolved in nuclease-free water according to the pellet size. The concentration of RNA was then measured using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) at an absorbance of 260 nm and 280 nm.
LncRNAs PCR array
The Human LncRNA PCR Array kit, termed the Human LncRNA Profiler, which includes 94 lncRNAs associated with numerous human diseases, was designed and produced by Qiagen China Co., Ltd. (Shanghai, China). The sample preparation and Human LncRNA PCR Array kit was prepared according to the manufacturer's instructions. Purified RNA was reverse transcribed into cDNA with human LncRNA Profiler cDNA synthesis buffer. qPCR and PCR array amplification was conducted using an IQ5 machine (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and the conditions were set as follows: 15 sec at 95°C, followed by 40 cycles of 15 sec at 95°C, 30 sec at 65°C and 30 sec at 72°C. Subsequently, a melt curve analysis was performed to determine the specificity of the PCR reaction. Relative gene expression was calculated using the ΔΔCt method (20). Primers used for qPCR are summarized in Table I.
Table I.
Primer sequences used for quantitative polymerase chain reaction.
| LncRNAs | Primer sequence |
|---|---|
| Lnc-7SK | F: 5′-AAACAAGCTCTCAAGGTC-3′ |
| R: 5′-CCTCATTTGGATGTGTCT-3′ | |
| Lnc-SRA1 | F: 5′-TTACAGAGATTAGAACCACATT-3′ |
| R: 5′-GGCAAGTCAGAGTTACAAT-3′ | |
| Lnc-IGF2-AS | F: 5′-CGCCACTGTGTTACCATT-3′ |
| R: 5′-TTGCCCATCCCAGATAGAA-3′ | |
| LncRNA-SChLAP1 | F: 5′-ACACAAGGTCCACCTTCCAG-3′ |
| R: 5′-GGTGGTATCTGCATCCCCAG-3′ |
F, forward; R, reverse; IGF2, insulin-like growth factor 2; SRA1, steroid receptor RNA activator 1; lncRNAs, long non-coding RNAs.
Reverse transcription-qPCR
Reverse transcription of RNA was performed according to the manufacturer's instructions of the Reverse Transcription System (Promega Corporation, Madison, WI, USA). The primers were designed using the NCBI primer blast tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The PCR system contained 2 µl of diluted cDNA, 12.5 µl of Fast SYBR Green Master mix (Invitrogen Life Technologies) and 10 µM of each gene specific primer in a total volume of 25 µl. qPCR reactions were performed in triplicate in 96-well plates under the following conditions: 15 sec at 95°C, followed by 40 cycles of 15 sec at 95°C, 30 sec at 65°C and 30 sec at 72°C.
siRNA transfection
Monolayers of Huh7 cells with 70% confluence in 6-well plates were transfected with siRNA lncRNA-7SK, siRNA lncRNA-SRA1, siRNA lncRNA-IGF2-AS and siRNA lncRNA-SChLAP1 or with the siRNA negative control. Transfection was performed using Lipofectamine™ 2000 (Invitrogen Life Technologies) according to the manufacturer's instructions. In brief, Huh7 cells (in serum free DMEM) were transfected using different quantities of Lipofectamine™ 2000 according to the quantity of siRNA. At 8 h post infection, the cells were harvested to measure the viral gene mRNA level by qPCR or PI4P-kinase level by western blot analysis. Four pairs of siRNA sequences were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The negative control sequence was a missense sequence. The sequences of the four siRNAs are provided in Table II.
Table II.
siRNA sequences of the four lncRNAs.
| LncRNAs | siRNA sequences |
|---|---|
| LncRNA-7SK | 5′-GCTCTCAAGGTCCATTTGTAGGAGA-3′ |
| LncRNA-SRA1 | 5′-CCACACAAGGAAGCAGGTATGTGAT-3′ |
| LncRNA-IGF2-AS | 5′-CCTCTGGTTATTACCTCAATTCTGA-3′ |
| LncRNA-SChLAP1 | 5′-CACCATGTACCTGGAAGCAACATCT-3′ |
siRNA, small interfering RNA; lncRNAs, long non-coding RNAs; IGF2, insulin-like growth factor 2; SRA1, steroid receptor RNA activator 1.
Western blot analysis
Cells were collected by trypsinization. Total protein was extracted using RIPA plus phenylmethyl-sulfonyl fluoride according to the manufacturer's instructions (Beyotime Institute of Biotechnology, Shanghai, China). The protein samples were diluted in 5X loading buffer, boiled for 10 min, subjected to a 10% polyacrylamide gel (Sangon Biotech Co., Ltd., Shanghai, China) and then underwent electrophoresis. The proteins were transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Following transfer, the membranes were blocked with 5% (w/v) non-fat dry milk in Tris-buffered saline and Tween 20 (TBST; 10 mM Tris-HCl, pH 8.0, 150 mM NaCl containing 0.1% v/v Tween 20) for at least 1 h and then the membranes were incubated overnight at 4°C with the following monoclonal antibodies from Abcam (Cambridge, MA, USA) in blocking buffer: Mouse monoclonal STAT3 (ab119352; 1:5,000), mouse monoclonal HCV-E2 (ab20852; 1:100) and rabbit monoclonal PI4K-β (ab109418; 1:1,000). Following being washed three times with TBST, the membranes were incubated with the following secondary antibodies (CST) from Abcam: Goat anti-mouse(ab131368; 1:5,000), goat anti-rabbit (ab7085; 1:1,000) for 30 min and were revealed using an ECL PLUS detection kit (Biomiga, Inc., San Diego, CA, USA). β-actin was obtained from Sigma-Aldrich.
Results
Overexpression of STAT3
In order to identify the lncRNAs regulated by STAT3, a stable transfection cell line of STAT3 was established. qPCR demonstrated that the quantity of STAT3 mRNA in the cells transfected with GV166-STAT3 was significantly higher than that in the GV166-control transfected cells. The western blot assay of STAT3 demonstrated that the concentration of STAT3 in the GV166-STAT3 transfected cells was higher than that of the control cells (Fig. 1).
Figure 1.
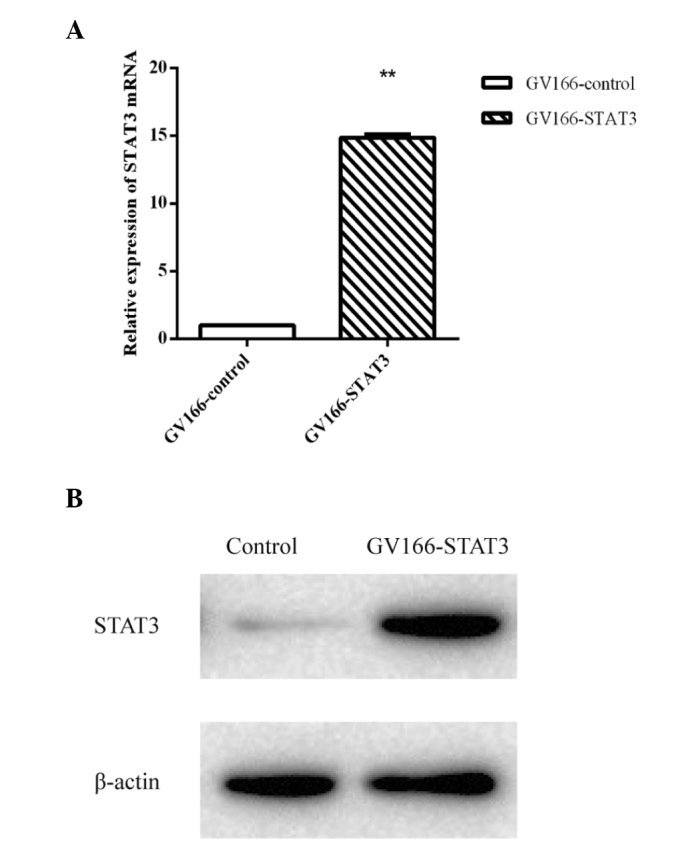
STAT3 stable transfection verified by qPCR and western blot analysis. (A) The mRNA level of STAT3 was detected by qPCR. Results were analyzed by the relative quantification method (ΔΔCt method). Data are expressed as the mean ± standard deviation from three independent experiments. P-values were calculated by Student's t-test and statistical significance is indicated. **P<0.005, compared with the control. (B) STAT3 expression level was confirmed through western blot analysis. β-actin was used as an internal control. qPCR, quantitative polymerase chain reaction; STAT3, signal transducer and activator of transcription 3.
STAT3 upregulates four lncRNAs
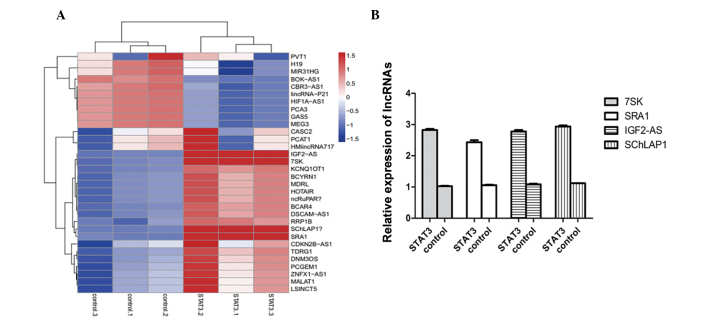
Following identification of the stable transfection of STAT3, lncRNA PCR array method was used to detect the different expression levels of lncRNAs in STAT3 overexpressing and control cells. As shown in Fig. 2A, lnc-IGF2-AS, lnc-7SK, lnc-SChLAP1 and lnc-SRA1 were upregulated in STAT3 overexpressing cells compared with the control cells. To further certify the preliminary result, qPCR was performed. The histogram in Fig. 2B confirmed that STAT3 is able to increase these four lncRNAs.
Figure 2.
LncRNA-7SK, lncRNA-SRA1, lncRNA-IGF2-AS and lncRNA-SChLAP1 are the most significantly altered lncRNAs in Huh7 cells stimulated by STAT3. (A) Heat maps and hierarchical clustering demonstrated lncRNAs differential expression profile between STAT3 overexpressing and control cells. Heat maps and hierarchical clustering are the most widely used clustering methods for analyzing lncRNAs or mRNA expression data. Red indicates a high relative expression and blue indicates a low relative expression. (B) LncRNA-7SK, lncRNA-SRA1, lncRNA-IGF2-AS and lncRNA-SChLAP1 expression changes were further confirmed by quantitative polymerase chain reaction. STAT3, signal transducer and activator of transcription 3; IGF, insulin-like growth factor; SRA1, steroid receptor RNA activator 1; lncRNAs, long non-coding RNAs.
LncRNA-7SK and lncRNA-IGF2-AS siRNA inhibits HCV replication
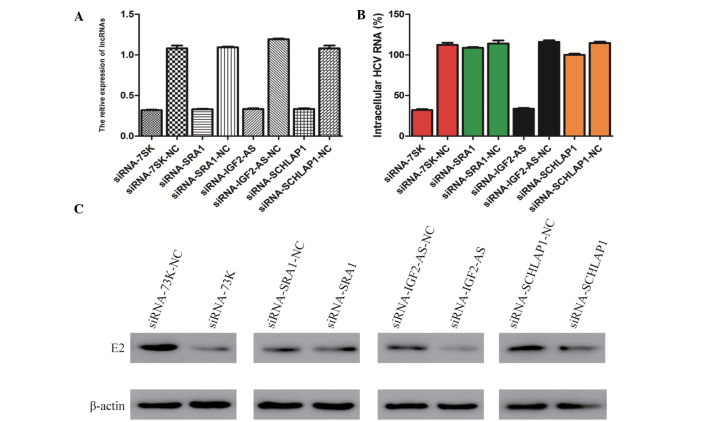
As these four lncRNAs can be upregulated by STAT3, the present study aimed to determine whether these four lncRNAs have a direct association with HCV replication. To verify the involvement of these four lncRNAs in HCV replication, the Huh7 cells were transfected with siRNA lncRNA-IGF2-AS, siRNA lncRNA-7SK, siRNA lncRNA-SChLAP1 and siRNA lncRNA-SRA1, and the expression of all these four lncRNAs were analyzed at the mRNA level. The qPCR data shown include at least three independent experiments. As shown in Fig. 3A, the relative expression of the four lncRNAs in Huh7 cells transfected with siRNA lncRNA-IGF2-AS, siRNA lncRNA-7SK, siRNA lncRNA-SChLAP1 and siRNA lncRNA-SRA1, respectively, were significantly lower than cells transfected with the control siRNA (P<0.005).
Figure 3.
Knockdown of lnc-7SK and lnc-IGF2-AS by siRNA inhibit HCV replication. (A) Silencing efficiency of lnc-7SK, lnc-SRA1, lnc-IGF2-AS and lnc-SChLAP1 were verified through quantitative polymerase chain reaction. (B) Effects of lnc-7SK, lnc-SRA1, lnc-IGF2-AS and lnc-SChLAP1 knockdown on the mRNA level of the envelope glycoprotein E2. The results are presented as the mean ± standard deviation from three independent experiments performed in triplicate. Relative expression is presented in arbitrary units. (C) Effects of lnc-7SK, lnc-SRA1, lnc-IGF2-AS and lnc-SChLAP1 knockdown on the expression level of E2 were verified through western blot analysis. β-actin was used as an internal control. IGF, insulin-like growth factor; SRA1, steroid receptor RNA activator 1; siRNA, small interfering RNA; HCV, hepatitis C virus; lncRNAs, long non-coding RNAs; NC, negative control.
Subsequently, the siRNA-transfected Huh7 cells were infected with the HCV JFH-1 strain and the cell lysates were analyzed by qPCR and western blot analysis to measure the expression level of the envelope glycoprotein E2. Notably, it was observed that only following transfection of siRNA lncRNA-7SK and siRNA lncRNA-IGF2-AS, the mRNA level of E2 was reduced by 71.5 and 70.9%, respectively, compared with the negative control. While the transfection of siRNA lncRNA-SAR1 and siRNA lncRNA-SChLAP1 had no pronounced effect on the E2 mRNA level (Fig. 3B). Subsequently, the western blot assay also demonstrated that knockdown of lncRNA-7SK and lncRNA-IGF2-AS affects the expression of envelope glycoprotein E2 (Fig. 3C). These results suggest that lncRNA-7SK and lncRNA-IGF2-AS rather than lncRNA-SChLAP1 and lncRNA-SRA1 are associated with the replication of HCV.
LncRNA-7SK and lncRNA-IGF2-AS siRNA inhibit HCV replication by reducing the expression of PI4P
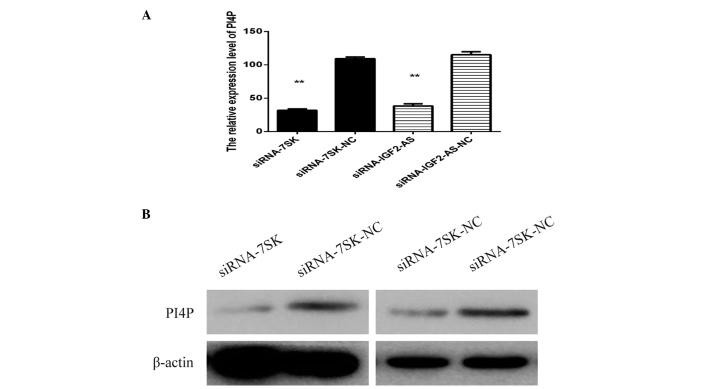
In order to investigate the potential mechanism by which siRNA lncRNA-7SK and siRNA lncRNA-IGF2-AS inhibited HCV replication, the expression level of PI4P-kinase, which has been verified to be involved in HCV replication, was measured by western blot analysis. Huh7 cell lysates were obtained following transfection with siRNA lncRNA-IGF2-AS, siRNA lncRNA-7SK as well as infected JFH-1. As expected, the expression of PI4P-kinase was reduced in the cells transfected with siRNA lncRNA-7SK and siRNA lncRNA-IGF2-AS as compared with the negative control. The results were verified through qPCR and western blot analysis (Fig. 4).
Figure 4.
Knockdown of lnc-7SK and lnc-IGF2-AS leads to the reduction of PI4P expression level. (A) Effect of lnc-7SK and lnc-IGF2-AS knockdown on the mRNA level of PI4P. The results are presented as the mean ± standard deviation from three independent experiments performed in triplicate. Relative expression is presented in arbitrary units. (B) Effects of lnc-7SK and lnc-IGF2-AS knockdown on PI4P expression level were further confirmed through western blot analysis. β-actin was used as an internal control. IGF, insulin-like growth factor; PI4P, phosphatidylinositol 4-phosphate; siRNA, small interfering RNA; NC, negative control. **P<0.005.
Discussion
In the present study, two novel findings were reported. Overexpression of STAT3 upregulated lnc-IGF2-AS, lnc-7SK, lnc-SChLAP1 and lnc-SRA1 in Huh7 cells and among these four types of lncRNAs, only lnc-IGF2-AS and lnc-7SK regulated HCV replication through affecting the expression of PI4P. A previous study demonstrated that genetic or pharmacological activation of STAT3 in glioma cells enhanced oncolytic herpes simplex virus replication and cytotoxicity (21). In addition, McCartney et al also verified that HCV replication increases STAT3 activation and in turn activation of STAT3 increases HCV replication by modulating microtubule dynamics (22). Activation or overexpression of STAT3 is relevant for various types of human cancer, including Epstein-Barr virus (EBV)-associated cancer. The expression of STAT3 rapidly increases upon EBV infection, which mediates relaxation of the intra-S phase cell-cycle checkpoint; this facilitates viral oncogene-driven cell proliferation (23). HCV core is capable of upregulating the expression of NANOG, which is a member of the homeobox family of DNA binding transcription factors and was identified as a pluripotency promoting gene. In addition, this core-induced NANOG expression enhanced cell growth and cell cycle progression through increased expression of phosphorylated STAT3 protein (24).
LncRNAs are RNAs >200 bp in length. Previous findings have revealed that lncRNAs are involved in several steps of cancer development (25,26). These lncRNAs interact with DNA, RNA and proteins, acting as an essential regulator of cell differentiation and organogenesis. Their misexpression may affect epigenetic information and may result in the progressive and uncontrolled growth of a tumor (11,27). Lnc-DC, which was exclusively expressed in human conventional DCs was involved in DC differentiation and stimulated T-cell activation through interacting with STAT3 (28). The lncRNA-HULC, highly upregulated in liver cancer, is markedly upregulated in HCC, which functions through suppressing the tumor suppressor gene p18 (29). Prensner et al initially identified lnc-SChLAP1 and found that it contributes to the development of lethal prostate cancer at least in part by antagonizing the tumor-suppressive functions of the SWI/SNF chromatin-modifying complex (30). Castelo-Branco et al indicated that the snRNA 7SK, a single non-coding RNA, is a gatekeeper of transcriptional termination and bidirectional transcription in embryonic stem cells and mediates transcriptional poising through a mechanism independent of chromatin bivalency (31).
In the present study, a stable expression cell line of STAT3 was constructed. Subsequently, a human lncRNA PCR array kit was used. Human LncRNA Profiler was used to compare the LncRNA expression profile differences between Huh7 cells and control cells. It was found that four lncRNAs, including lnc-7SK, lnc-SRAI, lnc-IGF2-AS and lnc-SChLAP1, can be upregulated by STAT3. Among the four lncRNAs, only lnc-7SK and lnc-IGF2-AS are associated with the replication of HCV as the transfection of siRNA lnc-7SK and siRNA lnc-IGF2-AS reduced the expression level of mRNA coding the envelope glycoprotein E2. The expression level of PI4P was then measured in cells following transfection with siRNA lnc-7SK and siRNA lnc-IGF2-AS. Compared with cells transfected with the siRNA-control, the expression level of PI4P in cells transfected with siRNA lnc-7SK and siRNA lnc-IGF2-AS were highly reduced.
In conclusion, these findings increase our understanding of the molecular mechanisms of HCV replication and the effect of STAT3 on HCV replication. Furthermore, a preliminary study was performed regarding the mechanism underlying the function of lnc-7SK and lnc-IGF2-AS. Future studies are required to determine whether lnc-7SK and lnc-IGF2-AS are potential diagnostic or even therapeutic targets for HCV infection.
Acknowledgments
This study was supported by the Natural Science Foundation of China (grant nos. 30872230 and 81171643).
References
- 1.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Kim CW, Chang KM. Hepatitis C virus: Virology and life cycle. Clin Mol Hepatol. 2013;19:17–25. doi: 10.3350/cmh.2013.19.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi QR, Yang ZM. Regulation and function of signal transducer and activator of transcription 3. World J Biol Chem. 2014;5:231–239. doi: 10.4331/wjbc.v5.i2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen AV, Wu YY, Lin EY. STAT3 and sphin-gosine-1-phosphate in inflammation-associated colorectal cancer. World J Gastroenterol. 2014;20:10279–10287. doi: 10.3748/wjg.v20.i30.10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabhavathy D, Prabhakar BN, Karunagaran D. HPV16 E2-mediated potentiation of NF-κB activation induced by TNF-α involves parallel activation of STAT3 with a reduction in E2-induced apoptosis. Mol Cell Biochem. 2014;394:77–90. doi: 10.1007/s11010-014-2083-6. [DOI] [PubMed] [Google Scholar]
- 6.Tacke RS, Tosello-Trampont A, Nguyen V, Mullins DW, Hahn YS. Extracellular hepatitis C virus core protein activates STAT3 in human monocytes/macrophages/dendritic cells via an IL-6 autocrine pathway. J Biol Chem. 2011;286:10847–10855. doi: 10.1074/jbc.M110.217653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, Valadkhan S. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42:10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 post-transcriptional expression. MBio. 2013;4:e00596–e00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou W, Bonkovsky HL. Non-coding RNAs in hepatitis C-induced hepatocellular carcinoma: Dysregulation and implications for early detection, diagnosis and therapy. World J Gastroenterol. 2013;19:7836–7845. doi: 10.3748/wjg.v19.i44.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Pu R, Du Y, Han Y, Su T, Wang H, Cao G. Non-coding RNAs in hepatitis B or C-associated hepatocellular carcinoma: Potential diagnostic and prognostic markers and therapeutic targets. Cancer Lett. 2012;321:1–12. doi: 10.1016/j.canlet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Tu ZQ, Li RJ, Mei JZ, Li XH. Downregulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 13.Bishè B, Syed G, Siddiqui A. Phosphoinositides in the hepatitis C virus life cycle. Viruses. 2012;4:2340–2358. doi: 10.3390/v4102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishé B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Zhang S, Chen J, Jiang K, Zhang Q, Guo K, Liu Y. The transcriptional profiling of glycogenes associated with hepatocellular carcinoma metastasis. PLoS One. 2014;9:e107941. doi: 10.1371/journal.pone.0107941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okemoto K, Wagner B, Meisen H, Haseley A, Kaur B, Chiocca EA. STAT3 activation promotes oncolytic HSV1 replication in glioma cells. PLoS One. 2013;8:e71932. doi: 10.1371/journal.pone.0071932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCartney EM, Helbig KJ, Narayana SK, Eyre NS, Aloia AL, Beard MR. Signal transducer and activator of transcription 3 is a proviral host factor for hepatitis C virus. Hepatology. 2013;58:1558–1568. doi: 10.1002/hep.26496. [DOI] [PubMed] [Google Scholar]
- 23.Koganti S, Hui-Yuen J, McAllister S, Gardner B, Grasser F, Palendira U, Tangye SG, Freeman AF, Bhaduri-McIntosh S. STAT3 interrupts ATR-Ch k1 signaling to allow oncovirus-mediated cell proliferation. Proc Natl Acad Sci USA. 2014;111:4946–4951. doi: 10.1073/pnas.1400683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou JJ, Chen RF, Deng XG, Zhou Y, Ye X, Yu M, Tang J, He XY, Cheng D, Zeng B, et al. Hepatitis C virus core protein regulates NANOG expression via the stat3 pathway. FEBS Lett. 2014;588:566–573. doi: 10.1016/j.febslet.2013.11.041. [DOI] [PubMed] [Google Scholar]
- 25.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 28.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 29.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly upregulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via downregulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castelo-Branco G, Amaral PP, Engström PG, Robson SC, Marques SC, Bertone P, Kouzarides T. The non-coding snRNA 7SK controls transcriptional termination, poising and bidirectionality in embryonic stem cells. Genome Biol. 2013;14:R98. doi: 10.1186/gb-2013-14-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]